Abstract
Introduction: Long non coding RNAs (lncRNAs) have emerged recently as major players in tumor biology and may be used for cancer diagnosis, prognosis, and potential therapeutic targets. The lncRNA HMlincRNA717, a newly identified lncRNA, was demonstrated to be down-regulated in gastric cancer. However, little is known about its role in non small cell lung cancer (NSCLC). Methods: Expression of lncRNA HMlincRNA717 in tumor and their matched non-tumor tissues was determined by quantitative real-time PCR (qRT-PCR) in NSCLC patients. Then, we analyzed the potential relationship between lncRNA HMlincRNA717 expression levels in tumor tissues and clinicopathological features of NSCLC, and clinical outcome. Results: lncRNA HMlincRNA717 expression level was significantly decreased in NSCLC tissues in comparison to adjacent non-tumor tissues. It was also proved that HMlincRNA717 expression was to be associated with NSCLC histological grade, and lymph node metastasis. In addition, survival analysis proved that down-regulated HMlincRNA717 expression was associated with poor overall survival of NSCLC patients. Multivariate survival analysis also proved that HMlincRNA717 was an independent prognostic factor for NSCLC patients. Conclusions: The present study showed the down-regulation of HMlincRNA717 and its association with tumor progression in human NSCLC. It also provided that HMlincRNA717 expression was an independent prognostic factor for patients with NSCLC, which might be a potential prognostic biomarker and therapeutic target for NSCLC.
Keywords: Long non coding RNA, HMlincRNA717, non small cell lung cancer, prognosis
Introduction
Lung cancer is the leading cause of cancer-related mortality, with 1.4 million deaths worldwide annually [1]. Almost 80% of lung cancers are non small cell lung cancer (NSCLC) [2]. Surgical resection, when possible, remains the only curative treatment for early stage NSCLC. However, nearly 50% of resected patients experience recurrence [3]. The prognosis for NSCLC is still dismal, and the overall 5-year survival of only 15% [4]. Therefore, identifying more accurate predictive biomarkers is of great clinical value to further understand NSCLC cell biology and develop novel therapeutic strategies.
Long non coding RNA is an RNA molecule that is longer than 200 nucleotides and is not translated into a protein [5]. Although these long non coding transcripts were once considered to be simply transcriptional “noise” or cloning artifacts [6]. Increasing evidence has suggested that lncRNAs participate in a spectrum of biological function, including cell differentiation, proliferation, apoptosis, migration and invasion [7-9]. In this regard, highlighting the potentially widespread functional roles of lncRNAs in human cancer is important. For example, Takahashi’s study demonstrated that PVT1 expression levels in colorectal cancer tissues were significantly higher than that in non-cancerous tissue, and patients with high PVT1 expression had a significantly poorer prognosis, what’s more, knockdown PVT1 expression could promotes apoptosis in colorectal cancer cells [10]. Geng showed that HOTAIR (HOX transcript antisense RNA) gene was significantly over-expressed in hepatocellular carcinoma tissues compared with adjacent non-tumour tissues and patients with high HOTAIR gene expression in their tumors had an increased risk of recurrence after hepatectomy [11]. Mourtada-Maarabouni identified that GAS5 transcript levels were significantly reduced in breast cancer samples relative to adjacent unaffected normal breast epithelial tissues and GAS5 as critical to the control of mammalian apoptosis and cell population growth [12]. Unfortunately, the emerging functional role of lncRNAs in lung cancer remains unclear.
LncRNA HMlincRNA717 is a long intergenic non coding RNA (lincRNA) with 818 nucleotides in length and the gene which is located at 18p11.228. LncRNA HMlincRNA717 was first found to be associated with gastric cancer, the committee has named them as “gastric cancer associated transcript 2 (GACAT2)” [13]. Shao et al found that LncRNA HMlincRNA717 to be down-regulated in gastric cancer cell lines and tissues, their results also indicated that HMlincRNA717 expression levels were correlated with cancer distal metastasis, venous invasion, and nervous invasion [14]. However, the role of lncRNA HMlincRNA717 in NSCLC has not yet been elucidated.
In the present study, we have investigated the expression level of lncRNA HMlincRNA717 in clinical NSCLC specimens or cell lines compared with adjacent non-tumor tissues or normal cell line, as well as analyzing its association with overall survival of patients.
Materials and methods
Cell culture
The human lung cell lines A549, H157, HEK-293T and normal bronchial epithelial cell line 16HBE were purchased from the American Type Culture Collection (ATCC, USA). All cell lines were routinely maintained in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 100U/ml penicillin sodium, and 100mg/ml streptomycin sulfate at 37°C in a humidified air atmosphere containing 5% CO2. Cells were used when they were in the logarithmic growth phase.
Patients and specimens
118 paired NSCLC tissues and matched adjacent non-tumor tissues were obtained from Xin Hua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Shanghai, China) between Jan 2006 and Jan 2008. All patients recruited in this study were not subjected to preoperative radiotherapy and/or chemotherapy and were diagnosed with NSCLC based on histopathological evaluation. Clinical and pathological variables analyzed are shown in Table 1. All collected tissue samples were immediately stored at -80°C until use. The study was approved by the Research Ethics Committee of Shanghai Jiao Tong University School of Medicine, China. Written informed consent was obtained from all patients.
Table 1.
Relationship between HMlincRNA717 expression and clinicopathological variables in NSCLC patients
| Variable | lncRNA HMlincRNA717 | |||
|---|---|---|---|---|
|
|
||||
| Number | High | Low | P value | |
| Age (years) | ||||
| <60 | 69 | 30 | 39 | |
| ≥60 | 49 | 19 | 30 | 0.609 |
| Gender | ||||
| Male | 65 | 27 | 38 | |
| Female | 53 | 22 | 31 | 0.997 |
| Tumor size (cm) | ||||
| <3 | 51 | 23 | 28 | |
| ≥3 | 67 | 26 | 41 | 0.492 |
| Histology | ||||
| Adeno | 45 | 20 | 25 | |
| Squamous | 73 | 29 | 44 | 0.613 |
| Histological grade | ||||
| I | 40 | 28 | 12 | |
| II-III | 78 | 21 | 57 | 0.000 |
| Lymph nodes metastasis | ||||
| No | 54 | 33 | 21 | |
| Yes | 64 | 16 | 48 | 0.000 |
RNA extraction and qRT-PCR analyses
Total RNA was extracted from tissues or cultured cells with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. qRT-PCR assays were performed to detect HMlincRNA717 expression using the Prime Script RT reagent Kit and SYBR Premix ExTaq (Takara) according to the manufacturer’s instructions. Results were normalized to the expression of GAPDH. The primers used were as follows: HMlincRNA717 sense, 5’-TGGATGCTTA CAAAGGACTGG-3′ and anti-sense, 5′-CTGCAATTACGGAAAGAGCTG-3′; GAPDH sense, 5′-GGGAGCCAAAAGGGTCAT-3′ and anti-sense, 5′-GAGTCCTTCCACGATACCAA-3′. qRT-PCR and data collection were performed on an ABI 7900. qRT-PCR results were analyzed and expressed relative to CT (threshold cycle) values, and then converted to fold changes.
Statistical analysis
All statistical analyses were performed using SPSS 18.0 software (IBM). The statistical significance between groups was determined using the Student’s t test. Association between expression level of lncRNA HMlincRNA717 and each clinicopathologic parameter was evaluated using Pearson’s Chi-square test. Patient survival was evaluated using the Kaplan-Meier method and compared using log-rank test. Univariate and multivariate Cox regression analyses were performed to analyze the survival data. The data are shown as the mean ± SD from at least three independent experiments. Results were considered to indicate a statistically significant difference at values of P < 0.05.
Results
Expression of HMlincRNA717 in NSCLC tissues and cell lines
The first aim of the present study was to investigate whether HMlincRNA717 is detectable and altered in NSCLC tissues compared with adjacent non-tumor tissues. Using RNA isolated from tissues, we performed qRT-PCR to detect the expression levels of HMlincRNA717 using GAPDH as normalization control. Our results indicated that lncRNA HMlincRNA717 expression was significantly lower in tumor tissues compared with adjacent non-tumor tissues (P < 0.05, Figure 1A). qRT-PCR assays were further developed to quantify HMlincRNA717 in lung cancer cell lines, including A549, H157, HEK-293T, and normal bronchial epithelial cell line 16HBE. A significant low expression of HMlincRNA717 was found in lung cancer cell lines (A549, H157, HEK-293T) compared to normal bronchial epithelial cell line (16HBE) (P < 0.05, Figure 1B).
Figure 1.
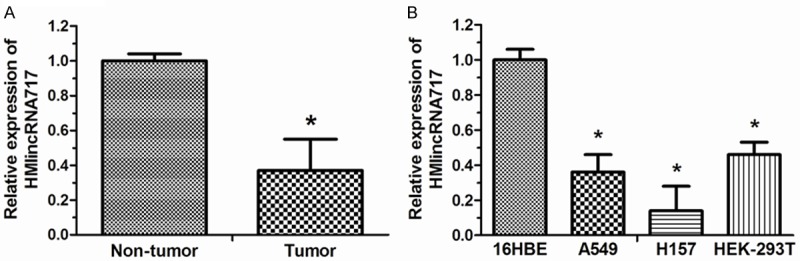
Relative expression of HMlincRNA717 in NSCLC tissues and cell lines. A. Expression of HMlincRNA717 in NSCLC tissues and matched non-tumor tissues was examined by qRT-PCR. B. Expression of HMlincRNA717 in 3 NSCLC cell lines, A549, H157, and HEK-293T, and human bronchial epithelial cell line (16HBE) was measured by qRT-PCR. *P < 0.05.
HMlincRNA717 expression and clinicopathologic factors in NSCLC
To assess the correlation of lncRNA HMlincRNA717 expression with clinicopathologic data, the expression levels of lncRNA HMlincRNA717 in tumor tissues were categorized as low or high in relation to the mean value. As shown in Table 1, the HMlincRNA717 level was associated with histological grade (P < 0.05), and lymph node metastasis (P < 0.05). However, there was no significant correlation between HMlincRNA717 expression and other clinicopathological features, such as age, gender, tumor size, or histology (P > 0.05).
Relationship of HMlincRNA717 to overall survival of NSCLC patients
Kaplan-Meier analysis was applied to examine the prognostic value of lncRNA HMlincRNA717 expression to overall survival of patients with NSCLC. Results proved that patients with NSCLC of low lncRNA HMlincRNA717 expression tended to have worse overall survival (log rank test, P < 0.05, Figure 2). As shown in Table 2, Univariate survival analysis showed that patients with NSCLC of low HMlincRNA717 expression had a 2.885-fold higher risk of death (95% CI: 1.531-5.921; P < 0.05). As far as clinicopathological characteristics were considered, histological grade, lymph node metastasis were also proved to be associated with overall survival (P < 0.05). However, age, gender, tumor size, and histology had no prognostic value on overall survival of patients with NSCLC (P > 0.05). As lncRNA HMlincRNA717 expression was proved to be associated with overall survival of patients in univariate survival analysis, we further investigated whether lncRNA HMlincRNA717 could serve as an independent prognostic marker for patients with NSCLC. As shown in Table 2, Multivariate analysis revealed that lncRNA HMlincRNA717 expression, histological grade, and lymph node metastasis were independent prognostic markers for NSCLC (P < 0.05). Taken together, these data indicated that lncRNA HMlincRNA717 was an independent prognostic factor of overall survival for patients with NSCLC.
Figure 2.
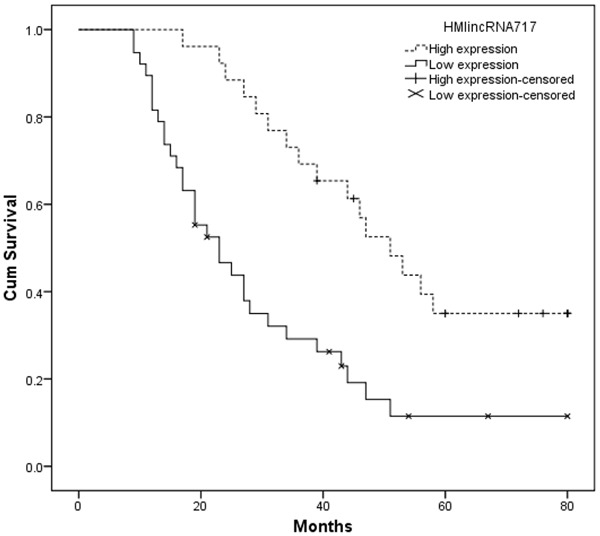
Kaplan-Meier postoperative survival curve for patterns of patients with NSCLC and HMlincRNA717 expression.
Table 2.
Prognostic factors in Cox proportional hazards model
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Risk ratio | 95% CI | P | Risk atio | 95% CI | P | |
| Age (years) | ||||||
| ≥60 vs <60 | 0.977 | 0.716-1.821 | 0.513 | |||
| Gender | ||||||
| Male vs Female | 1.612 | 0.817-2.313 | 0.286 | |||
| Tumor size | ||||||
| ≥3 cm vs <3 cm | 1.493 | 0.618-2.713 | 0.306 | |||
| Histology | ||||||
| Adeno vs Squamous | 0.891 | 0.533-1.873 | 0.184 | |||
| Histological grade | ||||||
| II, III vs I | 2.716 | 1.476-6.425 | 0.012 | 2.403 | 1.318-5.771 | 0.015 |
| Lymph nodes metastasis | ||||||
| Yes vs No | 3.319 | 2.072-9.221 | 0.007 | 2.847 | 1.832-7.837 | 0.009 |
| HMlincRNA717 | ||||||
| low vs high | 2.885 | 1.531-5.921 | 0.006 | 2.473 | 1.484-5.107 | 0.011 |
Discussion
NSCLC ranks among the most common and lethal malignant diseases. Poor prognosis of early stage NSCLC is crucially linked to the onset of tumor metastasis [15]. So finding new molecular targets for its diagnosis, prognosis and treatment has the potential to improve the clinical strategies and outcomes of this disease [16]. In recent years, studies have shown that ~18 % of the protein coding genes that produce lncRNAs are associated with cancer, whereas only 9% of all human protein coding genes are associated with cancer [17]. Due to their great importance in the regulation of gene expression, it has been widely accepted that lncRNAs are involved in multiple cellular functions including proliferation, apoptosis and differentiation, thus, have been implemented in diverse physiological and pathological processes ranging from development to cancer [7,18]. So identification of tumor associated lncRNAs is critical for understanding the roles of lncRNAs in tumorigenesis and may be important for novel therapeutic targets [19]. In the present study, our attention focused on the lncRNA HMlincRNA717.
In the present study, we have investigated lncRNA HMlincRNA717 expression by qRT-PCR assay in 118 cases of NSCLC from patients who had not received radiotherapy or chemotherapy. Based relative expression calculation, we analyzed the association of lncRNA HMlincRNA717 with clinicopathological characteristics as well as prognosis of patients. Results showed that HMlincRNA717 expression was decreased in NSCLC tissues compared with that in adjacent non-tumor tissues for low expression of HMlincRNA717 was more likely to be detected in NSCLC specimens, which indicating its possible participation on carcinogenesis. It is also found that HMlincRNA717 expression was closely related to NSCLC histological grade, and lymph node metastasis for low expression of HMlincRNA717 was more frequently to be detected in tumors with advanced histological grade, and lymph node metastasis, suggesting the possible participation of HMlincRNA717 on NSCLC invasion and metastasis. Together with the above evidence, it was thus proposed that HMlincRNA717 may play important roles in NSCLC carcinogenesis and progression.
As HMlincRNA717 expression was found to be associated with NSCLC invasion and metastasis, considering the invasion and metastasis are crucial factors affecting the prognosis of patients, HMlincRNA717 might be a potential prognostic marker for patients with NSCLC. In order to investigate the prognostic role of HMlincRNA717 on NSCLC, we performed Kaplan-Meier analysis of overall survival. Results showed that patients with NSCLC of low HMlincRNA717 expression tend to have worse overall survival in comparison to patients with tumor of high HMlincRNA717 expression, which suggested that HMlincRNA717 expression was a prognostic marker for patients with NSCLC. To further evaluate the prognostic value of HMlincRNA717 in NSCLC, we performed Cox proportional hazards model. Results proved that decreased HMlincRNA717 expression was an independent marker of poor overall survival of NSCLC patients. These data indicated that HMlincRNA717 could constitute a molecular prognostic marker for patients with NSCLC, identifying high risk individuals who are more likely to have tumor relapse in clinical practice, thus, good candidates to receive more aggressive treatment. These results were in consistent with investigations focused on gastric cancer, indicating the consistence of HMlincRNA717 function in these types of tumor. Thus, the positive linkage between HMlincRNA717 down-regulation and poor prognosis may not only be used for identifying NSCLC patients with higher risk of early tumor relapse but also for providing valuable clues to understand the possible mechanism of NSCLC invasion and metastasis.
In conclusion, we have proved that HMlincRNA717 expression was significantly decreased in NSCLC tissues and cell lines. A lower expression of HMlincRNA717 was detected in tumor of advanced histological grade, and with lymph node metastasis. In addition, the down-regulation expression of HMlincRNA717 was associated with poor prognosis. These results demonstrated that HMlincRNA717 might be a novel prognostic indicator in NSCLC and may be a potential target for diagnosis and gene therapy.
Acknowledgements
This study was supported by two grants from the National Natural Science Foundation of China (No. 81071924; No. 81372520).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 3.Cataldo VD, Gibbons DL, Perez-Soler R, Quintas-Cardama A. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med. 2011;364:947–955. doi: 10.1056/NEJMct0807960. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, Lam WL. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, Yamamoto H, Doki Y, Mori M, Mimori K. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large Intervening Non-coding RNA HOTAIR is Associated with Hepatocellular Carcinoma Progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 12.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Li P, Xiao B, Guo J. Long noncoding RNA HMlincRNA717 and AC130710 have been officially named as gastric cancer associated transcript 2 (GACAT2) and GACAT3, respectively. Tumour Biol. 2014;35:8351–2. doi: 10.1007/s13277-014-2378-y. [DOI] [PubMed] [Google Scholar]
- 14.Shao Y, Chen H, Jiang X, Chen S, Li P, Ye M, Li Q, Sun W, Guo J. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 2014;35:9591–5. doi: 10.1007/s13277-014-2243-z. [DOI] [PubMed] [Google Scholar]
- 15.Crino L, Weder W, van Meerbeeck J, Felip E. Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v103–115. doi: 10.1093/annonc/mdq207. [DOI] [PubMed] [Google Scholar]
- 16.Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J. Clin. Oncol. 2013;31:1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]


