Abstract
Upon encystment induction, Azotobacter vinelandii produces the phenolic lipids alkylresorcinols (ARs) that are structural components of the cysts. The enzymes responsible for the ARs synthesis are encoded in the arsABCD operon, whose expression is activated by ArpR. The transcription of arpR is initiated from an RpoS dependent promoter. The nitrogen-related phosphotransferase system (PTSNtr) is a global regulatory system present in Gram negative bacteria. It comprises the EINtr, NPr and EIIANtr proteins encoded by ptsP, ptsO and ptsN genes respectively. These proteins participate in a phosphoryl-group transfer from phosphoenolpyruvate to protein EIIANtr via the phosphotransferases EINtr and NPr. In A. vinelandii, the non-phosphorylated form of EIIANtr was previously shown to repress the synthesis of poly-ß-hydroxybutyrate. In this work, we show that PTSNtr also regulates the synthesis of ARs. In a strain that carries unphosphorylated EIIANtr, the expression of arpR was reduced, while synthesis of ARs and transcription of arsA were almost abrogated. The expression of arpR from an RpoS-independent promoter in this strain restored the ARs synthesis. Taken together these results indicate that unphosphorylated EIIANtr negatively affects activation of arpR transcription by RpoS.
Introduction
Azotobacter vinelandii is a soil bacterium that undergoes a differentiation process resulting in the formation of a desiccation resistant cyst. A mature cyst consists of a contracted cell, known as the central body, which is surrounded by a capsule made up of a laminated outer layer called the exine and an inner layer called the intine [1]. The polysaccharide alginate is a major component of the capsule layers. Other components of the cysts are the reserve polyester poly-ß-hydroxybutyrate (PHB), that is present in the central body forming large granules, and the phenolic lipids alkylresorcinols (ARs), which replace the membrane phospholipids in the cyst and are also components of the exine [2]. Encystment can be induced by transferring log-phase vegetative cells to Burk’s minimal medium with either n-butanol or ß-hydroxybutyrate as the sole carbon source [3].
ARs play a structural role in the cyst, and strains carrying mutations in ARs biosynthetic genes produce cysts with a defective exine [4]. The arsABCD gene cluster encodes the enzymes that synthesize these lipids [5]. These genes are specifically expressed in encystment induction medium [4]. The transcriptional activator ArpR positively regulates transcription of the arsABCD operon, by direct binding to the arsA promoter region [6]. The mutational inactivation of rpoS impairs ARs synthesis [7] because this sigma factor is needed for the transcription of arpR [6].
The ptsP, ptsO and ptsN genes encode EINtr, NPr and EIIANtr proteins, respectively, that are components of the nitrogen-related phosphotransferase system (PTSNtr), which is homologous to the carbohydrate transport PTS. The PTSNtr proteins participate in a phosphoryl transfer chain from phosphoenolpyruvate, where EIIANtr appears to be the terminal phosphoryl acceptor [8]. The PTSNtr regulates a wide variety of processes in bacteria; in Legionella pneumophila, a ptsP mutation, negatively affected its virulence in guinea pigs [9]; in Rhizobium species, the PTSNtr is associated to melanin synthesis, nitrogen fixation and regulation of ABC transport activation [10,11]. In Escherichia coli, the EIIANtr protein controls the potassium transport by interacting with the Trk transporter subunit TrkA and the sensor kinase KdpD (that controls the expression of high affinity potassium transporter system KdpFABC) [12,13]. The response of E. coli to phosphate starvation is also activated by EIIANtr due to an interaction with the sensor kinase PhoR [14].
In the A. vinelandii UW136, the non-phosphorylated form of EIIANtr was shown to impair PHB production, by exerting a negative effect on expression of phbR, the gene encoding the transcriptional activator of the PHB biosynthetic operon phbBAC [15].
In this work we report the effect of mutations in the genes coding for the proteins of the PTSNtr on alkylresorcinol synthesis and show that the non-phosphorylated EIIANtr protein has a negative effect on the transcriptional activation of arpR by RpoS.
Materials and Methods
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used are listed in S1 Table. A. vinelandii was cultured at 30°C in Burk’s nitrogen-free salts medium [16] supplemented with 2% sucrose (BS) for vegetative growth or 0.2% n-butanol (BBOH) for encystment induction. For determination of β-glucuronidase activity of transcriptional phbR-gusA and phbB-gusA fusions, the cells were grown in peptone yeast medium supplemented with 2% sucrose (PY). Liquid cultures were carried out in 250-mL or 125-ml flasks containing 50 or 25 ml of medium, respectively, in a rotatory shaker at 200 rpm and 30°C. Inocula for all experiments were grown on BS, washed three times with sterile 10mM MgSO4, and transferred to BBOH medium.
E. coli strain DH5α was grown in Luria-Bertani medium (LB) at 37°C. Transformation of A. vinelandii were carried out as previously described [16].
Nucleic acid procedures
DNA purification and cloning procedures were carried out as previously described [17]. Total RNA extraction was performed as reported by Barry et al. [18]. DNA sequencing was done with a Perkin Elmer/Applied Biosystems DNA Sequencer. The sequences of oligonucleotides used in this work are described in the S2 Table.
Constructions of transcriptional and translational fusions of arpR and arsA with gusA reporter
The pUMATc plasmid [19] was digested with EcoRI and HindIII to clone the gusA reporter gene obtained from pAHFUTs-Tc [20], resulting in the plasmid pUMATcgusAT.
The plasmids pUMATcgusAT and pUMATcgusAPT [19], unable to replicate in A. vinelandii and used for transcriptional and translational fusions, respectively, were digested with SacI and KpnI restriction enzymes to remove the tetracycline cassette. The ends of the plasmids were made blunt by treatment with Klenow fragment and used for cloning a blunted MluI gentamicin cassette obtained from pBSL98 [21]. The new plasmids pLM2 and pLM3 (S1 Table) were used to construct the transcriptional and translational fusions, respectively.
For the construction of transcriptional arsA-gusA and arpR-gusA fusions, DNA fragments of 1.0 and 0.99 Kb, containing the promoter region of arsA and arpR, respectively, were amplified using the primers FwarsA and RvarsAtrans and FwarpR and RvarpRtrans (S2 Table). The fragments were gel-purified, digested with XbaI and PstI and ligated to XbaI-PstI pLM2 vector to construct the plasmids pLM4 (arsA-gusA) and pLM6 (arpR-gusA). These plasmids were digested with NdeI and ScaI, respectively, and used to transform A. vinelandii strains for the selection of transformants carrying transcriptional arsA-gusA or arpR-gusA fusions integrated into the melA gene by a double recombination event. Digestion of the plasmids was carried out in order to avoid the selection of strains with plasmids integrated into the chromosome generated by single recombination events. The melA gene has been previously used as a neutral site to introduce gene fusions [19]. These strains are described in S1 Table.
For the construction of translational arsA´-´gusA and arpR´-´gusA fusions, DNA fragments of 1.3 and 1.1 Kb (containing the promoter region, the 5’ untranslated region and the first five codons of each gene) were amplified with FwarsA and RvarsAtrad and FwarpR and RvarpRtrad primers for arsA and arpR, respectively. The PCR products were purified, digested with XbaI and PstI enzymes and ligated to XbaI-PstI pLM3 resulting in the plasmids pLM5 (arsA´-´gusA) and pLM7 (arpR´-´gusA). The plasmids pLM5 and pLM7 were digested with NdeI and ScaI, respectively, and used to transform A. vinelandii strains for the selection of the transformants carrying translational arsA´-´gusA or arpR´-´gusA fusions described in S1 Table. The presence of all fusions in the strains was confirmed by PCR analysis (data not shown).
Construction of plasmid pBpgyrA-arpR to express arpR from RpoS-independent promoter
First, we constructed the plasmid pJET-pgyrA cloning a 0.3 Kb DNA fragment containing the promoter region of gyrA gene (pgyrA) into vector pJET1.2/blunt (Thermo Scientific). A DNA fragment of 1.0 Kb containing the encoding region of arpR was amplified using the oligonucleotides arpRFw2 and arpRRv2 (S2 Table) and cloned into pJET-pgyrA downstream and the same direction of pgyrA. The fusion pgyrA-arpR was excised by digestion with BglII enzyme, gel purified, made blunt and cloned into SmaI-digested plasmid pBBR1MCs-5 [22], resulting in the plasmid pBpgyrA-arpR, which was transferred by conjugation into strain UW136::pALA8a.
Quantitative Real Time PCR (q-RT-PCR)
Expression levels of arsA and arpR was measured by qRT-PCR as previously reported [15]. The primers used for the assays (S2 Table) were as follows: arsA-RT-F and arsA-RT-R for arsA, arpR-RT-F and arpR-RT-R for arpR, and fw-gyrA and rev-gyrA for gyrA. The level of gyrA was used as internal control to normalize the results. All assays were performed in triplicate. The data was analyzed by the 2-Δ,ΔCT method reported by Livak and Shmittgen [23].
Determination of alkylresorcinol production
The production of ARs was measured as previously described [24]. Briefly, the lipids were extracted with acetone for 1h at room temperature. The acetone extract was removed, and a second extraction was done for 12 h at room temperature. The resulting extracts were mixed and used for spectrophotometric determination of alkylresorcinols by the use of Fast Blue B as previously described [24]. Orcinol was used as a standard. The protein content of the cells used for AR determination was measured by the method of Lowry et al [25].
Quantification of β-glucuronidase activity
The β-glucuronidase activity was measured as described previously [26] from encystment-induced cells in BBOH medium harvested to 72 hours of incubation. 1 U corresponds to 1 nmol of p-nitrophenyl-β-D-glucuronide hydrolyzed per minute per mg of protein.
Results
Effect of ptsP, ptsO and ptsN mutations on ARs synthesis
Strain UW136 is unable to produce alginate due to an insertion within the algU gene [27] therefore this strain is unable to produce genuine mature cysts, but under encystment induction medium produces ARs [4].
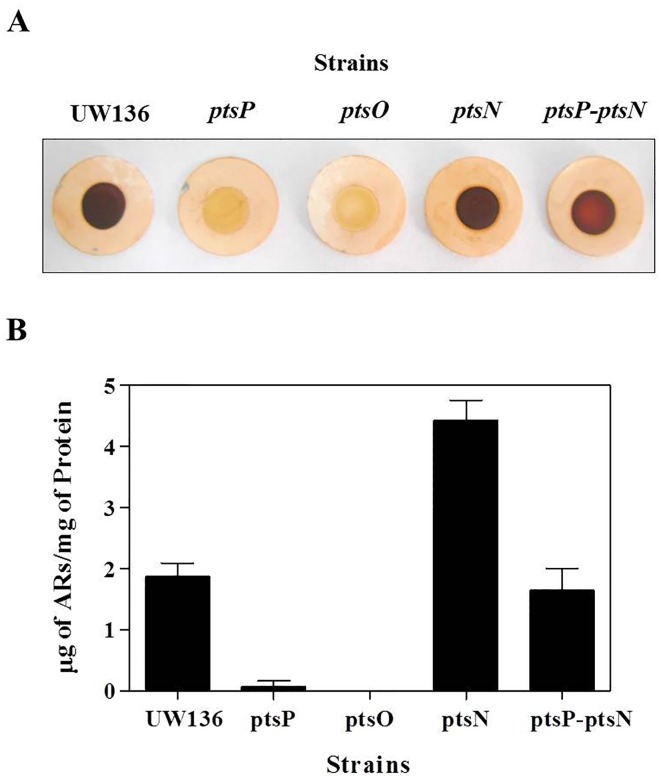
To determine if PTSNtr is involved in the regulation of the ARs synthesis, we analyzed ARs production in encystment-induced cells of pts mutants, by staining these lipids with dye Fast Blue B [4]. The ptsN mutant and the UW136 wild type strain developed a red color indicative of ARs synthesis, while the ptsP and ptsO mutants remained white (Fig. 1A). The quantification of ARs production in these strains confirmed the observed phenotype in plates; no ARs were detected in the ptsP and ptsO mutants, while the ptsN mutant presented a significant increase in ARs production relative to the UW136 strain (Fig. 1B). According to the phosphorylation cascade proposed for the PTSNtr [15] the ptsP and ptsO inactivations are expected to impair the phosphorylation of EIIANtr, therefore, the unphosphorylated form of EIIANtr could be involved in the negative effect observed on ARs synthesis. In agreement with this hypothesis, inactivation of ptsN, in the ptsP mutant background (ptsP-ptsN double mutant) restored the ARs synthesis (Fig. 1B).
Fig 1. PTSNtr controls the ARs synthesis.
Staining (A) and quantification (B) of alkylresorcinols produced by strains of A. vinelandii in BBOH medium to 120 hours of incubation. These data are mean of three independent experiments, error bars, SD.
Effects of PTSNtr mutations on arsA expression
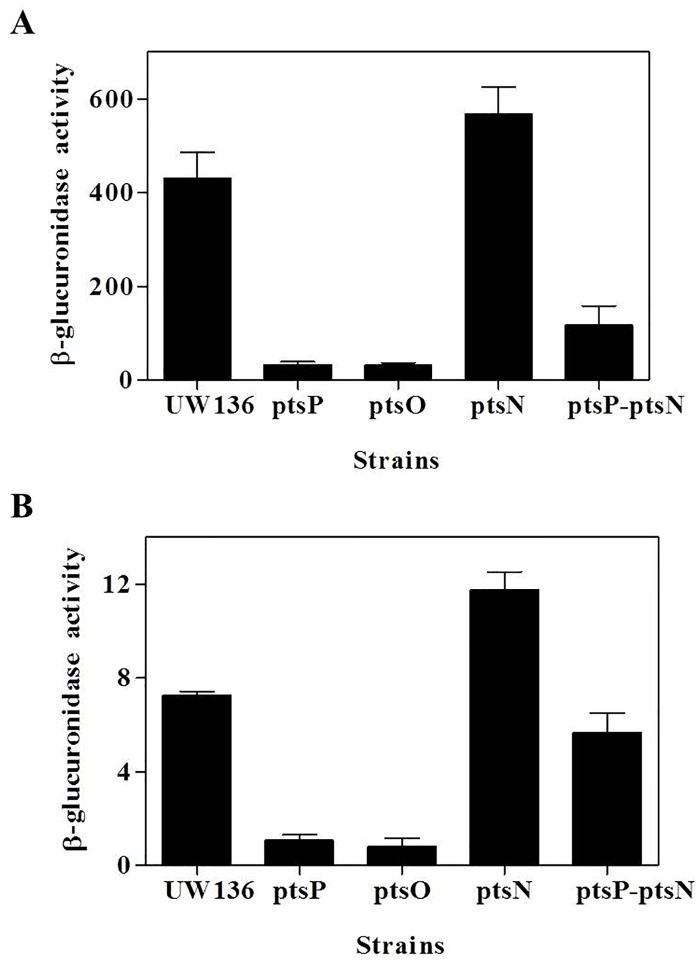
In order to determine if PTSNtr affected ARs synthesis through an effect on arsABCD expression, transcriptional arsA-gusA and translational arsA´-´gusA gene fusions were used. The transcription and translation levels of arsA were determined by measuring β-glucuronidase activity in derivatives of the wild type UW136 strain and the ptsP, ptsO, pstN and ptsP-ptsN mutants carrying the gene fusions (S1 Table). We observed that ptsP and ptsO inactivations caused a similar decrease in the β-glucuronidase activity of both fusions (Fig. 2A and 2B), while in the ptsN mutant the activity increased in the transcriptional and translational fusions by 27% and 60% respectively. In the double mutant ptsP-ptsN, the β-glucuronidase activity of transcriptional fusion was partially restored, while the translational fusion showed a similar level to that observed in the wild type strain (Fig. 2A and 2B).
Fig 2. Effect of PTSNtr on arsA expression.
β-glucuronidase activity in UW136 wild type strain and pts mutants carrying transcriptional arsA-gusA (A), or translational arsA´-´gusA (B) gene fusions. The cells were grown for 72 h in BBOH medium at 30°C. The data represent the mean of three independent experiments. Error bars, SD.
The level of arsA transcripts in the pts mutants was also evaluated by qRT-PCR. Table 1 shows that the arsA mRNA levels in ptsP and ptsO mutants were very low when compared to those observed in the wild type strain. In contrast, the arsA mRNA level was higher in the ptsN mutant and the double mutant ptsP-ptsN than in the UW136 strain. These results support the hypothesis that the non-phosphorylated form EIIANtr negatively affects arsA expression at the transcriptional level.
Table 1. Relative mRNA levels of arsA and arpR in UW136 and pts mutants strains.
| Relative mRNA levels* | ||
|---|---|---|
| Strain | arsA | arpR |
| UW136 | 1.0 ± 0.0 | 1.0 ± 0.0 |
| PtsP | 0.043 ± 0.007 | 0.021 ± 0.003 |
| PtsO | 0.099 ± 0.003 | 0.005 ± 0.003 |
| PtsN | 2.07 ± 0.24 | 2.7 ± 0.16 |
| ptsP-ptsN | 1.62 ± 0.10 | 1.3 ± 0.11 |
*The mRNA levels of arsA and arpR in pts mutants are relative to showed by UW136 strain, which are assumed to be 1.0. The values are the mean of two independent experiments.
Effects of PTSNtr mutations on arpR expression
The results shown above suggest that PTSNtr controls the ARs synthesis trough the regulation of expression of arsABCD. We recently reported that ArpR, a LysR-type regulator, directly actives the transcription of arsABCD [6]. Therefore, the question of whether the PTSNtr affected the transcription of arsA through an effect on the arpR expression was raised.
To study the effects of pts mutations on arpR expression, we used the UW136, ptsP, ptsO, ptsN and ptsP-ptsN strains carrying transcriptional and translational fusions of arpR (S1 Table). Fig. 3A shows that transcription of arpR, measured as β-glucuronidase activity, decreased about 40% in the ptsP and ptsO mutants relative to the wild type strain, while the ptsN inactivation had no affect on arpR transcription in the UW136, nor in the ptsP strains (Fig. 3A). The β-glucuronidase activity in the wild type and pts mutants carrying the translational arpR´-´gusA fusion (Fig. 3B), showed that the ptsP and ptsO mutations reduced about 50% the translation of arpR, as compared to the UW136 strain. In contrast, in the ptsN and ptsP-ptsN mutants a significant increase in the β-glucuronidase activity, relative to UW136 and ptsP strains respectively, was observed (Fig. 3B).
Fig 3. Effect of PTSNtr on arpR expression.
β-glucuronidase activity in UW136 wild type strain and pts mutants carrying transcriptional arpR-gusA (A) or translational arpR´-´gusA (B) gene fusions. The cells were grown for 72 h in BBOH medium at 30°C. The data represent the mean of three independent experiments. Error bars, SD.
Using qRT-PCR, we found that the ptsP and ptsO mutations diminished the arpR mRNA level, while the ptsN inactivation increased it, in both the wild type UW136 and ptsP strains (Table 1). These results suggest that the non-phosphorylated EIIANtr of the PTSNtr negatively controls arpR expression at the transcriptional and posttranscriptional levels.
H68A mutation in the phosphorylation site of EIIANtr impairs the transcription of arpR
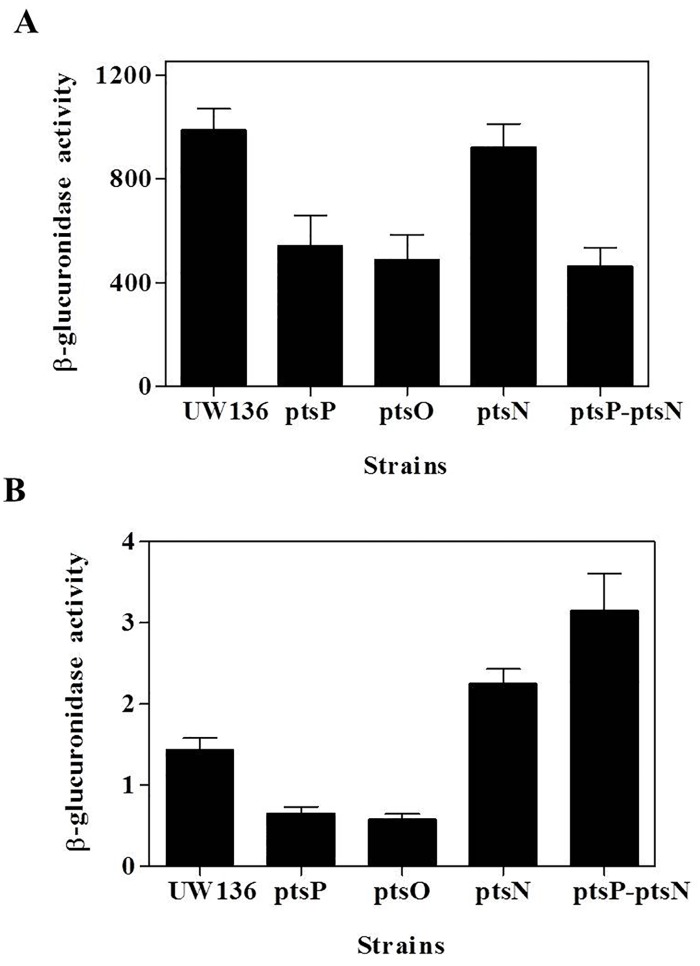
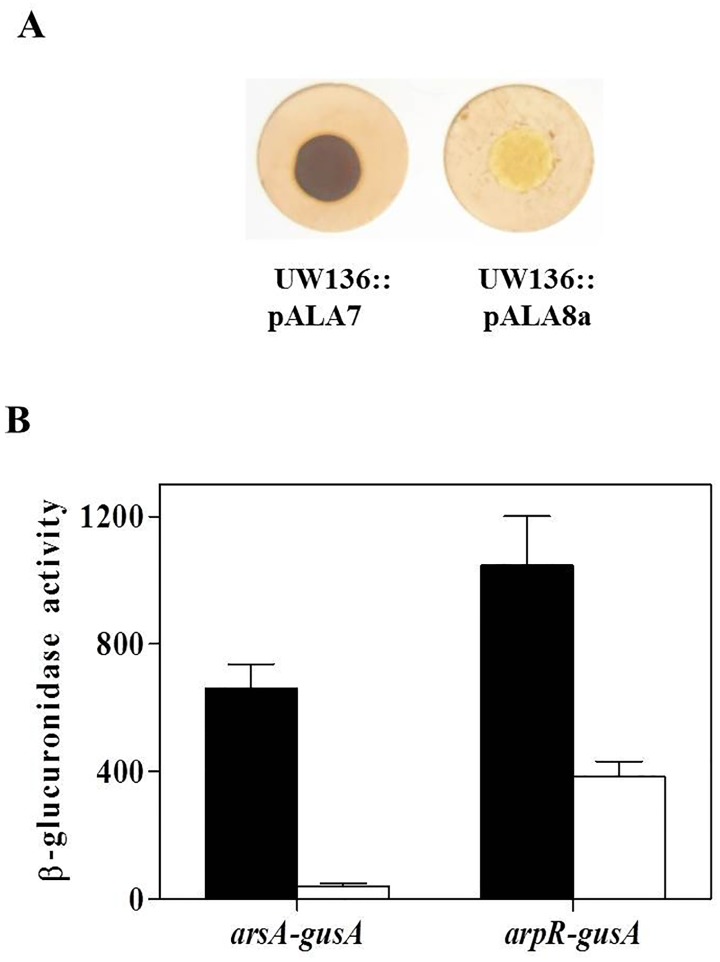
The results presented above imply that the unphosphorylated form of EIIANtr negatively controls the expression of arpR, affecting the transcription of arsA and, in turn, the synthesis of ARs. Thus, we tested the effect of a point mutation in ptsN (H68A), which produces a non-phosphorylatable EIIANtr, on ARs production and on transcription of arpR and arsA. For this experiment we used the strain UW136::pALA8a, which carries the ptsN-H68A mutation [15]. As shown in Fig. 4A, this strain showed a negative ARs production, similar to that observed in the ptsP and ptsO mutants (compare Figs. 1A and 4A). In contrast, the strain UW136::pALA7, which carries a wild type ptsN gene [15] presented a phenotype of ARs production, similar to UW136 wild type strain. As shown in Fig. 4B, the ptsN-H68A mutation almost abrogated the β-glucuronidase activity in the strain carrying the transcriptional arsA-gusA fusion, and reduced by 60% the activity of the arpR-gusA fusion relative to the strain UW136::pALA7. These results indicate that the unphosphorylated EIIANtr protein represses the transcription of arpR.
Fig 4. The unphosphorylated EIIANtr negatively affects the ARs synthesis.
Effect of H68A mutation on EIIANtr on ARs production (A), and activity of transcriptional arsA-gusA and arpR-gusA fusions (B). The strains UW136::pALA7 (black bars) and UW136::pALA8a (white bars) carry an EIIANtr and H68A EIIANtr, respectively. The cells were grown for 120 h for (A) and 72 h for (B) in BBOH medium at 30°C. The data represent the mean of three independent experiments. Error bars, SD.
The negative effect of unphosphorylated EIIANtr H68A on arpR transcription is through RpoS
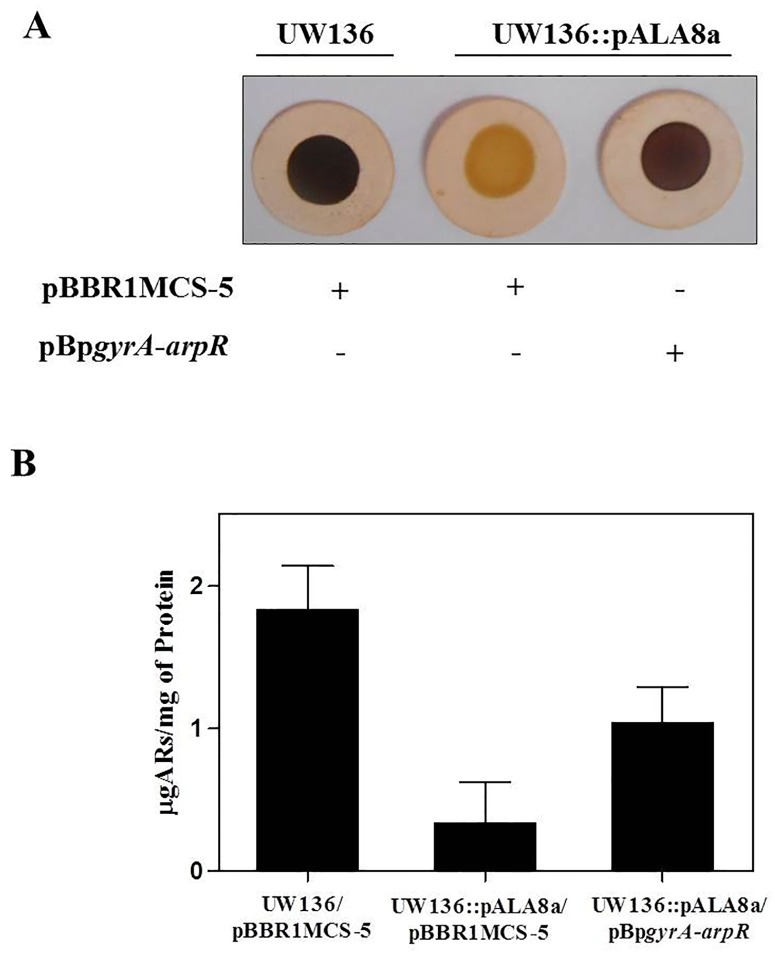
The data presented above indicate that transcription of arpR is negatively regulated by the unphosphorylated EIIANtr protein. Since arpR transcription is dependent on RpoS [6], we wanted to determine if the unphosphorylated EIIANtr affects arpR expression through this sigma factor. For this, we determined the capacity of AR synthesis in the strain UW136::pALA8a (expressing the unphosphorylatable EIIANtr) carrying the plasmid pBpgyrA-arpR, which expresses the arpR gene from an RpoS-independent promoter (gyrA promoter). As shown in the Fig. 5A, this strain was able to synthesize ARs in BBOH plates. In contrast, a negative phenotype of ARs production was shown by the strain UW136::pALA8a when was transformed with the empty plasmid pBBR1MCS-5. A similar effect was observed in BBOH liquid medium; the RpoS-independent expression of arpR increased the AR levels in the strain UW136::pALA8a (Fig. 5B). These results suggest that negative effect of unphosphorylated EIIANtr on arpR expression is due to a negative effect on its transcriptional activation by RpoS.
Fig 5. Effect of arpR expression from RpoS-independent promoter in the strain that carries the nonphosphorylatable EIIANtr H68A protein.
(A) Staining of ARs produced by UW136 and U136::pALA8a strains, transformed with plasmid PBpgyrA-arpR, carrying a constitutively expressed arpR gene or the empty plasmid pBBR1MCS-5 as negative control. (B) Quantification of ARs levels produced by the strains of the panel A. The data represent the mean of three independent experiments. Error bars, SD.
Discussion
The alkylresorcinols are exclusively synthesized during the encystment in A. vinelandii, since the expression of arsABCD operon is specifically activated in this condition [4]. Here, we identified that PTSNtr regulates the expression of ARs biosynthetic operon, through regulation of its transcriptional activator ArpR.
The PTSNtr is present in many bacterial genus and controls diverse physiological processes through the phosphorylation state of EIIANtr [8]. Since mutations on ptsP or ptsO impair the phosphoryl-group transfer to EIIANtr, we hypothesized that absence of ARs synthesis in ptsP and ptsO mutants was mainly due to the presence of the unphosphorylated EIIANtr. This was confirmed by two approaches. First, the inactivation of ptsN was sufficient to restore the ARs levels in the ptsP mutant (Fig. 1B), and second, the strain that harbors an unphosphorylatable EIIANtr H68A (which presents a replacement on the phosphorylation site histidine by an alanine) showed a negative ARs production phenotype (Fig. 4A).
Unexpectedly, in the ptsP-ptsN double mutant the ARs levels were lower than in the ptsN mutant (Fig. 1B). Additionally, the ptsP mutation produced a stronger negative effect on ARs production than the mutation producing an unphosphorylatable EIIANtr H68A protein (compare Figs. 1B and 5B), suggesting a secondary regulatory role of EINtr and/or NPr proteins on ARs synthesis independent of its role in the phosphorylation of EIIANtr. Additional experiments are necessary to validate this hypothesis.
The difference in ARs production between the ptsN and the ptsP-ptsN mutants could also be explained by the presence of an EIIANtr paralog that partially complements the ptsN mutation. However a single ptsN gene was found in the A. vinelandii genome.
The transcription of arsA was reduced when EIIANtr was present in its unphosphorylated form (Fig. 4B). Recently, we reported that both arsABCD and arpR transcription are directly activated by ArpR and acetoacetyl Coenzyme A (acetoacetyl-CoA) as coinducer [6]. Because the unphosphorylated EIIANtr also reduced the arpR transcription (Fig. 4B), we concluded that the negative effect on arsABCD expression was due to a reduction of arpR expression. The negative effect of the EIIANtr on expression of arpR could be explained by a reduction of the acetoacetyl-CoA pool. However this does not seems to be the case, since the presence of 5 and 50 μM of acetoacetyl-CoA did not restore the AR synthesis in ptsP, ptsO and ptsN H68A mutants (S1 Fig.). In contrast, an increase of ARs production phenotype dependent of acetoacetyl-CoA concentration was observed in the strains UW136, ptsN and ptsP-ptsN (S1 Fig.).
EIIANtr has been shown to indirectly regulate the expression of several genes. For example, in E. coli, the interactions between EIIANtr and kinase sensors KdpD and PhoR, increase the phosphorylation of response regulators KdpE and PhoB, resulting in increased expression of kdpFABC and the pho regulon, respectively [13,14]. Another interesting example is present in Salmonella, where EIIANtr interacts with the SsrB response regulator, reducing the expression of Salmonella pathogenicity island 2 (SPI-2) [28]. Additionally, a relationship between EIIANtr and the activity of sigma factors RpoS and RpoD has been previously described in E. coli [29]. In the absence of EIIANtr (in a ptsN mutant), the potassium levels increase (by derepression of activity of K+ Trk transporter) resulting in preferential binding of the core RNA polymerase to RpoS instead of RpoD, and therefore, affecting the transcription of sigma regulons [29]. Here, we found that in A. vinelandii the negative effect of the unphosphorylated EIIANtr on arpR transcription is through RpoS, since the expression of arpR from an RpoS-independent promoter was sufficient to restore ARs synthesis in the presence of unphosphorylated EIIANtr (Figs. 5A and 5B). Further evidence supporting the participation of RpoS in the regulation exerted by EIIANtr includes previous results showing that transcription of phbR, the gene encoding the transcriptional activator of PHB, and transcription of promoter pB2 of phbBAC are also RpoS dependent [30,31] and repressed by unphosphorylated EIIANtr [15]. We carried out additional experiments to confirm the negative effect of unphosphorylated EIIANtr protein on the phbB and phbR RpoS-dependent promoters (S2A and S2B Fig.). Indeed, the β-glucuronidase activity of transcriptional phbR-gusA and phbB-gusA fusions is reduced in the ptsP mutant (S2A and S2B Fig.). The mechanism by which the nonphosphorylated EIIANtr affects the RpoS activity in A. vinelandii remains to be elucidated.
Nonphosphorylated EIIANtr also seems to control the expression of arpR at a posttranscriptional level since the ptsN mutation increased the activity of the translational arpR-gusA fusion in the wild type and ptsP strains (Fig. 3B). Additionally, mutations of ptsP and ptsO diminished about twofold the activity of the transcriptional arpR-gusA fusion (Fig. 3A), while the arpR mRNA levels, measured by qRT-PCR, were even lower in the ptsP and ptsO mutants (Table 1). A similar effect was shown on the expression of ilvBN in E. coli, where a ptsN mutation reduced about 50% the activity of a transcriptional ilvB-lacZ fusion, while the ilvB mRNA levels (detected by RT-PCR) were more drastically reduced [32]. The mechanism by which nonphosphorylated EIIANtr negatively affects the arpR expression at posttranscriptional level remains to be determined. However, as the translational arpR fusion contains the 5’ untranslated region of arpR mRNA (including the Shine-Dalgarno sequence), this mechanism could be related to a reduction of mRNA stability and/or to a blockage of translation.
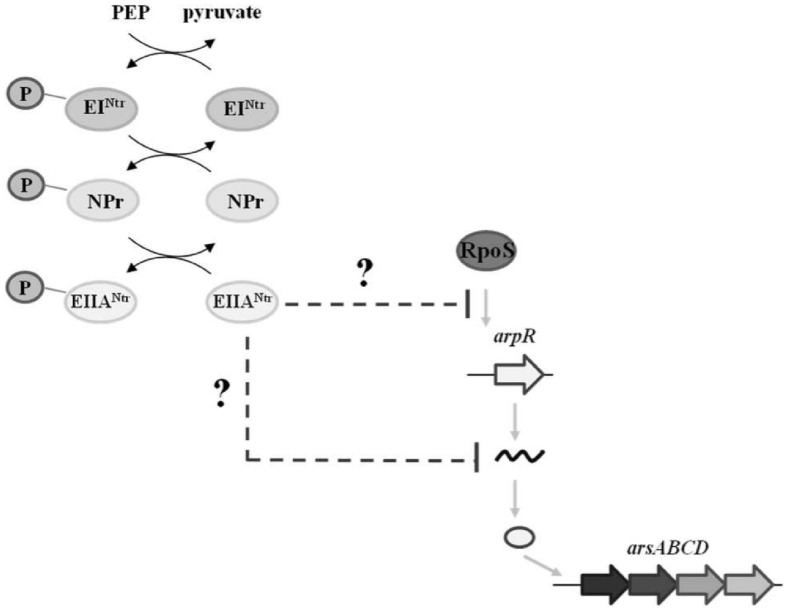
In summary, a regulatory model for the control of ARs synthesis by PTSNtr is proposed (Fig. 6). The EIIANtr protein in its nonphosphorylated state inhibits the activation of the transcription of arpR by RpoS. The repression of arpR expression impairs the transcriptional activation of biosynthetic arsABCD operon. Additionally, EIIANtr negatively affects the arpR mRNA levels by an unknown mechanism. The elucidation of the molecular mechanisms that link PTSNtr with RpoS and posttranscriptional regulation of arpR will allow us understand the role of PTSNtr in A. vinelandii.
Fig 6. Model for the control of ARs synthesis by PTSNtr in A. vinelandii.
The EIIANtr protein in its unphosphorylated form represses the arpR expression both transcriptional (RpoS activity) and posttranscriptional levels. The dashed lines and gray arrows indicate negative effect and activation, respectively. PEP: Phosphoenolpyruvate, P: Phosphoryl group.
Supporting Information
The strains were grown in BBOH medium in absence or presence of 5 and 50 μM acetoacetyl-CoA (coinducer) for 72 h at 30°C.
(TIF)
β-glucuronidase activity of transcriptional phbR-gusA (A) and phbB-gusA (B) fusions in UW136 and ptsP strains. The cells were grown in PY solid medium for 48 h at 30°C. The data represent the mean of two independent experiments. Error bars, SD.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank Paul Gaytán, Jorge Yanez, Eugenio López and Santiago Becerra for DNA synthesis and sequencing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by grant DGAPA-PAPIIT 209814. LFMM received financial support by CONACyT during his PhD studies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sadoff HL (1975) Encystment and germination in Azotobacter vinelandii . Microbiol Mol Biol Rev 39: 516–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reusch RN, Sadoff HL (1983) D-(-)-poly-beta-hydroxybutyrate in membranes of genetically competent bacteria. J Bacteriol 156: 778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin LP, Sadoff HL (1968) Encystment and Polymer Production by Azotobacter vinelandii in the Presence of B-Hydroxybutyrate. J Bacteriol 95: 2336–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Segura D, Vite O, Romero Y, Moreno S, Castaneda M, et al. (2009) Isolation and characterization of Azotobacter vinelandii mutants impaired in alkylresorcinol synthesis: alkylresorcinols are not essential for cyst desiccation resistance. J Bacteriol 191: 3142–3148. 10.1128/JB.01575-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Funa N, Ozawa H, Hirata A, Horinouchi S (2006) Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii . Proc. Natl. Acad. Sci. USA 103: 6356–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romero Y, Moreno S, Guzmán J, Espín G, Segura D (2013) Sigma factor RpoS controls alkylresorcinol synthesis through ArpR, a LysR-type regulatory protein, during encystment of Azotobacter vinelandii . J Bacteriol 195: 1834–1844. 10.1128/JB.01946-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cocotl-Yañez M, Sampieri A, Moreno S, Núñez C, Castañeda M, et al. (2011) Roles of RpoS and PsrA in cyst formation and alkylresorcinol synthesis in Azotobacter vinelandii . Microbiology 157: 1685–1693. 10.1099/mic.0.046268-0 [DOI] [PubMed] [Google Scholar]
- 8. Pflüger-Grau K, Görke B (2010) Regulatory roles of the bacterial nitrogen-related phosphotransferase system. Trends Microbiol 18: 205–214. 10.1016/j.tim.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 9. Higa F, Edelstein PH (2001) Potential virulence role of the Legionella pneumophila ptsP ortholog. Infect Immun 69: 4782–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Untiet V, Karunakaran R, Krämer M, Poole P, Priefer U, et al. (2013) ABC transport is inactivated by the PTSNtr under potassium limitation in Rhizobium leguminosarum 3841. PLOS ONE 8: e64682 10.1371/journal.pone.0064682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michiels J, Van Soom T, D´hooghe I, Dombrecht B, Benhassine T, et al. (1998) The Rhizobium etli rpoN locus: DNA sequence analysis and phenotypical characterization of rpoN,ptsN, and ptsA mutants. J Bacteriol 180: 1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang-Ro Lee S-HC, Mi-Jeong Yoon, Peterkofsky Alan, Yeong-Jae Seok (2007) Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc Natl Acad Sci USA 104: 4124–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lüttmann D, Heermann R, Zimmer B, Hillmann A, Rampp IS, et al. (2009) Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIANtr in Escherichia coli . Mol Microbiol 72: 978–994. 10.1111/j.1365-2958.2009.06704.x [DOI] [PubMed] [Google Scholar]
- 14. Lüttmann D, Göpel Y, Görke B (2012) The phosphotransferase protein EIIANtr modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli . Mol Microbiol 86: 96–110. 10.1111/j.1365-2958.2012.08176.x [DOI] [PubMed] [Google Scholar]
- 15. Noguez R, Segura D, Moreno S, Hernandez A, Juarez K, et al. (2008) Enzyme INtr, NPr and IIANtr are involved in regulation of the poly-β-Hydroxybutyrate biosynthetic genes in Azotobacter vinelandii . J Mol Microbiol Biotechnol 15: 244–254. [DOI] [PubMed] [Google Scholar]
- 16. Bali A, Blanco G, Hill S, Kennedy C (1992) Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol 58: 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 18. Barry T, Geary S, Hannify S, MacGearailt C, Shalloo M, et al. (1992) Rapid mini-preparations of total RNA from bacteria. Nucl Acids Res 20: 4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cocotl-Yañez M, Moreno S, Encarnación S, López-Pliego L, Castañeda M, et al. (2014) A small heat-shock protein (Hsp20) regulated by RpoS is essential for cyst desiccation resistance in Azotobacter vinelandii . Microbiology 160: 479–487. 10.1099/mic.0.073353-0 [DOI] [PubMed] [Google Scholar]
- 20. Hernandez-Eligio A, Moreno S, Castellanos M, Castañeda M, Nuñez C, et al. (2012) RsmA post-transcriptionally controls PhbR expression and polyhydroxybutyrate biosynthesis in Azotobacter vinelandii . Microbiology 158: 11. [DOI] [PubMed] [Google Scholar]
- 21. Alexeyev MF, Shokolenko IN, Croughan TP (1995) Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160: 63–67. [DOI] [PubMed] [Google Scholar]
- 22. Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, et al. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔC(T) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 24. Segura D, Cruz T, Espin G (2003) Encystment and alkylresorcinol production by Azotobacter vinelandii strains impaired in poly-β-hydroxybutyrate synthesis. Arch Microbiol 179: 437–443. [DOI] [PubMed] [Google Scholar]
- 25. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent J Biol Chem 193: 265–275. [PubMed] [Google Scholar]
- 26. Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans ADL, et al. (1995) β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology 141: 1691–1705. [DOI] [PubMed] [Google Scholar]
- 27. Martínez-Salazar JM, Moreno S, Najera R, Boucher JC, Espín G, et al. (1996) Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J Bacteriol 178: 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi J, Shin D, Yoon H, Kim J, Lee C-R, et al. (2010) Salmonella pathogenicity island 2 expression negatively controlled by EIIANtr-SsrB interaction is required for Salmonella virulence. Proc Natl Acad Sci USA 107: 20506–20511. 10.1073/pnas.1000759107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee C-R, Cho S-H, Kim H-J, Kim M, Peterkofsky A, et al. (2010) Potassium mediates Escherichia coli enzyme IIANtr-dependent regulation of sigma factor selectivity. Mol Microbiol 78: 1468–1483. 10.1111/j.1365-2958.2010.07419.x [DOI] [PubMed] [Google Scholar]
- 30. Peralta-Gil M, Segura D, Guzman J, Servin-Gonzalez L, Espin G (2002) Expression of the Azotobacter vinelandii poly-β-hydroxybutirate biosynthetic phbBAC operon is driven by two overlapping promoters and is dependent on the transcriptional activator PhbR. J Bacteriol 184: 5672–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hernandez-Eligio A, Castellanos M, Moreno S, Espin G (2011) Transcriptional activation of the Azotobacter vinelandii polyhydroxybutyrate biosynthetic genes phbBAC by PhbR and RpoS. Microbiology 157:3014–3023. 10.1099/mic.0.051649-0 [DOI] [PubMed] [Google Scholar]
- 32. Lee C-R, Koo B-M, Cho S-H, Kim Y-J, Yoon M-J, et al. (2005) Requirement of the dephospho-form of enzyme IIANtr for derepression of Escherichia coli K-12 ilvBN expression. Mol Microbiol 58: 334–344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The strains were grown in BBOH medium in absence or presence of 5 and 50 μM acetoacetyl-CoA (coinducer) for 72 h at 30°C.
(TIF)
β-glucuronidase activity of transcriptional phbR-gusA (A) and phbB-gusA (B) fusions in UW136 and ptsP strains. The cells were grown in PY solid medium for 48 h at 30°C. The data represent the mean of two independent experiments. Error bars, SD.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.