Abstract
The correction of disease-causing mutations by single-strand oligonucleotide-templated DNA repair (ssOR) is an attractive approach to gene therapy, but major improvements in ssOR efficiency and consistency are needed. The mechanism of ssOR is poorly understood but may involve annealing of oligonucleotides to transiently exposed single-stranded regions in the target duplex. In bacteria and yeast it has been shown that ssOR is promoted by expression of Redβ, a single-strand DNA annealing protein from bacteriophage lambda. Here we show that Redβ expression is well tolerated in a human cell line where it consistently promotes ssOR. By use of short interfering RNA, we also show that ssOR is stimulated by the transient depletion of the endogenous DNA mismatch repair protein MSH2. Furthermore, we find that the effects of Redβ expression and MSH2 depletion on ssOR can be combined with a degree of cooperativity. These results suggest that oligonucleotide annealing and mismatch recognition are distinct but interdependent events in ssOR that can be usefully modulated in gene correction strategies.
Keywords: DNA repair, gene correction, mismatch repair, Redβ, single-strand oligonucleotide
INTRODUCTION
Many inherited genetic disorders are caused by simple mutations in single disease genes. Correction of these mutations is an attractive approach to gene therapy with several advantages over gene addition approaches (Humbert et al., 2012; Yáñez and Porter, 1998). Gene correction can be achieved by homologous recombination (HR) with a double stranded DNA (dsDNA) repair template including several kilobase pairs of homology to the target locus. This general approach is widely used for site-specific mutagenesis in mammalian cells (gene targeting), frequencies of which are typically in the region of 10−5 to 10−7 events per transfected cell (Deng and Capecchi, 1992). Fortunately frequencies can be greatly stimulated by use of customised endonucleases to make a double strand break (DSB) in the target locus (Gaj et al., 2013; Urnov et al., 2005), or vectors based on Adeno-Associated Virus (Khan et al., 2011). Limitations remain, however, including off-target cleavage by the nucleases, competing DSB repair by mutagenic nonhomologous end-joining, and the challenges of constructing and efficiently delivering both custom nucleases and large DNA templates.
Single-stranded oligonucleotides can also be used as templates for chromosomal gene modification and have the attraction of being relatively easy to make and deliver. Initial work with chimeric RNA/DNA oligonucleotides (RDOs) (Andersen et al., 2002; Igoucheva et al., 2004; Liu et al., 2003) was difficult to reproduce (Manzano et al., 2003; Taubes, 2002) and has been superseded by gene modifications mediated by single-stranded oligodeoxyribonucleotides (ssOs) (Aarts and te Riele, 2011). Nevertheless, unaided frequencies of ssO-mediated gene repair/modification (ssOR) are still variable and often very low (Aarts et al., 2006; Hu et al., 2005; Igoucheva et al., 2001; Kenner et al., 2002; Nickerson and Colledge, 2003; Pierce et al., 2003).
Although ssOR, like gene targeting with dsDNA templates, can be greatly enhanced by nuclease-mediated cleavage of the target locus (Chen et al., 2011; Majumdar et al., 2008; Radecke et al., 2006; 2010), and seems to involve some components of the HR pathway of DSB repair (Ferrara and Kmiec, 2004; McLachlan et al., 2009; Morozov and Wawrousek, 2008), an attractive model for ssOR involves annealing of the ssO to regions of target ssDNA transiently exposed during DNA replication (Aarts and te Riele, 2011; Papaioannou et al., 2012). Consistent with this, procedures likely to promote or stabilise ssO annealing at replication forks can enhance ssOR without the need to risk introducing a chromosomal break. For example, ssOR can be stimulated by increasing the proportion of cells with replication forks and so increase opportunities for ssO annealing (Brachman and Kmiec, 2005; Olsen et al., 2005; Wu et al., 2005). Furthermore, cells deficient in the DNA mismatch repair (MMR), a key protein of the DNA mismatch repair (MMR) pathway (Jiricny, 2006), support greatly elevated levels of ssOR (Dekker et al., 2003), most likely because annealed ssOs, which must have some mismatches in order to modify the chromosome, are no longer removed by MMR. To exploit the latter observation it is important to show that transient depletion of MSH2 also stimulates ssOR.
Another approach that may promote ssO annealing, and therefore ssOR, is expression of Redβ, a single strand DNA annealing protein encoded by bacteriophage lambda (Erler et al., 2009; Iyer et al., 2002). Expression in E. coli of Redβ, and its partner Redα, a 5′-3′ exonuclease (or the equivalent rac prophage proteins, RecE and RecT), is the basis of a range of genetic engineering methods termed “recombineering” that require only short regions of homology between recombining dsDNA partners (Court et al., 2002; Muyrers et al., 2000). Recombineering also encompasses the use of ssOs to introduce defined sequence alterations into a target duplex, a procedure known to be particularly efficient and to depend on Redβ but not on Redα (Ellis et al., 2001; Swaminathan et al., 2001; Zhang et al., 2003). In common with ssOR in mammalian cells, Redβ-mediated ssOR in E. coli is impaired by the MMR system (Costantino and Court, 2003; Li et al., 2003). Furthermore, both mammalian ssOR (Igoucheva et al., 2001) and Redβ-mediated ssOR in E. coli (Ellis et al., 2001; Swaminathan et al., 2001; Zhang et al., 2003) can display a strand preference. These parallels between ssOR in mammalian cells and Redβ-mediated ssOR in bacteria suggest that Redβ might be able to promote ssOR in mammalian cells, a possibility that was supported by preliminary analyses in mouse ES cells (Zhang et al., 2003).
To characterise the effects of Redβ in mammalian cells in detail we describe here a human cell line in which nuclear expression of Redβ can be induced by removal of tetracycline from the medium. We show that such induction does indeed stimulate ssOR, that this effect can be enhanced by concomitant MSH2 depletion and that these stimulations are co-operative.
MATERIALS AND METHODS
Cell culture
The human fibrosarcoma HT1080 cell line was from American Tissue Culture Collection, grown as previously described (Itzhaki et al., 1997) in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal calf serum (GIBCO BRL, USA). Where appropriate the following antibiotics were added to the medium: zeocin (200 μg/ml), hygromycin (200 μg/ml), G418 (300 μg/ml) and tetracycline (1 μg/ml) (Sigma-Aldrich Inc., USA). Rht14 cells are zeocin-resistant HT1080 derivatives, stably transfected with a plasmid encoding a tetracycline transactivator protein. Unless stated otherwise, Rht14 cells transfected with pTRE-Tight-Redβ were maintained with tetracycline in the medium. Cell plating efficiencies were measured by placing 500 cells in a 9 cm diameter Petri dish and counting colonies formed after 10 days. To measure proliferation rates, cells growing in medium with or without tetracycline were passaged every 2–3 days; at each passage cells were counted and re-plating at 0.2 million cells per 9 cm diameter dish.
Plasmids and oligonucleotides
pcDNA/PGK-neo* and pcDNA-Redβ/PGK-neo* have been described (Zhang et al., 2003). A derivative of the latter, pcDNA-nlsRedβ/PGK-neo*, was made by adding AGGATCCAACC ATG GGC CCT AAA AAG AAG CGT AAA GTC GCC AGT in place of AGGAATTCACC ATG AGT at the N-terminus of Redβ. To make pTRE-Tight-Redβ, the nls-Redβ open-reading-frame was removed from pcDNA-nlsRedβ/PGK-neo* as an 825 bp BamHI/HindIII fragment and cloned into the multiple cloning site of pTRE-Tight (BD Biosciences, USA). Oligonucleotides were commercially synthesised by Sigma-Aldrich Inc.
Stable transfection
Stable transfection was performed by electroporation at 400 V and 250 μF on a BIO-RAD (USA) Gene Pulser as described (Itzhaki et al., 1997). HT1080 cells containing humanised transactivator (htTA) (8 × 106 cells) were co-transfected with pTRE-Tight-Redβ and pcDNA-Hyg-PGK-neo* (10 μg total, 20-fold molar excess of pTRE-Tight-Redβ) by electroporation; selection in hygromycin started 48 h after transfection and continued for 10–14 days until hygromycin-resistant colonies appeared.
Immunoblots
Immunoblots were as described (Liu et al., 2012). Briefly, cell pellets were prepared in lysis buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, sodium pyrophosphate, The β-glycerophosphate, EDTA, Na3VO4, leupeptin). Lysates were electrophoresed on SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked for 1 h at room temperature with 5% milk protein, 0.1% Tween 20 in PBS, then were probed with antibodies. Redβ was detected with a rabbit polyclonal antibody used at a 1:1000 dilution. MSH2 was detected with mouse monoclonal antibody (Calbiochem, USA), used a 1:100 dilution. Actin was detected with rabbit polyclonal antibody (SIGMA-Aldrich), used at a 1:1000 dilution. Secondary antibodies were horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (DAKO, USA), used at a 1:2000 dilution, or HRP-conjugated goat anti-mouse antibody (DAKO), used at a 1:1000 dilution.
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed with 2% (v/v) paraformaldehyde in PBS for 15 min, and then permeabilised with 0.4% (v/v) Triton X-100 in PBS for 15 min. Coverslips were washed with PBS and blocking buffer (5% (v/v) fetal bovine serum, 5% (v/v) normal goat serum in PBS) and then incubated for 1 h each with primary (Redβ polyclonal) and secondary (FITC-conjugated donkey anti-rabbit IgG; Autogen Bioclear, U K) antibodies, diluted (1:100) in blocking buffer. Coverslips were washed with PBS and mounted with Vectashield mounting medium containing DAPI (VECTOR laboratories, USA). Slides were viewed with a fluorescence microscope (Leica DM RB, USA).
Single-strand oligonucleotide repair
The neo*-carrying plasmid and ssO were co-transfected by lipof ectamine 2000 (Invitrogen, USA) according to manufacturer’s instructions. Three days before ssO transfection, tetracycline was removed from (− Tet, Redβ+), or maintained in (+ Tet, Redβ−) the medium; absence or presence of tetracycline in the medium was maintained for the following four days before restoring tetracycline to all media. The day before ssO-transfection, either untreated cells, or cells that had been transf ected with siRNA for 48 h, (next section), were seeded at ∼ 50% confluence in 15 cm diameter dishes. The following day, a mixture (3 ml) containing pcDNA-Hyg-PGK-neo* (5 pmole, 24 μg), ssO (95 pmole, 2 μg) and lipofectamine 2000 (60 μl) in OptiMEM was prepared according to manufacturer’s instructions, and added to fresh growth medium (9 ml) in each 15 cm diameter dish of cells, now approaching confluence (3 × 106 per dish). After 24 h, the cells of each dish were detached by trypsinisation and distributed into three similar dishes in medium containing G418 and tetracycline. On occasion, a sample of cells was taken at this stage for immunoblot analysis of Redβ expression. Fresh medium containing G418 and tetracycline was added every 2–3 days for 10–14 days until colonies appeared. Colonies were stained and counted as described (Porter and Itzhaki, 1993).
siRNA transfection
Sequences of siRNA duplexes (Dharmacon Inc, USA) used are shown in Supplementary Table S1. Four of these were components of a “SMARTpool” mixture of duplexes based on the MSH2 coding sequence. The siRNAs were delivered by oligofectamine (Invitrogen) according to manufacturer’s instructions. Cells grown in medium with tetracycline were plated at ∼50% confluence, (2 million cells per 15 cm diameter dishes) maintaining tetracycline. The following day, a mixture (1.8 ml) containing siRNA (90 μl of 20 μM stock) and oligofectamine (90 μl) in OptiMEM was prepared according to manufacturer’s instructions, and added to each 15 cm diameter dish of cells, now approaching confluence, containing 8 ml medium (with or without tetracycline), to give a final siRNA concentration of 180 nM. After 48 h, the cells of each dish were trypsinised and distributed into two 15cm diameter dishes (for ssOR) and/or at a similar density into a single well of a 6-well plate (for immunoblot analysis). After a further 24 h, cells were processed for ssOR (see previous section) or harvested for immunoblots.
RESULTS
Inducible Redβ expression in HT1080 cells
Because Redβ expression could be deleterious in mammalian cells, we expressed Redβ stably but inducibly. The Redβ open-reading-frame was cloned, with an N-terminal nuclear localisation signal (nls), downstream of a tetracycline responsive element (TRE) to generate pTRE-Redβ (Fig. 1A). To provide a target gene for ssOR, as well as a drug resistance marker for the selection of stable transfectants, a second plasmid, pcDNA/PGKneo* was used (Fig. 1B). It carries two expression cassettes, one conferring resistance to hygromycin, the other (PGKneo*) capable of conferring resistance to G418 only after correction of a 4 bp insertion in its neomycin phosphotransferase (neo) coding sequence. The DNA sequence of this region, and of repair and control ssOs used in ssOR assays, are shown in Figs. 1C and 1D. Plasmids pTRE-Redβ and pcDNA/PGKneo* were co-transfected, the former in molar excess, into an HT1080 derivative [Rht14; (Brough et al., 2011)] expressing an improved tetracycline transactivator protein capable of driving transcription from the TRE. Hygromycin-resistant colonies were expanded and analysed for Redβ expression by immunoblotting. Several clones showing tetracycline-regulated Redβ expression were identified and immunoblots for some are shown in Fig. 1E.
Fig. 1.
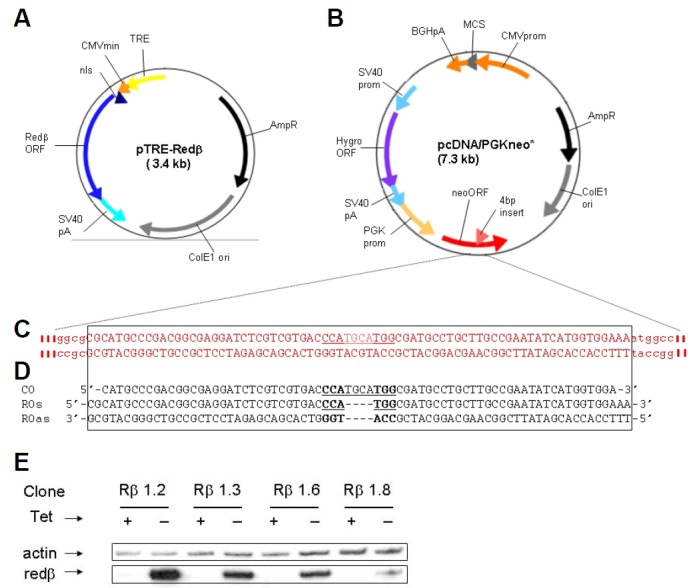
Reagents used for Redβ expres sion and ssOR assays. (A) Structure of Redβ expression plasmid. (B) Structure of plasmid carrying the neo* target for gene correction. (C) Sequence surrounding the 4 bp insertion (bold underlined) in neo*. (D) Sequence of control (CO) and repair (RO) ssOs used in ssOR assays. (E) Immunoblot analysis of Redβ expression in hygroR clones isolated after cotransfection of HT1080 cells with the plasmids shown in (A) and (B). Clones were expanded and grown continually with tetracycline (Tet) in the growth medium (+), or for 3 days after the removal of tetracycline (−), before analysis.
A detailed analysis of one of these clones (Rβ1.6) showed that, after addition of tetracycline to the growth medium, almost all immunoblot-detectable Redβ expression was lost within a period of 4 days (Fig. 2A). Removal of tetracycline from such cells resulted in abundant Redβ re-expression within 2 days and close to maximal expression within 3 days. Expression of Redβ had no effect on the doubling time of clone Rβ1.6 over a period of 17 days (Fig. 2B), at the end of which Redβ expression was still abundant in the culture that had been grown without tetracycline throughout (Fig. 2C). There were thus no acutely adverse effects of Redβ expression on cellular proliferation. Immunofluorescence on clone Rβ1.6 confirmed that Redβ was located in the nucleus (Fig. 2D).
Fig. 2.
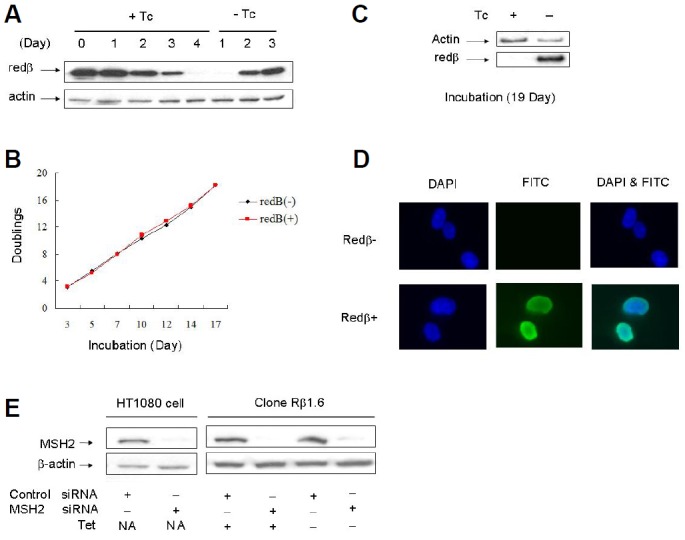
Analyses of Redβ expression and cell proliferation in clone Rβ1.6. (A) Cells that had been grown for 4 days without tetracycline (Tet) were transferred to medium with Tet (+Tc) for 4 days and then back into medium without Tet (−Tc) for 3 days, passaging whenever necessary to avoid confluence. Samples were taken at 24 h intervals, starting at the time of Tet addition, for immunoblot analysis. (B) Measurement of doubling times for cell grown with or without Tet. (C) Immunoblot analysis of cell samples taken at day 19 from cultures analysed in (B). (D) Nuclear location of Redβ in clone Rβ1.6. Cells grown with (−Redβ) or without (+Redβ) Tet for > 3 days were stained with DAPI to detect DNA, and analysed for Redβ (FITC-signal) by fluorescence microscopy. The pattern of staining shown is typical of all cells analysed. (E) MSH2 expression in siRNA-transfected cells. Immunoblot analyses of MSH2 in parental HT1080 or clone Rβ1.6 are shown. Rβ1.6 cells were grown with or without Tet, as indicated. Samples of HT1080 and Rβ1.6 cultures were taken at the time of ssO transfection in the ssOR experiments shown in Figs. 3B and 3C, respectively.
Stimulation of gene correction by Redβ
To measure ssOR of a chromosomal neo* target, cells from each of three clones with inducible Redβ (Rβ1.2, Rβ1.6 and Rβ1.8) were transfected with a 70nt repair ssO (RO, Fig. 1D). Correction of the neo* gene should generate G418r colonies, but none was obtained from a total of 16 million transfected Rβ1.6 cells, and 3 million transfected Rβ1.2 or Rβ1.8 cells. The frequency of gene correction for the chromosomal neo* was therefore well below 1 event per 106 cells. For all further measurement of ssOR we therefore measured gene correction of an episomal neo* gene by co-transfection of pcDNA/PGKneo* with RO or control ssO (CO, Fig. 1D). This approach was used in clones Rβ1.2, Rβ1.6 and Rβ1: all behaved similarly and together showed an average 1.6-fold stimulation of ssOR by Redβ (Fig. 3A). Control transfections with CO generated no G418R colonies.
Fig. 3.
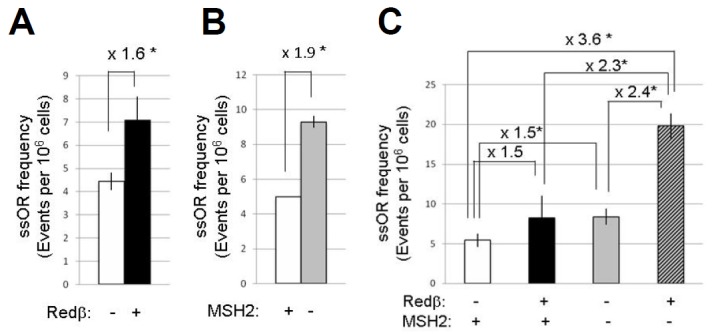
Effect of Redβ induction and MSH2 depletion on ssOR. (A) Effect of Redβ induction on neo* correction by RO in clones Rβ1.2, Rβ1.6 and Rβ1.8 (each clone assayed once). (B) Effect of MSH2 depletion on neo* gene correction by RO in HT1080 cells with or without the induction of Redβ (n = 3). (C) The effects of Redβ induction and MSH2 depletion, separately and together, on neo* gene correction by RO in clone Rβ1.6. For all experiments: repair frequencies were calculated from frequencies of G418 colonies as described in “Materials and Methods”; parallel control experiments using CO in place of RO generated no G418-resistant colonies; means and standard deviations of ssO frequencies are shown for three independent experiments; numerical fold increases and significance (* indicates p-values of < 0.05; Student’ t-test) are shown.
To determine whether transient Redβ expression could be used to stimulate ssOR, parental HT1080 cells were cotransfected with a neo* expression vector and ssO with or without Redβ expression vector (Supplementary Table S2). Although immunoblotting confirmed Redβ expression in amounts similar to stably transfected cells, any Redβ-mediated increase in ssOR was too small to detect in this experiment.
Stimulation of gene correction by MSH2 knockdown with and without Redβ expression
Control or MSH2-specific siRNAs were delivered to parental HT1080 cells with oligofectamine 72 h before co-transfecting the target neo* gene with repair or control ssOs. Cell samples taken for immunoblot analysis at the time of the second transfection showed clear MSH2 depletion specifically by the MSH2 siRNA (Fig. 2E). Subsequent selection in G418 and scoring for G418R colonies revealed a nearly 2-fold increase in the frequency of ssOR in MSH2-depleted cells (Fig. 3B).
To gain further evidence that transient MSH2 depletion can stimulate ssOR, and to test whether this could be enhanced by the simultaneous induction of Redβ expression, three independent MSH2 depletion experiments were carried out in clone Rβ1.6 cells, with or without the induction of Redβ, measuring neo* correction throughout. Again, immunoblot analysis of samples taken at the time of the neo* and ssO co-transfection confirmed MSH2 depletion (results for one experiment are shown in Fig. 2E). The frequencies of G418R colonies obtained are summarised in Fig. 3C. In these experiments the individual effects of Redβ expression and MSH2 depletion were observed again, though were slightly less pronounced than in the previous experiments (Figs. 2A and 2B). Notably, however, the stimulation of ssOR by Redβ expression became more pronounced when MSH2 was depleted than when it was not (2.4-fold vs. 1.5-fold). Similarly, the stimulation of ssOR by MSH2 depletion became more pronounced when Redβ was induced than when it was not (2.3-fold vs. 1.5-fold). Furthermore, the overall stimulation of ssOR caused by the combination of Redβ expression and MSH2 depletion was 3.6-fold, a figure considerably larger than the product of the individual stimulations (2.3 = 1.5 × 1.5). These observations suggest that the two stimulatory mechanisms may in fact work co-operatively.
DISCUSSION
The use of Redβ expression for genetic manipulations in bacteria is well established, but the suggestion (Swaminathan et al., 2001; Zhang et al., 2003) that this may be extended to eukaryotic organisms has received little attention. We have therefore begun to study the effects of nuclear Redβ expression in a human cell line concentrating, in the present study, on ssOR. We find that Redβ carrying an N-terminal NLS is well tolerated in human fibrosarcoma cells and able to stimulate ssOR on co-transfected episomes. Although the degree of stimulation is small (< 2-fold), it is highly reproducible and, in common with Redβ-mediated ssOR in bacteria (Costantino and Court, 2003; Li et al., 2003), it can be enhanced by inactivation of the MMR system. The ssOR activity of Redβ in mammalian cells therefore appears to be qualitatively similar to that in bacterial cells raising the possibility that quantitatively useful stimulations may yet be achievable in mammalian cells.
The mechanism of ssOR is believed to involve annealing of the ssOs to regions of ssDNA in the target locus transiently exposed during replication (Court et al., 2002; Dekker et al., 2003; Li et al., 2003; Zhang et al., 2003) or transcription (Igoucheva et al., 2003). We suggest that Redβ is able to promote such ssO annealing, albeit modestly, in mammalian cells. MMR has previously been proposed to suppresses ssOR by recognising and removing imperfectly annealed ssOs (Costantino and Court, 2003; Dekker et al., 2003). The fact that we observed some co-operativity between Redβ expression and MSH2 depletion, suggests that annealing and mismatch recognition may be concerted events during ssOR. It is possible, for instance, that Redβ protects annealed ssOs from the MMR system, while impaired MMR slows replication forks allowing Redβ to promote further annealing.
There are several reasons why the extent of Redβ-dependent ssOR activity may be limited in mammalian cells. First, one or more cellular proteins may compete or otherwise interfere with the ability of Redβ to access ssDNA. Proteins that, like Redβ, have single strand DNA binding or annealing activity may be particularly relevant here, e.g. Replication Protein A (Bochkarev and Bochkareva, 2004), Rad52 (Lisby and Rothstein, 2009). Transient depletion of such candidate proteins by RNA interference could be used to explore this possibility. Second, ssO instability may be a limiting factor. Although DNA stability is not a major factor for recombineering with ssOs in bacteria, it is limiting for recombineering with duplex DNA where expression of Redβ is required to inactivate host nuclease activities. To stabilise ssOs during ssOR experiments in mammalian cells, 2′-O-methyl-uracil residues are often added to 5′ or 3′ ends of ssOs, but their efficacy in this context is questionable although other modifications may help (Andrieu-Soler et al., 2005; Dekker et al., 2003). Other aspects of ssO design, such as length and mismatch position, are also known to be important variables. It may therefore be that larger stimulations can be achieved after ssO design has been re-optimised in the context of Redβ-mediated ssOR. Alternatively, cellular nucleases with known specificity for ssDNA (Shevelev and Hübscher, 2002) might usefully be depleted by RNA interference. One approach that may circumvent both ssO instability and preferential binding of endogenous proteins may be to co-express Redα with Redβ and to use dsOs in place of ssOs. This is suggested by the fact that Redβ is thought to physically interact with Redα so that the generation of 3′-ssDNA tails by exonuclease digestion occurs cooperatively with the formation of Redβ-ssDNA filaments (Court et al., 2002; Muyrers et al., 2000). Lastly, it is conceivable that bacterial host proteins interact with Redβ to generate maximum ssOR activity, and that similar proteins are either absent in mammalian cells or too poorly conserved to interact effectively. Although we cannot discount this possibility, it is notable that strand annealing (and invasion) reactions are catalysed by Redβ in vitro without the need for accessory proteins (Karakousis et al., 1998; Li et al., 1998).
Our results go beyond a previous study (Zhang et al., 2003) in which mouse ES cells were stably transfected with pcDNA-redβ/PGK-neo*; in contrast to cells transfected with vector only, these gave rise to G418R colonies at low frequency when transfected with a repair ssO. However subsequent analysis showed that Redβ was predominantly cytoplasmic in these cells and attempts at similar experiments involving nls-Redβ (i.e. using pcDNA-nlsredβ/PGK-neo*) failed to generate clones with nuclear Redβ, presumably because nuclear Redβ is not tolerated by ES cells. It is unclear at present why HT1080 cells should tolerate Redβ expression in the nucleus better than ES cells. ES cells have a short doubling time and it is possible that this makes them more sensitive to perturbations in the DNA repair machinery. Alternatively, a checkpoint mechanism may be inactive in HT1080 cells that, in ES cells, is responsible for sensitivity to Redβ expression. Redβ does appear to be tolerated in yeast where delivery of a Redβ expression construct stimulated ssOR nine-fold (Brachman and Kmiec, 2003).
Our results with MSH2 depletion extend a previous study (Dekker et al., 2003) showing that ES cells in which both MSH2 alleles are disrupted support up to 100-fold greater levels of ssOR than control cells. Here we have shown that transient MSH2 depletion by RNA interference can be used to promote ssOR, a necessary advance if efforts to stimulate ssOR are to be used more widely and, especially, for therapeutic purposes. The stimulations achieved were relatively modest but might be improved by the use of more efficient siRNA, or by the combined use of siRNA specific for more than one DNA mismatch repair protein.
Acknowledgments
This work was supported by the Volkswagen Stiftung (78784) to AP, National Natural Science Foundation of China (81372519) to KX, and Key Project of Tianjin Municipal Science and Technology Commission (14JCZDJC33800) to KX.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aarts M., te Riele H. Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Ther. 2011;18:213–219. doi: 10.1038/gt.2010.161. [DOI] [PubMed] [Google Scholar]
- Aarts M., Dekker M., de Vries S., van der Wal A., te Riele H. Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells. Nucleic Acids Res. 2006;34:e147. doi: 10.1093/nar/gkl896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M.S., Sørensen C.B., Bolund L., Jensen T.G. Mechanisms underlying targeted gene correction using chimeric RNA/DNA and single-stranded DNA oligonucleotides. J. Mol. Med. (Berl) 2002;80:770–781. doi: 10.1007/s00109-002-0393-8. [DOI] [PubMed] [Google Scholar]
- Andrieu-Soler C., Casas M., Faussat A.-M., Gandolphe C., Doat M., Tempé D., Giovannangeli C., Behar-Cohen F., Concordet J.-P. Stable transmission of targeted gene modification using single-stranded oligonucleotides with flanking LNAs. Nucleic Acids Res. 2005;33:3733–3742. doi: 10.1093/nar/gki686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A., Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Brachman E.E., Kmiec E.B. Targeted nucleotide repair of cyc1 mutations in Saccharomyces cerevisiae directed by modified single-stranded DNA oligonucleotides. Genetics. 2003;163:527–538. doi: 10.1093/genetics/163.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachman E.E., Kmiec E.B. Gene repair in mammalian cells is stimulated by the elongation of S phase and transient stalling of replication forks. DNA Repair (Amst) 2005;4:445–457. doi: 10.1016/j.dnarep.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Brough R., Frankum J.R., Costa-Cabral S., Lord C.J., Ashworth A. Searching for synthetic lethality in cancer. Curr. Opin. Genet. Dev. 2011;21:34–41. doi: 10.1016/j.gde.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Chen F., Pruett-Miller S.M., Huang Y., Gjoka M., Duda K., Taunton J., Collingwood T.N., Frodin M., Davis G.D. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino N., Court D.L. Enhanced levels of lambda Redβ-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D.L., Sawitzke J. a, Thomason L.C. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- Dekker M., Brouwers C., te Riele H. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res. 2003;31:e27. doi: 10.1093/nar/gng027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Capecchi M.R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H.M., Yu D., DiTizio T., Court D.L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler A., Wegmann S., Elie-Caille C., Bradshaw C.R., Maresca M., Seidel R., Habermann B., Muller D.J., Stewart A.F. Conformational adaptability of Redbeta during DNA annealing and implications for its structural relationship with Rad52. J. Mol. Biol. 2009;391:586–598. doi: 10.1016/j.jmb.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Ferrara L., Kmiec E.B. Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004;32:5239–5248. doi: 10.1093/nar/gkh822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. a, Barbas C.F. 2012 ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Parekh-Olmedo H., Drury M., Skogen M., Kmiec E.B. Reaction parameters of targeted gene repair in mammalian cells. Mol. Biotechnol. 2005;29:197–210. doi: 10.1385/MB:29:3:197. [DOI] [PubMed] [Google Scholar]
- Humbert O., Davis L., Maizels N. Targeted gene therapies: tools, applications, optimization. Crit. Rev. Biochem. Mol. Biol. 2012;47:264–281. doi: 10.3109/10409238.2012.658112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoucheva O., Alexeev V., Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8:391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- Igoucheva O., Alexeev V., Pryce M., Yoon K. Transcription affects formation and processing of intermediates in oligonucleotide-mediated gene alteration. Nucleic Acids Res. 2003;31:2659–2670. doi: 10.1093/nar/gkg360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoucheva O., Alexeev V., Yoon K. Oligonucleotide-directed mutagenesis and targeted gene correction: a mechanistic point of view. Curr. Mol. Med. 2004;4:445–463. doi: 10.2174/1566524043360465. [DOI] [PubMed] [Google Scholar]
- Itzhaki J.E., Gilbert C.S., Porter A.C. Construction by gene targeting in human cells of a “conditional” CDC2 mutant that rereplicates its DNA. Nat. Genet. 1997;15:258–265. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- Iyer L.M., Koonin E. V, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Karakousis G., Ye N., Li Z., Chiu S.K., Reddy G., Radding C.M. The beta protein of phage lambda binds preferentially to an intermediate in DNA renaturation. J. Mol. Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- Kenner O., Kneisel A., Klingler J., Bartelt B., Speit G., Vogel W., Kaufmann D. Targeted gene correction of hprt mutations by 45 base single-stranded oligonucleotides. Biochem. Biophys. Res. Commun. 2002;299:787–792. doi: 10.1016/s0006-291x(02)02749-3. [DOI] [PubMed] [Google Scholar]
- Khan I.F., Hirata R.K., Russell D.W. AAV-mediated gene targeting methods for human cells. Nat. Protoc. 2011;6:482–501. doi: 10.1038/nprot.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Karakousis G., Chiu S.K., Reddy G., Radding C.M. The beta protein of phage lambda promotes strand exchange. J. Mol. Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- Li X., Costantino N., Lu L., Liu D., Watt R.M., Cheah K.S.E., Court D.L., Huang J.-D. Identification of factors influencing strand bias in oligonucleotide-mediated recombination in Escherichia coli. Nucleic Acids Res. 2003;31:6674–6687. doi: 10.1093/nar/gkg844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 2009;8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Parekh-Olmedo H., Kmiec E.B. The development and regulation of gene repair. Nat. Rev. Genet. 2003;4:679–689. doi: 10.1038/nrg1156. [DOI] [PubMed] [Google Scholar]
- Liu B., Wu X., Liu B., Wang C., Liu Y., Zhou Q., Xu K. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim. Biophys. Acta. 2012;1822:1692–1704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Majumdar A., Muniandy P. a, Liu J., Liu J., Liu S., Cuenoud B., Seidman M.M. Targeted gene knock in and sequence modulation mediated by a psoralen-linked triplex-forming oligonucleotide. J. Biol. Chem. 2008;283:11244–11252. doi: 10.1074/jbc.M800607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano A., Mohri Z., Sperber G., Ogris M., Graham I., Dickson G., Owen J.S. Failure to generate atheroprotective apolipoprotein AI phenotypes using synthetic RNA/DNA oligonucleotides (chimeraplasts) J. Gene Med. 2003;5:795–802. doi: 10.1002/jgm.403. [DOI] [PubMed] [Google Scholar]
- McLachlan J., Fernandez S., Helleday T., Bryant H.E. Specific targeted gene repair using single-stranded DNA oligonucleotides at an endogenous locus in mammalian cells uses homologous recombination. DNA Repair (Amst) 2009;8:1424–1433. doi: 10.1016/j.dnarep.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Morozov V., Wawrousek E.F. Single-strand DNA-mediated targeted mutagenesis of genomic DNA in early mouse embryos is stimulated by Rad51/54 and by Ku70/86 inhibition. Gene Ther. 2008;15:468–472. doi: 10.1038/sj.gt.3303088. [DOI] [PubMed] [Google Scholar]
- Muyrers J.P., Zhang Y., Buchholz F., Stewart A.F. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- Nickerson H.D., Colledge W.H. A comparison of gene repair strategies in cell culture using a lacZ reporter system. Gene Ther. 2003;10:1584–1591. doi: 10.1038/sj.gt.3302049. [DOI] [PubMed] [Google Scholar]
- Olsen P.A., Randol M., Krauss S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005;12:546–551. doi: 10.1038/sj.gt.3302454. [DOI] [PubMed] [Google Scholar]
- Papaioannou I., Simons J.P., Owen J.S. Oligonucleotide-directed gene-editing technology: mechanisms and future prospects. Expert Opin. Biol. Ther. 2012;12:329–342. doi: 10.1517/14712598.2012.660522. [DOI] [PubMed] [Google Scholar]
- Pierce E.A., Liu Q., Igoucheva O., Omarrudin R., Ma H., Diamond S.L., Yoon K. Oligonucleotide-directed single-base DNA alterations in mouse embryonic stem cells. Gene Ther. 2003;10:24–33. doi: 10.1038/sj.gt.3301857. [DOI] [PubMed] [Google Scholar]
- Porter A.C., Itzhaki J.E. Gene targeting in human somatic cells. Complete inactivation of an interferon-inducible gene. Eur. J. Biochem. 1993;218:273–281. doi: 10.1111/j.1432-1033.1993.tb18375.x. [DOI] [PubMed] [Google Scholar]
- Radecke F., Peter I., Radecke S., Gellhaus K., Schwarz K., Cathomen T. Targeted chromosomal gene modification in human cells by single-stranded oligodeoxynucleotides in the presence of a DNA double-strand break. Mol. Ther. 2006;14:798–808. doi: 10.1016/j.ymthe.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Radecke S., Radecke F., Cathomen T., Schwarz K. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol. Ther. 2010;18:743–753. doi: 10.1038/mt.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelev I. V, Hübscher U. The 3′ 5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Ellis H.M., Waters L.S., Yu D., Lee E.C., Court D.L., Sharan S.K. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Taubes G. Gene therapy. The strange case of chimeraplasty. Science. 2002;298:2116–2120. doi: 10.1126/science.298.5601.2116. [DOI] [PubMed] [Google Scholar]
- Urnov F.D., Miller J.C., Lee Y.-L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Wu X.-S., Xin L., Yin W.-X., Shang X.-Y., Lu L., Watt R.M., Cheah K.S.E., Huang J.-D., Liu D.-P., Liang C.-C. Increased efficiency of oligonucleotide-mediated gene repair through slowing replication fork progression. Proc. Natl. Acad. Sci. USA. 2005;102:2508–2513. doi: 10.1073/pnas.0406991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez R.J., Porter A.C. Therapeutic gene targeting. Gene Ther. 1998;5:149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Muyrers J.P.P., Rientjes J., Stewart A.F. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol. Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


