Abstract
We provide the first demonstration that isopeptide ligation, a non-canonical activity of the enzyme sortase A, can be used to modify recombinant proteins. This reaction was used in vitro to conjugate small molecules to a peptide, an engineered targeting protein, and a full-length monoclonal antibody with an exquisite level of control over the site of conjugation. Attachment to the protein substrate occurred exclusively through isopeptide bonds at a lysine ε-amino group within a specific amino acid sequence. This reaction allows more than one molecule to be site-specifically conjugated to a protein at internal sites, thereby overcoming significant limitations of the canonical native peptide ligation reaction catalyzed by sortase A. Our method provides a unique chemical ligation procedure that is orthogonal to existing methods, supplying a new method to site-specifically modify lysine residues that will be a valuable addition to the protein conjugation toolbox.
Keywords: protein modification, site-specific bioconjugation, sortase, biotechnology
Protein conjugates are generally heterogeneous due to the presence of multiple reactive sites in the protein; this heterogeneity is undesirable, as conjugation at these sites can interfere with protein function and stability and also leads to regulatory complexity.[1,2] Improved conjugation methods that precisely specify the site of protein modification are therefore highly desirable and several have been developed that utilize unnatural amino acids,[3,4] enzymatic reactions,[5–7] cysteine engineering,[8] glycan remodeling,[9] and site-selective small molecule crosslinking agents.[10]
We demonstrate herein a new approach to site-specifically modify proteins by a ligation reaction catalyzed by sortase A (SrtA) from Staphylococcus aureus (S. aureus) in which the two moieties are covalently ligated by an isopeptide bond. This function of SrtA, termed isopeptide ligation, is distinct from its well known canonical function of ligating two moieties through a native peptide bond, as isopeptide ligation occurs at the ε-amino group of a lysine residue. Because the reactive site is positioned within a defined peptide sequence, this approach provides a new method for protein modification that allows precise control of the number of conjugated molecules as well their location. We characterized the site-specificity of this method and applied it to attach a payload of multiple small molecules to both a model targeting protein and a full-length monoclonal antibody. This isopeptide, sortase-catalyzed ligation site-specifically modifies lysine residues, thus making it a valuable addition to the protein conjugation toolbox that is orthogonal to the majority of available methods.
SrtA recognizes the primary sequence LPXTG (where X is any amino acid) in a protein and cleaves the peptide bond between threonine and glycine, forming a stable intermediate that joins the catalytic thiol in SrtA to the carboxyl group of threonine in a thioester bond. In its canonical function in S. aureus, this intermediate undergoes nucleophilic attack by the α-amino group of an oligoglycine branch in the peptidoglycan, generating a native peptide bond that anchors the substrate protein to the cell wall (Supp. Fig. 1).[11] This reaction has been used to attach a diverse array of compounds to proteins by linking them to synthetic oligoglycine or LPXTG peptides, making them suitable nucleophiles or SrtA substrates, respectively.[12–16] However, this ligation reaction has two major limitations: (1) only a protein’s termini can be modified, and (2) only one molecule can be attached to each polypeptide chain.
Certain Gram-positive bacteria utilize homologs of S. aureus SrtA to assemble pili—fibrous polymers of structural proteins—that extend from the bacterial surface and are implicated in adhesion and biofilm formation.[17] The initial step for pilin formation is the same as that used to anchor proteins to the cell wall. Extension of a pilin polymer proceeds when the thioester undergoes nucleophilic attack by the ε-amino group of a lysine residue in another sortase-linked pilin monomer (Supp. Fig. 2). In Corynebacterium diphtheriae and Actinomyces naeslundii, the nucleophilic lysine used in polymer extension is contained in a “pilin domain” with the sequence WX3VXVYPKH, where X is a non-conserved residue.[17,18]
Isopeptide ligation catalyzed by the S. aureus SrtA has been demonstrated using a model peptide, but it has never been applied to modify proteins, nor has its site-specificity been investigated.[19] We first confirmed the ability of SrtA to carry out isopeptide ligation by reacting a LPETGRAGG peptide containing an amino-terminal biotin with a pilin domain peptide (VGGSWLQDVHVYPKHGGSGR). SrtA was produced as a fusion protein with an elastin-like polypeptide (ELP)—ELPs are peptide polymers composed of repeats of the pentapeptide VPGXG where X is any amino acid except proline. ELPs and their fusions phase separate in aqueous solution when heated above a characteristic transition temperature (Supp. Fig. 3) to form micron-size aggregates that can be isolated from host cell proteins by centrifugation.[20,21] ELPs are inert—they impart no new bioactivity beyond phase transition behavior to their fused peptide or protein partner—and provide efficient purification tags that allow easy recovery of ELP fusions without column chromatography by exploiting their phase transition behavior. After incubating these two peptides with the SrtA-ELP overnight, matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry indicated the formation of a species that corresponded to the isopeptide-linked biotin-LPET and the pilin domain peptide (Supp. Fig. 4).
Recombinant protein modification is a logical application of this method as it provides a stringent test of the site-specificity of the reaction due to the presence of multiple, potentially cross-reactive lysine residues in most proteins of interest. We hypothesized that a small molecule bearing a sortase LPXTG recognition peptide could be conjugated to a recombinant protein engineered to contain the pilin domain, and that these two sequence elements would together provide a means of site-specific attachment. Additionally, by incorporating multiple pilin domains into the protein, we hypothesized that a stoichiometry of more than one small molecule per protein could be achieved, which would represent a significant advantage over the canonical, native peptide ligation reaction catalyzed by this enzyme.
We initially used the fibronectin type III (Fn3) domain as a model substrate. Fn3 domains are of interest as an alternative to antibodies because they can be affinity matured against a target but do not contain the complex quaternary structure, disulfide bonds, or glycosylation found in antibodies.[22,23] We generated a Fn3 domain fused to an ELP with 3 intervening copies of the pilin domain (Fn3-PLN3-ELP), and a Fn3-ELP fusion lacking any pilin domains as a control. We incorporated multiple copies of the pilin domain into the Fn3 fusion protein between the Fn3 domain and the ELP for two reasons: (1) to confirm that the isopeptide ligation can be carried out at an internal site, and (2) because a payload of several small molecules per protein is typically necessary to attain a therapeutically relevant dose in protein-drug conjugates.
The Fn3-ELP and Fn3-PLN3-ELP fusions were incubated with SrtA-ELP and biotin-LPETGRAGG peptide overnight (Figure 1a) and the product was analyzed by Western blot using a streptavidin-Cy5 conjugate to detect biotinylated protein. SDS-PAGE and the corresponding Western blot are shown in Figure 1b. Only the reaction of biotin-LPETGRAGG peptide with Fn3-PLN3-ELP resulted in biotinylated target protein, which suggested that biotinylation was specific for the pilin domain. No biotinylation of the Fn3-ELP without pilin domains was observed—no band was observed in lane 1 of the Western blot at the expected molecular weight of 36 kDa—despite the fact that this protein contains 3 lysine residues and a terminal primary amine that offer sites for off-target conjugation. Only SrtA-ELP was observed in this reaction (at approximately 120 kDa) because the thioester-linked SrtA-biotin intermediate is stable in SDS-PAGE. In contrast, biotinylated Fn3-PLN3-ELP appeared as a 43 kDa band in lane 2 of the Western blot, along with the thioester-linked SrtA-ELP intermediate at 120 kDa. Lanes 3 and 4 show control reactions containing Fn3-ELP and Fn3-PLN3-ELP, respectively, where the biotin-LPETGRAGG peptide was excluded. As expected, no bands were observed in the Western blot for the reactions in lanes 3 and 4.
Figure 1.
Sortase A catalyzes protein-small molecule isopeptide ligation. (a) Overview of the reaction used to conjugate biotin to the pilin domain in the Fn3-PLN3-ELP fusion protein, along with the expected tryptic peptides of the product (boxed). SrtA-ELP and biotin-LPETGRAGG were used at 2- and 10-fold molar excess to Fn3-PLN3-ELP, respectively. (b) SDS-PAGE of biotinylation reactions with Fn3-ELP (36 kDa) and Fn3-PLN3-ELP (43 kDa) and Western blot with streptavidin-Cy5 indicate that only the reaction containing both the pilin domain and biotin-modified LPETG peptide yields biotinylated target protein. (c) MALDI-TOF spectrum of the unpurified Fn3-PLN3-ELP biotinylation reaction product, corresponding to lane 2 of the Western blot in panel (b), after trypsinization. Biotin is installed specifically at the pilin domain lysine (ions 1 and 2), with all other ions corresponding to unmodified peptides from the Fn3-PLN3-ELP and SrtA-ELP fusion proteins. (d) MALDI-TOF spectrum of the tryptic digest of SrtA-ELP reacted with Fn3-PLN3-ELP without biotin-LPETGRAGG. All peaks corresponding to panel (c) are present except those corresponding to biotinylated ions 1 and 2.
In order to determine the biotinylation site and the selectivity of the reaction for the pilin domain, the unpurified product was digested with trypsin and analyzed by MALDI-TOF mass spectrometry. The spectrum indicated the presence of peptides with a missed digestion at the pilin domain lysine and an additional mass of 666.8 Da (matching the mass of biotin-LPET) in the reaction of Fn3-PLN3-ELP with biotin-LPETGRAGG (ions 1 and 2 in Figure 1c). These ions corresponded to m/z values of both 2763.5 and 3871.2 because the tryptic peptide of the amino-terminal copy of the pilin domain contained some residues from the Fn3 sequence. These ions were not observed when biotin-LPETGRAGG peptide was not included in the reaction (Figure 1d). All other ions in the spectra mapped to expected, unmodified segments of the Fn3-PLN3-ELP or SrtA-ELP fusion proteins (Supp. Fig. 5). Importantly, no ions corresponding to biotinylation at any of the 3 lysines in the Fn3 domain or at the Fn3 amino-terminus were observed in the MALDI-TOF spectra (Supp. Fig. 5). Moreover, no biotinylated peptides were observed for reactions with Fn3-ELP lacking the pilin domain (Supp. Fig. 6).
The same SrtA linked to a hexahistidine tag—rather than an ELP—and purified by immobilized metal affinity chromatography also biotinylated the Fn3-PLN3-ELP through isopeptide bonds at pilin domain lysines, as assessed by Western blotting and MALDI-TOF (Supp. Fig. 7). This rules out the potential influence of the ELP tag on the enzyme’s activity and suggests that protein modification by isopeptide ligation is a general in vitro function of SrtA. We also attached fluorescein isothiocyanate (FITC) coupled to an LPETGRAGG peptide to the Fn3-PLN3-ELP by isopeptide ligation (Supp. Fig. 8), and confirmed conjugation to the pilin domain lysine by in-gel fluorescence detection and MALDI-TOF mass spectrometry. This demonstrates that the reaction is not restricted to biotin and can be used with bulkier small molecule substrates.
We purified the biotinylated Fn3-PLN3-ELP from the reaction mixture using cation exchange chromatography (Supp. Fig. 9), then trypsinized the protein and analyzed the resulting peptides by liquid chromatography-electrospray ionization tandem mass spectrometry (LC-MS/MS). This achieved two aims: (1) conclusive identification of the isopeptide bond in the linker between biotin and the pilin domain and (2) quantitation of the average number of biotin molecules per protein by comparing the relative amounts of peptides with modified and unmodified pilin domain lysines.
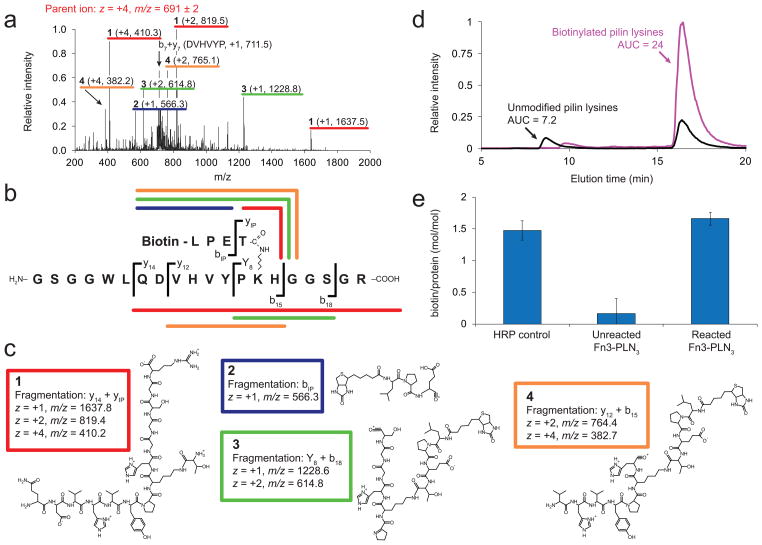
The MS1 spectrum (Supp. Fig. 10) showed the presence of biotinylated pilin domain peptides and other, unmodified peptides from the Fn3-PLN3-ELP sequence, with biotinylation only observed for peptides containing the pilin domain lysine, in agreement with the results obtained using MALDI-TOF mass spectrometry. To provide direct evidence for the isopeptide conjugation of biotin to the pilin domain lysine, we fragmented the biotinylated pilin domain peptide ion by low-energy collisions with helium. The MS2 spectrum for the parent ion with z = +4 (Figure 2a) contains daughter ions formed by breaking peptide bonds in the protein backbone (the fragmentation pattern is outlined in Figure 2b). Several of these daughter ions correspond to peptides with an isopeptide junction between biotin-LPET and the pilin domain lysine (ions 1-4, with the chemical structures shown in Figure 2c). A similar fragmentation pattern was observed for the parent ion with z = +3 (Supp. Fig. 11). The presence of ions 1-4 in the MS2 spectra for multiple charge states conclusively shows that the sortase-mediated reaction installs biotin-LPET at the pilin domain lysine through an isopeptide bond, as the daughter ions produced by tandem-MS provide an unambiguous signature that specifically confirms the identity of the biotinylated parent ion.
Figure 2.
Tandem mass spectrometry (MS) of tryptic peptides of biotinylated Fn3-PLN3-ELP confirms biotinylation at the pilin domain lysine through an isopeptide bond. (a) MS2 spectrum of daughter ions produced by isolating the +4 charge state (m/z = 691) of a biotinylated pilin domain peptide. The spectrum shows four ions with multiple charge states whose masses confirm the expected chemical structures for biotin-LPET linked to the pilin domain lysine through an isopeptide bond (ions 1-4). These fragments correspond to y- and b-type ions produced by breaking peptide bonds along the pilin domain backbone (ions 1, 3, and 4) as well as within the LPET linker region (ions 1 and 2). (b) Outline of the observed fragmentation pattern and the nomenclature used to classify daughter ions, with colors corresponding to the daughter ion chemical structures in panel (c). Peptide bond cleavage results in a charged amino-terminal fragment (b ions) or a charged carboxy-terminal fragment (y ions). bIP and yIP correspond to daughter ions from fragmentation events within the isopeptide-linked LPET moiety. (d) Extracted ion chromatograms (EICs) for individual ions were summed for peptides containing biotinylated (
 ) or non-biotinylated (⚊) pilin domain lysines. (e) Quantitation of the biotin:protein molar ratio using a colorimetric assay measuring HABA displacement from avidin. A chemically-crosslinked biotin-horseradish peroxidase (HRP) conjugate was analyzed as a positive control.
) or non-biotinylated (⚊) pilin domain lysines. (e) Quantitation of the biotin:protein molar ratio using a colorimetric assay measuring HABA displacement from avidin. A chemically-crosslinked biotin-horseradish peroxidase (HRP) conjugate was analyzed as a positive control.
Extracted ion chromatograms (EICs) from MS1 for peptides with biotinylated or unmodified pilin domain lysines track the intensity of these ions over time as they elute from the chromatography column. This allows the relative abundance of biotinylated versus unmodified lysines to be determined by comparing the integrated areas of their corresponding EICs. By summing the EIC intensities for all identified ions containing either biotinylated or unmodified pilin domain lysine residues, we determined that 75% of the pilin domain lysines were biotinylated, which corresponds to an average of 2.2 biotinylated pilin domains per Fn3-PLN3-ELP protein (Figure 2d). To verify this result, we utilized a colorimetric 4′-hydroxyazobenzene-2-carboxylic acid (HABA) biotin quantitation assay to determine the molar ratio of biotin to protein, which yielded a molar ratio of biotin to protein of 1.7±0.2, whereas unreacted Fn3-PLN3-ELP gave no signal above background (Figure 2e).
Having characterized the isopeptide ligation reaction with the Fn3 domain, we next sought to demonstrate the generality of this reaction by modifying a more complex protein. To examine the feasibility of isopeptide ligation to generate antibody-drug conjugates, we cloned the 4D5 monoclonal antibody against the human epidermal growth factor receptor 2 (Her2) and genetically modified it to contain a pilin domain at the carboxy-termini of its heavy chains. The recombinant antibody was incubated with SrtA-ELP and biotin-LPETGRAGG peptide overnight (Figure 3a), and the product was compared to several control reactions run in parallel to assess the specificity of biotinylation of the pilin domain. Based on an anti-biotin Western blot (Figure 3b), SrtA-ELP did not biotinylate any sites on the anti-Her2 antibody when it did not contain the pilin domain. Additionally, no biotinylation of any of 4 murine IgG isotype controls (IgG1, IgG2a, IgG2b, or IgG3), or either class of murine light chain (kappa or lambda) was detected. Only the anti-Her2 heavy chain modified to contain the pilin domain was biotinylated by SrtA-ELP, which—consistent with our data for the Fn3 domain—strongly suggests that isopeptide ligation is specific for this amino acid sequence. The biotinylated antibody also retained antigen targeting, as it showed efficient labeling of Her2 on SK-OV-3 human ovarian adenocarcinoma cells (Figure 3c) compared to a biotinylated isotype control antibody (Figure 3d) in immunofluorescence microscopy. Additional images from both groups are available in Supp. Fig. 12. Using a HABA displacement assay, we determined that there were 1.8 ± 0.1 biotins conjugated per antibody out of 2 possible pilin domain attachment sites (Supp. Fig. 13). This equates to a conversion of 90%, which is consistent with reported yields for other enzymatic reactions at the carboxy-termini of antibody heavy chains.[24]
Figure 3.
Sortase-mediated isopeptide ligation can be used to modify monoclonal antibodies (mAbs). (a) An anti-Her2 mAb, genetically modified to contain the pilin domain at the heavy chain carboxy-terminus, was reacted with biotin-LPETGRAGG peptide and SrtA-ELP overnight. SrtA-ELP and biotin-LPETGRAGG were used at 2- and 100-fold molar excess to the mAb, respectively. (b) SDS-PAGE and an anti-biotin Western blot indicate that the pilin domain is required for biotinylation of antibodies. Non-reduced anti-Her2 containing the pilin domain on its heavy chain is biotinylated in the reaction and this modification maps exclusively to the heavy chain when the antibody’s interchain disulfide bonds are reduced prior to electrophoresis. No biotinylation is observed for reactions on a panel of antibodies including anti-Her2 without the pilin domain modification, as well as all murine IgG isotypes and both kappa and lambda murine light chain variants. (c) Immunofluorescence of the Her2-overexpressing cell line SK-OV-3 using biotinylated anti-Her2 containing a pilin domain on its heavy chain followed by secondary staining with streptavidin-FITC conjugate. (d) Immunofluorescence of SK-OV-3 using a biotinylated isotype control antibody in the primary staining step. Blue = nuclear stain, green = FITC, scale bars are 15 μm.
There are several factors that will likely control the reaction yield and kinetics. First, the location of the pilin domain within a protein will impact its solvent accessibility and may hence affect both kinetics and yield, as this is known to have a major effect on the reactivity of thiol groups in antibodies.[1,25] Second, the precise sequence of the pilin domain sequence is also likely to impact both parameters; we believe that optimization of this sequence through a library screening approach may well yield improvements in both kinetics and reaction yield. Finally, we expect that the pilin domain may tolerate truncations that will make it easier to engineer this sequence into a variety of solvent-accessible loops within proteins.[24] Each of these issues is currently under investigation.
In conclusion, we have demonstrated that the heretofore unexplored isopeptide ligation function of S. aureus SrtA can conjugate multiple small molecules to a protein at internal and terminal sites when their sequences are modified to contain one or more copies of the pilin domain. This method overcomes two major limitations of the canonical native peptide ligation reaction catalyzed by SrtA, which can only conjugate a single moiety to one terminus of a polypeptide chain, The specificity of this reaction is remarkable, as the proteins tested herein contain many potential off-target sites for conjugation (86 lysines in the case of the mAb). The precise control over the attachment site and stoichiometry, as well as the fact that this site-specific ligation reaction targets lysine residues, provides a new bioconjugation technique that is orthogonal to available strategies and will have a variety of research and therapeutic applications.
Supplementary Material
Footnotes
The authors acknowledge the contributions of Joel Bozekowski, Abigail Heiller, and Mitchell Zhang. This work was supported by the National Institutes of Health through grant R01 GM061232 to A.C. and by a University Scholars Fellowship awarded by the Duke University Graduate School and a pre-doctoral fellowship from NIH 5T32 GM008487 to J.J.B. We thank Dr. Dewey McCafferty (Department of Chemistry, Duke University) for the sortase A gene, and Dr. Rihe Liu (Eshelman School of Pharmacy, University of North Carolina at Chapel Hill) for the gift of the Fn3 gene.
Homepage: www.chilkotilab.pratt.duke.edu
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx.
For detailed experimental methods, please see the Supporting Information section.
References
- 1.Shen BQ, et al. Nat Biotechnol. 2012;30:184–189. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- 2.Junutula JR, et al. Nat Biotechnol. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 3.Lang K, Chin JW. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 4.Tian F, et al. Proc Natl Acad Sci USA. 2014;111:1766–1771. doi: 10.1073/pnas.1321237111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen I, Howarth M, Lin W, Ting AY. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 6.Jeger S, et al. Angew Chemie Int Ed. 2010;49:9995–9997. doi: 10.1002/anie.201004243. [DOI] [PubMed] [Google Scholar]
- 7.Hudak JE, et al. Angew Chemie Int Ed. 2012;51:4161–4165. doi: 10.1002/anie.201108130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cal PM, Bernardes GJ, Gois PM. Angew Chemie Int Ed. 2014 doi: 10.1002/anie.201405702. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Fang T, Boons GJ. Angew Chemie Int Ed. 2014;53:7179–7182. doi: 10.1002/anie.201402606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asano S, Patterson JT, Gaj T, Barbas CF. Angew Chemie Int Ed. 2014 doi: 10.1002/anie.201405924. [DOI] [PubMed] [Google Scholar]
- 11.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao H, Hart SA, Schink A, Pollok BA. J Am Chem Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 13.Ling JJ, Policarpo RL, Rabideau AR, Liao X, Pentelute BL. J Am Chem Soc. 2012;134:10749–10752. doi: 10.1021/ja302354v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willamson DJ, Fascione MA, Webb ME, Turnbull WB. Angew Chem Int Ed. 2012;51:9377–9380. doi: 10.1002/anie.201204538. [DOI] [PubMed] [Google Scholar]
- 15.Swee LK, et al. Proc Natl Acad Sci USA. 2013;110:1428–1433. doi: 10.1073/pnas.1214994110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antos JM, et al. J Am Chem Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ton-That H, Schneewind O. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 18.Maraffini LA, Dedent AC, Schneewind O. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dasgupta S, Samantaray S, Sahal D, Roy RP. J Biol Chem. 2011;286:23996–24006. doi: 10.1074/jbc.M111.247650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer DR, Chilkoti A. Nat Biotechnol. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 21.Bellucci JJ, Amiram M, Bhattacharyya J, McCafferty D, Chikoti A. Angew Chem Int Ed. 2013;52:3703–3708. doi: 10.1002/anie.201208292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards J, et al. J Mol Biol. 2003;326:1475–1488. doi: 10.1016/s0022-2836(03)00082-2. [DOI] [PubMed] [Google Scholar]
- 23.Duan J, Wi J, Valencia CA, Liu R. Biochemistry. 2007;46:12656–12664. doi: 10.1021/bi701215e. [DOI] [PubMed] [Google Scholar]
- 24.Drake PM, et al. Bioconjug Chem. 2014;25:1331–1341. doi: 10.1021/bc500189z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu P, et al. Proc Natl Acad Sci USA. 2009;106:3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.