Abstract
Industrial biotechnology and microbial metabolic engineering are poised to help meet the growing demand for sustainable, low-cost commodity chemicals and natural products, yet the fraction of biochemicals amenable to commercial production remains limited. Common problems afflicting the current state-of-the-art include low volumetric productivities, build-up of toxic intermediates or products, and byproduct losses via competing pathways. To overcome these limitations, cell-free metabolic engineering (CFME) is expanding the scope of the traditional bioengineering model by using in vitro ensembles of catalytic proteins prepared from purified enzymes or crude lysates of cells for the production of target products. In recent years, the unprecedented level of control and freedom of design, relative to in vivo systems, has inspired the development of engineering foundations for cell-free systems. These efforts have led to activation of long enzymatic pathways (>8 enzymes), near theoretical conversion yields, productivities greater than 100 mg L−1 hr−1, reaction scales of >100L, and new directions in protein purification, spatial organization and enzyme stability. In the coming years, CFME will offer exciting opportunities to (i) debug and optimize biosynthetic pathways, (ii) carry out design-build-test iterations without re-engineering organisms, and (iii) perform molecular transformations when bioconversion yields, productivities, or cellular toxicity limit commercial feasibility.
Keywords: cell-free metabolic engineering, biocatalysis, metabolic pathway debugging, synthetic biology, in vitro, enzyme cascade, reaction cascade, biotransformation, one-pot
1. Introduction
Today’s chemicals and materials are almost entirely derived from fossil fuels and their products [1]. For example, the petrochemical industry has historically focused on seven low-cost, high-volume commodity chemicals (methanol, ethylene, propene, butadiene, benzene, toluene, xylene), from which nearly all other materials are synthesized. The consequences of the petrochemical/materials paradigm and the resulting world dependence on fossil fuels include: (i) supply limitations due to increasing and fluctuating prices and finite availability [2], (ii) undesirable climate effects [3], and (iii) constraints on materials innovation stemming from a limited set of petrochemical building blocks [4]. While the petrochemical/materials paradigm will not be quickly replaced (if at all), advances in our foundations for engineering biology are poised to open access to novel chemicals and materials based on a potentially richer palette of starting points (i.e., more starting building blocks than the seven typically used today) [4]. This could lead to the production of chemicals and materials with new attractive properties and functions. In turn, novel, sustainable, and cost-effective technologies based on biological synthesis could be realized that help meet the world’s increasing need for fuels, pharmaceuticals, and commodity chemicals.
In recent decades, the growth of industrial biotechnology and metabolic engineering has shown a tremendous potential to help meet the need for new chemicals and materials [4]. Harnessing enzymes as robust, specific, and efficient catalysts of chemical reactions [5, 6], cellular factories are being constructed to make complex reaction cascades within living organisms to produce a variety of useful small molecules, peptides, and polymers [7–10]. In addition to the achievements of first-generation bioethanol [11], successful commercial bioprocesses have been recently developed for the production of 1,3 propanediol (Dupont Tate & Lyle) [12], polylactic acid (Cargill), and isoprenoids (Amyris) [13], among others. While the number of microbial metabolic engineering success stories is rapidly growing [14], the fraction of biochemicals amenable to economical production is still limited, requiring hundreds of person-years of work to bring a single pathway to market [15–17].
A major challenge to current microbial biomanufacturing practice lies with the inherent limitations imposed by cells. It is virtually impossible to balance intracellular fluxes to optimally satisfy a very active synthetic pathway while maintaining the growth and maintenance needs to the host. This leads to a variety of challenges afflicting the current state-of-the-art. Yields of target molecules are limited to non-toxic levels (e.g., ~2.5% v/v butanol) [18] and unwanted byproducts are common. Product excretion and/or separation are constrained by intracellular transport barriers (i.e., membranes). Scaling of lab-scale cultures to the industrial level is hampered by heterogeneous fermentation conditions [19]. The inability to control and direct resource allocation (e.g., electron flux or ATP) towards the exclusive synthesis of the target product can result in poor volumetric productivities that limit economic process viability. Additionally, the unwieldy complexity of cells makes rational design unpredictable and difficult to engineer [15, 20]. Put another way, current biomanufacturing efforts are limited by the need to balance the tug-of-war that exists between the cell’s physiological and evolutionary objectives and the engineer’s process objectives.
Many efforts are underway to address the aforementioned challenges. In one example, the growing field of synthetic biology offers new advanced tools and generalized capabilities to modify living organisms for process engineering objectives. For example, our newfound ability to rapidly read, write, and edit DNA is accelerating the pace of design-build-test loops for product development [21].
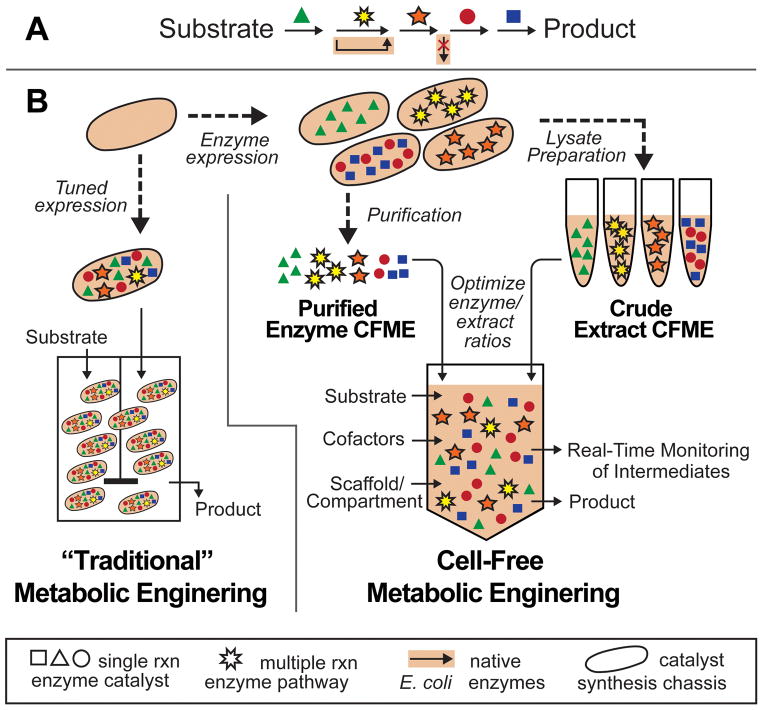
Cell-free metabolic engineering (CFME) offers an alternative, yet underutilized, approach. The foundational principle of CFME is that precise complex biomolecular synthesis can be conducted without using intact cells (Figure 1) [22, 23]. Instead, purified enzyme systems or crude cell lysates are used, which can be accurately monitored and modeled.
Figure 1.
Paradigms for metabolic engineering (A) sample pathway (B) traditional and cell-free approaches.
In this review, we focus on the emergence and growth of CFME. Many overlapping terms have been discussed to describe one-pot, cell-free, enzymatic pathways including “synthetic pathway biotransformations (SyPaB) [24],” “cell-free biosystems for biomanufacturing (CFB2) [25],” “multi-enzyme processes [26],” “synthetic metabolic engineering [27],” and “cell-free pathways/minimized reaction cascades [28]”. CFME encompasses many of these concepts while emphasizing long pathways (>4 enzymes), economical substrates, potential for industrial-scale production, and rapid pathway prototyping and debugging [23]. Other perspectives on tools for engineering complexity in cell-free systems [29], as well as a broader discussion of applications for cell-free biology [17, 23] are discussed elsewhere. We begin with a brief introduction of the technological capabilities of the field and its potential advantages for pathway debugging and biomanufacturing. In the next section, we discuss state-of-the-art cell-free systems for small molecule metabolite production and pathway optimization. Finally, we examine frontier applications, as well as associated challenges and opportunities.
2. Background
Activation of cell-free enzyme pathways has been possible for over a century. Indeed, seminal biochemical discoveries of glycolysis (by Eduard Buchner) [30] and the genetic code (by Nirenberg and Matthaei) [31] used cell extracts, or lysates, from yeast and Escherichia coli, respectively. In other examples, the catalytic proteins of metabolic pathways have been purified from living cells to elucidate and understand their biological activity, as well as to harness them for industrial purposes. To date, commercial usage of enzymes in vitro is primarily for single reactions (or very short pathways). Usage of these enzymes is often in a specialized context where enzymes have distinct advantages over chemical methods, including reduced reaction time, increased product yield, increased product specificity, reduced cost, and reduced environmental impact. Examples include isomerases in starch processing, proteases/lipases in clothing detergents, cellulases/amylases for bioethanol pretreatment, and many others [32].
Only within the last ~2 decades have long enzymatic pathways been activated in vitro with the specific intent to both maximize product yield from low-cost substrates as well as characterize the reaction elements. In this “engineering” context, two approaches to CFME have emerged: purified enzymes and crude extracts.
Purified systems use enzymes that have been overexpressed and purified individually, then recombined to assemble a pathway of interest (Figure 1). These systems allow exquisite control of reaction conditions and pathway fluxes since the concentration and activity of every component is known. However, cofactor cost and regeneration is a challenge. The majority of CFME research to date has utilized purified systems.
Crude extracts, on the other hand, are prepared by simply lysing the cell, using centrifugation to remove cellular debris, and collecting the supernatant as “lysate” or “extract” (Figure 1). Lysates are cheap to generate and contain the thousands of catalytic proteins naturally present in cell metabolism. The activation of these native pathways in the extract can regenerate cofactors or energy to provide the support system necessary to fuel highly active metabolic conversions. For example, Jewett et al. demonstrated that formation of inverted lipid vesicles during extract preparation facilitates oxidative phosphorylation which converts reducing equivalents from central metabolism into ATP [33]. While this native metabolism can be looked upon as an advantage, this “background” metabolism, if unwanted, can be difficult to characterize, eliminate, or accurately model. The most prominent examples of crude extract systems involve cell-free protein synthesis (CFPS). Yields of lysate-based systems (now >1 g L−1 for bacterial systems) [33–38] have exceeded purified ones (e.g. the PURE system) [39] and commercial applications have been established [40, 41]. CFPS will not be discussed further in this paper, but several other outstanding reviews are available [42–44].
In comparing emerging biotechnology platforms to established ones, purified and crude extract-based CFME systems have individual merits, however trade-offs between yield, cost, and other requirements must be carefully considered. That said, CFME systems provide a complement to in vivo technologies; yet offer several distinct advantages for the design, modification, and control of biological systems (Table 1). From a biomanufacturing perspective, cell-free systems separate catalyst synthesis (cell growth) from catalyst utilization (metabolite production). This contrasts the prevailing paradigm of enclosed, cell-based, microbial “reactors”. By eliminating the requirement of cell growth and diverting all carbon flux to product, CFME allows for higher theoretical yields and productivities, a wider range of products, and easier manipulation of reaction conditions. For example, cell-free production of 1,3-propanediol from glycerol (0.95 mol/mol) highlights CFME’s ability to avoid byproduct losses associated with traditional fermentation (0.6 mol/mol) [45]. Further, cell-free systems avoid many toxicity constraints arising from substrates, pathway intermediates, or products. It has been noted that biological processes (unlike most chemical reactions) experience heterogeneous (and often deleterious) conditions at larger scales [19]; cell-free systems may experience more chemistry-like scale-up. In one example, cell-free protein synthesis has achieved a yield and rate expansion factor of 106 with nearly identical performance fidelity at 100L [40]. Finally, CFME systems may enable a new opportunities in logistics and on-demand, point-of-use manufacturing.
Table 1.
Advantages (+) and disadvantages (−) of cell-free systems
| Metric | Living cells | Cell-free systems |
|---|---|---|
| Pathway Engineering | − Engineer’s goal (overproduction) is opposed to microbe’s goal (growth) − Endogenous regulation is difficult to predict and modify + ability to use directed evolution |
+ easy to use chimeric enzyme pathways sourced from multiple organisms + allow mixing with chemical catalysts/hybrid solvents that would be cytotoxic |
| Yield | − Carbon flux diverted to cell maintenance and byproducts | + All carbon/energy can be directed to product |
| Cell Wall | − Selective barrier; intercellular characterization and product excretion can be challenging + membrane proteins can be used |
+ Direct substrate addition and product removal; easy sampling |
| Effect of yield toxicity | − Viability constraints (e.g.,1–2% (v/v) isobutanol [124]) | + Fewer viability constraints (e.g. 12% (v/v) isobutanol for some pathway enzymes [49]) |
| Stability | − Fermentation conditions affect intercellular environment | + Reaction conditions controlled by engineer |
| Cost | + Low; pending high yields and low product separation cost. | − Must incorporate cost of “catalyst” synthesis (e.g. cell growth and/or enzyme purification) − Enzyme and cofactor costs dominate |
| Scale-up | − Fermentation conditions are heterogeneous at industrial scale − Contamination can be catastrophic |
+ More linear scale-up? |
| Maturity | + years of practical experience; well established methods | − Recently established |
From a prototyping perspective, cell-free systems are well suited to rapid design-build-test cycles because they do not require the re-engineering of the entire organism with each design; merely the exogenous addition of the desired protein, cofactor, or metabolite. Consequently, there is a high degree of flexibility to model the kinetics of individual enzymes, measure metabolite fluxes in multistep pathways, determine stability of catalysts, study the effects of redox potential on pathway performance, and experimentally isolate many other process properties that are confounded in living organisms [17]. One of the largest application of cell-free biology to date has been in the context of pathway debugging [46].
Although cell-free technology offers many exciting advantages, challenges remain that provide opportunity for improvement. For example, most cell-free systems are not yet commercially available as production platforms. In addition, costs of in vitro systems currently exceed those of in vivo approaches, which limit scale-up. Despite these challenges and others (more thoroughly described in Section 4 below), the benefits of CFME are inspiring new applications. In the next section, we examine achievements in the capacity to synthesize metabolites using CFME.
3. Achievements in Cell-Free Metabolic Engineering
One of the first examples of purified enzyme pathway reconstruction was by Welch and Scopes in 1985 [47]. Yeast glycolytic enzymes were isolated and ethanol produced at rate of 0.042 mmol L−1 hr−1; it was also shown that overloading of substrate (glucose) sequestered inorganic phosphate in the early reaction steps preventing subsequent ATP generation and effectively stopping ethanol synthesis. These seminal results led others to extrapolate that cell-free ethanol production is theoretically capable of higher productivities and yields than living cells [48]. While this has yet to be realized industrially, a range of other CFME pathways have since been explored. These efforts have illuminated general rules for achieving high metabolic yields and productivities in vitro. To briefly summarize, requirements for optimal metabolic conversion include: robust and stable enzymes, appropriate enzyme ratios and substrate/cofactor/buffer concentrations, and a means for providing a favorable thermodynamic push towards a product (i.e., cofactor regeneration, product removal, repeated substrate feeding). These requirements are characteristic properties of the in vivo state of a rapidly growing cell. Thus, a guiding principle that has emerged in the development of CFME systems is that cytoplasmic mimicry enables highly productive systems [17, 23, 33, 35, 42].
3.1 Purified Enzyme Systems
3.1.1 Ethanol and Isobutanol
A landmark paper for purified CFME describes the conversion of glucose to ethanol (and isobutanol) using 6 (or 9) purified enzymes in a “minimized reaction cascade” [49] (Figure 2A). Novel use of promiscuous enzymes from hyperthermophilic archaea shortened the ubiquitous 10-enzyme Embden–Meyerhof–Parnas (EMP) pathway to four enzymes using a modified non-phosphorylative Entner-Doudoroff pathway. Glucose was converted to pyruvate while generating two NADH; ATP and additional reducing equivalents generated by the EMP pathway are not needed in this specific pathway. The catalytic activity and tolerance to product concentration of each purified enzyme was assayed, a task not easily accomplished in vivo. The 20 mL complete reaction contained all 6 (or 9) enzymes normalized to an equivalent activity (10 μmol min−1); if all enzymes were active at this rate, the reaction would be theoretically capable of producing ~30 mmol L−1 hr−1 ethanol. The reaction produced an impressive 57% molar yield of ethanol from glucose at 1.51 mmol L−1 hr−1 productivity. This is one of the best-reported examples to date of using CFME as a stand-alone bioproduction strategy. Higher enzyme concentrations and pathways with multiple cofactors (beyond NAD+ alone) are future directions, but this study demonstrates the ease of characterization and the “plug-and-play” nature of CFME optimization.
Figure 2.
Selected Metabolic Pathways (A) glucose to ethanol/isobutanol [49] (B) sugar to hydrogen [61] (C) glucose to DHAP or TDHP [71].
3.1.2 Pathway Prototyping and Non-Oxidative Glycolysis
Despite significant advances in traditional metabolic engineering, most successes to date have been through combinatorial screening of numerous mutants as opposed to rational design. Indeed, the semi-synthetic production of the anti-malaria drug artemisinin has been one of the great success stories of the metabolic engineering and synthetic biology communities, yet the high cost and substantial work (>150 person years) was mostly needed for “pathway balancing” [16]. Cell-free platforms offer a “breadboarding” capability that can be easily monitor and control complex pathways; this can augment the lack of information regarding in vivo steady-state kinetics and metabolic flux imbalances. For instance, mRNA-enzyme fusions bound to DNA were used to explore enzyme ratios in the glucose to trehalose pathway [50]. This concept is extremely valuable for rational metabolic engineering, but is also of useful in building engineerable genetic circuits for other synthetic biology applications [46, 51].
Cell-free methods were also recently used in development of a novel non-oxidative glycolysis pathway [52]. The goal of this effort was to develop an alternative to the classic EMP pathway and produce a maximal carbon yield from glucose (i.e., three acetyl-CoA instead of two). This higher carbon efficiency comes at the expense of reducing equivalents (i.e., no NADH is produced) and would be extremely useful in applications that have an external electron source or no need for reducing power. The Liao group was able to achieve ~100% in vitro conversion of glucose to acetyl phosphate at relatively high titer (30mM) using seven His-tag purified enzymes plus seven purchased enzymes. This result allowed them to validate their proposed pathway and the efficacy a heterologous phosphoketolase enzyme prior to subsequent expression in vivo. The elimination of CO2 as a byproduct of central metabolism is a potentially game-changing result for the metabolic engineering community at large; both inside and outside the cell.
3.1.3 Isoprenoids
In light of intrinsic commercial value and the in vivo efforts regarding artemisinin highlighted above, several groups have reconstituted isoprenoid pathways in vitro to inform further engineering. Farnesene was produced from acetyl-CoA using nine purified enzymes plus two NADPH reducing equivalents and three ATP [53]. Optimization of in vitro enzyme concentrations informed in vivo addition of second copy of the idi gene that encodes isopentenyl diphosphate isomerase; this resulted in a doubling of in vivo titers. A similar experimental design had been used earlier to improve production of fatty acid-derived compounds [54]. Chen et al. exploited several unique capabilities of CFME to reconstitute a seven-enzyme pathway from mevalonate to amorpha-4,11-diene, an artemisinin precursor [55]. In this work, design of experiments via Taguchi orthogonal arrays were used to quickly explore a broad sample space of relative enzyme concentrations and revealed that the fifth enzyme (IspA, which encodes for farnesyl pyrophosphate synthase) had an inhibitory effect and a lowered concentration improved yields. However, the yield reduction was due to precipitation the IspA product farnesyl pyrophosphate (FPP); subsequent experiments also showed the importance of pH and Mg2+ in final titer. These results exemplify the flexibility of CFME to advantageously modify and tune the reaction conditions while also demonstrating CFME’s limitations as an in vivo prototyping model since pH and ion concentration are not engineerable parameters within the cell.
In another example of isoprenoid synthesis, isoprene was made from glycolytic intermediates phosphenolpyruvate (PEP) and pyruvate via a twelve enzyme pathway at ~100% molar yield [56]. The PEP substrate provided adequate ATP for isoprenoid synthesis; NADPH was recycled from glucose-6-phosphate using glucose-6-phosphate dehydrogenase (G6PDH). A productivity of ~0.2 mmol L−1 hr−1 demonstrates the successful activation of the three-subunit pyruvate dehydrogenase complex and points to the feasibility of a complete pathway from glucose if the glycolytic pathway can be constructed so as to regenerate NADP+ (whereas NAD+ is the more common glycolytic cofactor).
3.1.4 Specialty chemicals
CFME has biochemically innovative abilities for isotopic and chiral precision. The specificity of enzymatic catalysis has been leveraged to make radiolabeled nucleotides de novo in which specific atoms can be radiolabeled depending on the substrate isotope. In an exemplary tour de force example, a 28 enzyme pathway was implemented for the purines GTP and ATP [57], while 18 enzymes are required for the pyrimidines UTP and CTP [58]. Purified enzyme systems have also been used to make chiral molecules difficult to synthesize due a variety of possible enantiomers. Examples include D-fagomine [59] and ethyl (S)-2-ethoxy-3-(p-methoxyphenyl)propanoate (EEHP), a precursor to family of type 2 diabetes drugs, which required discovery/purification of necessary catalytic enzymes resulting in a 100-fold higher productivity compared to yeast whole cells [60].
3.1.5 Hydrogen and fuel cells from sugar
The Zhang group has made significant progress in sugar to hydrogen conversion. H2 has the potential to be a useful secondary energy carrier in a proposed hydrogen economy; however, it has a relatively low storage density as a gas. An alternative proposal would use the high storage density of sugar to power an efficient, enzymatic sugar-to-hydrogen process with subsequent fuel cell conversion to electricity. The hypothetical energy conversion efficiency of a robust, mobile, enzyme-based sugar-to-hydrogen-to-electricity system would be very high relative to conventional (combustion) and proposed (electric battery or fuel cell) energy systems [24]. Impressive molar conversion of hydrogen has been observed from starch [61], cellobiose [62], xylose [63], sucrose [64], and glucose/G6P [65] (Table 2). Notably, the ~12 mol H2 mol−1 glucose-equivalent triples the in vivo Thauer limit [66] of 4 mol H2 mol−1 glucose-equivalent. The typical continuous flow reaction pioneered by the Zhang group uses thirteen purchased or purified enzymes with H2 and CO2 monitored in the gas stream (Figure 2B). Two dehydrogenases are used to reduce NADP+ via two-step oxidation of G6P to Ru5P; the majority of remaining enzymes are used to rearrange the five carbon sugars to six carbons in order to repeat the cycle. Meanwhile, H2ase generates H2 from the NADPH. This work demonstrates the utility of a complex pathway, the flexibility to use cheap, five-carbon substrates (i.e., xylose), and the increased efficacy of thermostable enzymes [63, 65]. It should be noted, however, that the xylose system requires phosphorylation of xylulose to xylulose-5-phophate which requires ATP or less costly polyphosphate [63]. In general, comparably high molar productivities point to advantages of a gas-phase target molecule whose separation presumably drives thermodynamic equilibrium towards H2. Further flexibility was demonstrated when the H2ase was replaced by xylose reductase to make xylitol from xylose while consuming cellobiose for energy [67]. While achieving (near-) theoretical yields, a challenge for CFME highlighted in these works is the need for costly cofactors (see section 4.3). For example, to produce~60 mmol H2 over 120 hours, 2 mmol of NADP+ is needed (a turnover of ~30) [62].
Table 2.
Yields and productivities of selected cell-free systems
| Target Molecule | Substrate | Substrate Conc. | Rxn Time | Product Final Titer | Volumetric Productivity | Percent of Theor. Yield | Turnover | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|
| NAD(P)+ | ATP | |||||||||
| Purified enzyme-based systems | ||||||||||
| Ethanols | Glucose | 25 mM | 19 hr | 28.7 mM | 1.51 mmol L−1 hr−1 | 57% | 5.7 | - | [49] | |
| Isobutanol | Glucose | 19.1 mM | 23 hr | 10.3 mM | 0.45 mmol L−1 hr−1 | 53% | 2.1 | - | [49] | |
| 1,3-propanediol | Glycerol | 11.7 mM* | 20 hr | 6.3 MM* | 0.73 mmol L−1 hr−1 | 54% | 3.1 | - | [45] | |
| n-butanol | Glucose | 4.2 mM† | 7 hr | 3.5 mM | 0.49 mmol L−1 hr−1 | 83% | 7 | 17.5 | [81] | |
| Lactate/lactic acid | Glucose | 6 mM† | 10 hr | 12.5 mM | 1.25 mmol L−1 hr−1 | ~100% | 10.4 | 31.3 | [80] | |
| Malate/malic acid | Glucose | 2.4 mM† | 4 hr | 3 mM | 0.75 mmol L−1 hr−1 | 63% | 1.5 | 7.5 | [27] | |
| Amorpha-4,11-diene | Mevalonate | 10 mM | 6 hr | 1.7 mM | 0.28 mmol L−1 hr−1 | 50% | - | 0.3 | [55] | |
| Farnesene | Acetyl-CoA | 1 mM | 9 hr | ~39 μM | 0.004 mmol L−1 hr−1 | 35% | 0.07 | 0.03 | [53] | |
| Isoprene | PEP | 1.5 mM | ~4.3 hr | ~0.5 mM | 0.13 mmol L−1 hr−1 | ~100% | 0.5 | 2.5 | [56] | |
| Acetyl phosphate | F6P | 10 mM | 1.5 hr | ~30 mM | 20.0 mmol L−1 hr−1 | ~100% | - | - | [52] | |
| Hydrogen (H2) | Starch | 2 mM‡ | ~16 hr | - | 0.44 § mmol L−1 hr−1 | 43% | 3 | - | [61] | |
| Hydrogen (H2) | Cellobiose | 2 mM‡ | ~10 hr | - | 0.48 § mmol L−1 hr−1 | 93% | 10 | - | [62] | |
| Hydrogen (H2) | Cellopentaose | 8 mM‡ | ~6 hr | - | 3.92 § mmol L−1 hr−1 | 68% | 18 | - | [62] | |
| Hydrogen (H2) | Xylose | 2 mM | ~14 hr | - | 1.95 § mmol L−1 hr−1 | 95% | 5.3 | 0.66 || | [63] | |
| Xylitol (NADPH) | Xylose/Cellobiose | 100/3 mM‡ | 48 hr | ~18 mM | 0.38 mmol L−1 hr−1 | 18% | 9 | - | [67] | |
| Hydrogen (H2) | Sucrose | 2 mM | ~100 hr | - | 2.98 § mmol L−1 hr−1 | 97% | 15 | || | [64] | |
| Hydrogen (H2) | Glucose/G6P | 2/100 mM | ~1 hr | - | 8/157§ mmol L−1 hr−1 | - | 6/15 | || | [65] | |
|
Crude extract-based systems
| ||||||||||
| DHAP | Glucose | 11.1 mM | ~5 hr | 9.5 mM | 1.90 mmol L−1 hr−1 | 83% | 33.0 | 6.61 | [71] | |
| TDHP | Glucose | 55.5 mM | ~1.3 hr | 23 mM | 17.25 mmol L−1 hr−1 | 41% | 80.0 | 16.00 | [71] | |
| Ethanol | Glucose | 217 mM | ~8 hr | 86.8 mM | 10.85 mmol L−1 hr−1 | 20% | 1579 | 21.7 | [73] | |
F6P, Fructose-6-phosphate
DHAP, dihydroxyacetone phosphate (unstable intermediate)
TDHP, 5,6,7-trideoxy-D-threo-heptulose-1-phosphate (final product)
values adapted from “fed-batch” to “batch” system for comparison; conc. determined by final substrate/product amount (mmol) divided by total rxn volume (40mL)
calculated by multiplying feeding rate by time at final titer
mM sugar substrate refers to mM G6P-equivalent
Peak (not average) productivity
ATP and/or polyphosphate is added and consumed as a substrate
Further work by Zhang’s team to utilize recalcitrant feedstocks resulted in a simultaneous enzymatic biotransformation and fermentation system [68]. Here, a short four-enzyme pathway converted cellulose to starch and glucose where the free glucose was metabolized by living yeast to ethanol. This system promises to makes a nonfood biomass much more readily available for useful purposes.
In order to further develop the envisioned pathway of sugar to electricity, the pathway in Figure 2B was repurposed for direct electricity generation using a NADPH-dependent diaphorase enzyme capable of reducing vitamin K3 (menadione) which was immobilized to a fuel-cell anode coated with carbon nanotubes [69]. This allowed direct electricity production from maltodextrin in a batch experiment. The energy storage density of this system is an order of magnitude higher than traditional batteries, though at a lower voltage and current density. While a complete review of enzymatic fuel cell literature is available elsewhere [25, 70], this work demonstrates a promising application and direction for future CFME pathways.
In summary, purified systems to date exhibit high percent molar conversion but relatively low molar (mmol L−1 hr−1) and mass (g L−1 hr−1) productivities (Table 2). Cofactor turnover could be improved; it currently ranges from ~1–30 for both NAD(P)+ and ATP (Table 2). Finally, though cheaper techniques are in development, cost of cofactors and enzymes remain a barrier to commercialization. Crude extract-based CFME has fewer published examples, but offers an alternative approach that could help address some of these issues at the expense of added complexity.
3.2 Crude Cell Lysate Systems
While a majority of CFME approaches use purified enzymes, lysates of cells have been used in other applications (e.g., protein synthesis) to great effect [42]. One seminal paper for crude extract-based CFME involves the synthesis of dihydroxyacetone phosphate (DHAP) from glucose (Figure 2C) [71]. Here, engineered E. coli cells were pelleted and lysed by a high-pressure homogenizer; subsequently, cell debris, along with insoluble components, was removed via centrifugation. The resulting cell lysate was buffered and fed glucose, ATP, and NAD+ in a 10mL batch reaction. The deletion of triosephosphate isomerase (tpiA) in the source strain caused DHAP to accumulate in the reaction while the absence of AMP-nucleosidase (amn) prevented degradation of the catalytic ADP/ATP. Butanal was added to the reaction to generate 5,6,7-trideoxy-D-threo-heptulose-1-phosphate (TDHP) via an endogenous aldolase (FbaA). The subsequent high yield of DHAP and TDHP demonstrates the viability of the crude extract approach and the ability for strain modifications (tpiA and amn deletion) and fermentation conditions to affect CFME reactions. Additionally, selective addition of NAD+ (and not NADP+) funneled glycolysis to DHAP as the residual concentration of NADP+ in the extract was apparently too low to allow activation of the competing pentose phosphate pathway. In order to further optimize reaction conditions, Bujara et al. built a continuously stirred reaction where enzymes are sequestered behind a membrane and fifteen pathway intermediates in the effluent quantified in real time using ESI-MS [72]. Analysis of intermediate flux after combinatorial addition of commercial enzymes informed construction of an operon to express needed enzymes during the fermentation; this improved DHAP yield approximately 3-fold. While the pathway to DHAP is inherently limited by loss of metabolite flux to lactate in order to regenerate NAD+, the real time analysis and optimization shows the potential for precise control of metabolite flux despite the presence of unused enzymes in the lysate. Further, this shows a powerful approach that can be used to build detailed kinetic models for the metabolic pathways as defined by values of KM, kcat, and Ki for each enzyme; these models have utility both in vivo and in vitro.
Recently, a yeast cell-free lysate system to produce ethanol from glucose was demonstrated to be highly productive (10 mmol L−1 hr−1) and required no pathway tuning or strain modification [73]. Yeast cells growing in waste of beer fermentation broth were isolated, grown, lysed via bead beating, buffered, and mixed with glucose to generate 4 g L−1 (87 mM) ethanol. Notably, concentrations of ATP (1.8mM) and NAD+ (0.11mM) in the extract were high enough to sustain the reaction without supplementation. In other words, enzyme turnover was >1500 for NAD+ and >20 for ATP, thus highlighting a potential advantage of crude-extract CFME as compared to purified enzyme systems.
Crude extract pathways offer flexible target molecules. Keller et al. generated 3-hydroxypropionate from acetyl-CoA and CO2 using H2 for reducing power (this is subsection of a proposed, thirteen-enzyme pathway to fix CO2 to acetyl-CoA via hydrogen reducing equivalents) [74]. Expression of the novel pathway was done in P. furianosis which has optimal activity at 100 °C; when the cell-free reaction was run at 70 °C, only the recombinantly expressed enzymes were active at the lower temperature. Crude extracts have demonstrated the ability to synthesize high-value biologics and therapeutics; examples include single-step conversion of diketides to chiral triketide lactones (polyketide building blocks) [75], lysine to ε-poly-L-lysine (an antimicrobial) [76], and 7α-demethoxycefminox to cefminox (an antibiotic) [77]. In another study, 14C-labeled substrates were used to elucidate possible biosynthetic pathways for manufacture of the antitumor agent Azinomycin B [78]. These high-value products are promising directions for economically feasible CFME; particularly since crude extracts do not require costly purification steps.
In summary, extract based cell-free systems have so far seen relatively few examples for biomanufacturing and pathway prototyping. Thus, it is still early to comment on their utility in this arena, as compared to purified systems. However, the underlying metabolism is able to support high-level cofactor and energy regeneration. Moreover, the approach can enable rapid combinatorial iterations of enzymes during optimization of biosynthetic pathways. Thus, we expect to see substantial growth in the upcoming years.
4. Challenges and Opportunities in Cell-Free Metabolic Engineering
CFME is already showing tremendous value for pathway construction and prototyping. While many proof-of-concept pathways have been developed at the lab scale, it remains unclear to what extent CFME systems might also serve as commercially relevant production factories. Many barriers remain to commercializing cell-free synthetic systems. These include high costs of protein purification, stability and longevity of reactions, cofactor regeneration, limited efforts in modeling, and ability to scale. Following is discussion of current efforts to address these challenges.
4.1 Purification
Large-scale purified enzyme systems are limited by several major obstacles including enzyme cost and stability. Strategies to increase total turnover of enzymes would alleviate some of these issues [24, 67]; in addition, cost-effective strategies to purify multiple enzymes are desirable. Heat purification and immobilization, as well as the use of cheap scaffolds, are the two most active areas of research being employed to address this challenge.
The Ohtake lab has done significant work in the CFME space by expressing thermophilic enzymes in E. coli, lysing the cell, and heating the extract to inactivate all proteins except those needed for the pathway of interest [79]. This effectively “purifies” the relevant enzymes in a single step. This method was used to build a ten-enzyme “chimeric glycolysis” pathway to make lactate [80]. The GAPDH/PGK enzymes of traditional EMP glycolysis are replaced by GAPN; thus, no net ATP is generated by the pathway. In a fed-batch system with a slow feed rate, lactate was produced from glucose at a rate of ~1.25 mmol L−1 hr−1 and nearly ~100% molar conversion. In a similar fashion, a sixteen-enzyme pathway to produce n-butanol from glucose was constructed [81]; this specific project used a pyruvate decarboxylase and CoA-acylating dehydrogenase to bypass the large and difficult-to-express pyruvate dehydrogenase complex [82]. Directed enzyme evolution of malic enzymes was needed to optimize a glucose to malate system [27, 83]. Stability of cofactors is a tradeoff when using thermophilic enzymes in CFME; the reaction temperature of 50°C is a compromise between the high temperature needed for maximum enzyme activity and lower temperature needed for NAD+ and ATP stability.
“Heat” purification was also utilized during efforts to engineer a thermotolerant aldehyde dehydrogenase for glucose-to-ethanol CFME [49]. Here, reaction velocity and NAD+ affinity were improved via protein refolding [84] and evolution by random mutagenesis [85]. Shorter pathways have also leveraged “heat” purification in conjunction with ammonium sulfate precipitation, a technique where the salt concentration is increased until proteins begin sequentially precipitating [86].
In an effort to improve stability of the thermophilic enzymes themselves, glutaraldehyde was used to crosslink proteins so that the E. coli chassis cell remained intact after heat treatment [87]. The membrane in this state is permeable to small molecules and the “whole cells” could repeatedly convert fumarate to malate. This “permeabilized cell” concept, like crude extracts, can eliminate the need for enzyme purification [88]. A number of permeabilizing agents using a variety of mechanisms are available [88]; for example, Krauser et al. used Triton X-100 in efforts to produce flavolin [89]. However, more research is needed to characterize enzyme stability and kinetics under permeabilized conditions.
In another purification strategy, dockerin-tagged enzymes are used to bind cohesion attachment sites connected by a scaffoldin module. The scaffoldin can be attached to a magnetic nanoparticle [90] or a cellulose binding domain (which can adsorb to a low-cost regenerated amorphous cellulose) [91]. As an added benefit, the spatial proximity and/or added enzyme stability of these enzymes in the CFME reaction improved activity of a three-enzyme pathway by ~5-fold and ~40-fold, respectively. Thus, the purification technique lowers costs and can lead to higher yields and productivities, particularly if chemical reactions are diffusion limited.
4.2 Spatial Organization and Enzyme Stability
The reaction environment for CFME is inherently different than in vivo. Cells have organelles for spatial organization which can sequester pathways with overlapping intermediates, isolate pathways with toxic intermediates, and accelerate flux through slow or bottleneck enzymes by creating high local concentrations. Additionally, enzyme concentrations in CFME are typically 1–10 mg total protein mL−1 in crude extracts [35, 71] and lower still for purified systems, whereas total cytoplasmic protein concentration is ~200 mg mL−1. Enzymes were evolved for maximal activity in the crowded macromolecular environment in vivo. Thus, compartmentalization, scaffolding, and immobilization are but a few of the strategies being used to circumvent these challenges [92, 93].
For compartmentalization, CFME could utilize liposomes (lipids), polymersomes (amphiphilic block copolymers), layer-by-layer capsules (charged polyions), microfluidic encapsulation, and other approaches [94]. For example, polymersome nanoreactors can be engineered for selective porosity (dependent on pH and sugar concentration) as well as positional control of a three-enzyme cascade [94]. Also, in vivo scaffolding strategies [95] using proteins [96] or DNA [97] could be easily adapted to cell-free applications. The success of the scaffoldin-dockerin work by Zhang described above [90, 91] indicates the reaction is diffusion-limited or that scaffolding improves enzyme stability and activity.
Enzyme stability and longevity are challenges for CFME. With continuous removal of inhibitory byproducts and feeding of substrates, crude extract cell-free protein synthesis has been shown to last up to ~100 hours [98], yet most batch systems show decreased productivity over time (Table 2), possibly due to enzyme inactivation. Enzyme immobilization is a common industrial practice to increase the longevity of enzymes, often the expense of activity, yet many immobilization strategies for CFME have been proposed including particle entrapment, crosslinking to microchannel surfaces, and immobilization to membranes or nanoparticles [26]. Recent work to improve activity of alcohol dehydrogenase on a variety of solid supports points to a hypothetical reaction scheme in which all pathway enzymes are immobilized [99]. For small quantities of high-value products, chip-based immobilization strategies (such as fusion pro-tags) could be useful [100]. In general, the increased local concentration of substrates and catalysts via scaffolding/immobilization has the potential to increase both the longevity and productivity of cell-free systems.
4.3 Cofactor Engineering
While crude extract based CFME efforts have shown the potential for activating and controlling long-lived, cost-effective cofactor regeneration in lysate systems [33, 73] (Table 2), cofactor costs and instability remains a significant challenge for CFME and presently limits its commercial viability for low-value commodity chemicals [24]. Cofactor engineering could catalyze more cost-effective CFME efforts, especially for purified systems that must supplement the reaction with the necessary cofactor(s). The nicotinamide nucleotides NAD+ and NADP+ are ubiquitous electron carriers that are utilized by most redox enzymes. These molecules differ only in an additional phosphate group esterified at the 2′-hydroxyl group of NAD+. To activate de novo or existing enzyme pathways, cofactor balance can be crucial, especially without the regulation of a living cell. A number of protein engineering studies has successfully changed cofactor specificity from NADP+ to NAD+ [101], NAD+ to NADP+ [102] or made the enzyme active with either cofactor [103]. As mentioned earlier, NAD+/NADP+ are unstable in vitro and expensive; several groups have been examining the feasibility of biomimetic cofactors such as 1-benzyl-3-carbamoyl-pyridinium [104]. Other excellent reviews are available for discussion of cofactor-specificity and enzyme engineering [25, 105].
In specific pathways or applications, the ease of access to the reaction space could inspire integration of electronics with CFME where reducing power could be provided directly to enzymes by an external electric current (i.e., electrosynthesis) [106, 107]. These enzymatic bioelectrochemical systems (e-BES) could be extremely useful in adapting CFME to pathways that are not redox balanced [107].
4.4 Modeling
Prediction of intracellular concentrations and fluxes using computational modeling has greatly improved the cofactor management and product yields of conventional metabolic engineering efforts. In silico analysis of cell-free metabolism could thus shed light on pathway dynamics and inform optimization strategies. Removal of transport barriers and microbial growth objectives allows adaptation of in vivo models to cell-free systems; alternatively, new models can be made from scratch. For example, to analyze the cellobiose-to-hydrogen system described previously [62], Ardao and Zeng used a kinetic model incorporating nineteen mass balance equations to predict that modularizing the one-pot system into two reactors could improve productivity 2 to 8-fold [108]. As another illustrative example, a genetic algorithm for one-pot multi-objective optimization of the thirteen enzyme loadings plus initial concentrations of cellobiose, phosphate, and NADP+ showed the importance of hydrogenase loading and predicted a possible productivity up to 355 mmol L−1 hr−1. This paper also identified which enzymes would benefit most from engineering to improve activity or solubility. Metabolic flux analysis (MFA) and associated tools could also be adapted to CFME efforts by removing transport operations and boundaries and reconceptualizing objective functions [109–111]. For example, the well-studied flux balance analysis tool (COBRA) was modified to analyze crude extract production of DHAP [111]. This model was able to correctly predict a previously identified undesired pathway for DHAP synthesis from adenosine phosphate, as well as identify other potentially interfering pathways that could be feasibly modified to further insulate the desired metabolic pathway.
The development of computational design tools such as BNICE [112, 113], PathPred [114], UM-PPS [115], Retropath [116], Metabolic Tinker [117], Act [118], and others [110] has led to an explosion of proposed novel metabolic pathways and circuits. CFME will be crucial tool in the experimental validation of these pathways. CFME’s ability to adjust cofactor/enzyme concentrations and easily sample reaction intermediates/products are significant advantages when troubleshooting and optimizing these pathways.
4.5 Scalability
It is well-known in microbial fermentation processes that reaction scale affects molecular diffusion, gas transfer, hydrostatic pressure, mixing rates, temperature gradients, and other parameters [19]. To date, cell-free systems have relatively few examples of industrial scale production. However, published results from crude-extract cell-free protein synthesis recently demonstrated linear scalability for protein yields from the microliter to liter scale for aglycosylated antibodies (5 L scale) [41] and cytokines (100 L scale) [40]. Highlighting an expansion factor of 106, these findings suggest cell-free engineering considerations are more parallel to chemistry than a living system and give promise to the technical scalability of CFME pending economic viability.
5. Conclusion
CFME holds tremendous potential for rapidly developing metabolic systems capable of synthesizing useful small molecules. As the number of successfully demonstrated pathways grows, purification and cofactor costs are challenges that have yet to be resolved. By addressing these challenges, as well as others described in Section 4, we anticipate that cell-free systems will enable new opportunities. One such emerging opportunity made possible by recent advances is the ability to explore combinatorial assembly of pathways by mixing-and-matching lysates or individual enzymes (Figure 3, shown for lysates). For example, when optimizing a selected pathway, multiple cell extracts could be prepared, each with a different enzyme(s) overexpressed. The extracts provide supporting metabolic enzymes for highly active energy production and co-factor regeneration (e.g., to convert glucose to acetyl-CoA). Subsequently, extracts selectively enriched with these different enzyme(s) could be mixed in different combinations/ratios to build partial or full-length pathways that provide initial estimates of beneficial enzyme ratios, as well as indications of enzyme promiscuity to different starting substrates and the presence of unwanted side reactions. Notably, such an approach does not require the focus on flux balancing and delicate promoter tuning to maintain viability as for in vivo systems [119–121]. Moreover, strain modification techniques, such as MAGE [122], could be leveraged to add protease cleavage sites [23] or affinity tags [123] to essential enzymes; this would allow selective degradation or physical removal of unwanted pathways critical for growth. An additional possibility is mixing lysates from multiple organisms, i.e., organismal amalgams, to construct novel natural product pathways that would be impossible to express in one organism in vivo (Figure 3).
Figure 3.
New opportunities to use crude cell lysates for pathway optimization and debugging.
Looking forward, we expect that CFME will enable new frontiers for rapidly generating and evaluating new enzymes and metabolic pathways. It is unclear to what extent CFME will be used for prototyping systems for in vivo application (e.g., pathway balance or enzyme promiscuity) or as a large-scale biomanufacturing system in its own right. Small-scale, high-value synthesis in chip-like applications may also prove to be useful. Obviously, these questions will only be answered as viable commercial ventures emerge. That said, by avoiding toxicity constraints, CFME will offer novel ways to create hybrid molecule products composed of elements derived from both chemical and biological synthesis strategies. In sum, CFME holds the potential to expand the definition of biomanufacturing, allowing cell-free biosynthesis to penetrate into new industrial applications and make immediate and significant solutions to global challenges in materials, medicine, and sustainability.
Acknowledgments
We gratefully acknowledge the National Science Foundation (MCB-0943393), the Office of Naval Research (N00014-11-1-0363), the DARPA YFA Program (N66001-11-1-4137), the Army Research Office (W911NF-11-1-044), the NSF Materials Network Grant (DMR - 1108350), the DARPA Living Foundries Program (N66001-12-C-4211), the David and Lucile Packard Foundation (2011-37152), ARPA-E (DE-AR0000435), and the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust for support.. ASK is an NSF Graduate Fellow. QMD is funded, in part, by the Northwestern Molecular Biophysics Training Program supported by NIH via NIGMS (5T32 GM008382).
Abbreviations
- CFME
Cell-Free Metabolic Engineering
- CFPS
Cell-Free Protein Synthesis
- EMP pathway
Embden–Meyerhof–Parnas glycolytic pathway
- G6P
glucose 6-phosphate
- DHAP
dihydroxyacetone phosphate
Footnotes
Conflict of interest: The authors declare no conflict of interest
References
- 1.Moulijn JA, Makkee M, Van Diepen AE. Chemical process technology. John Wiley & Sons; 2013. [Google Scholar]
- 2.Höök M, Tang X. Depletion of fossil fuels and anthropogenic climate change—A review. Energy Policy. 2013;52:797–809. [Google Scholar]
- 3.Karl TR, Melillo JM, Peterson TC. Global climate change impacts in the United States. Cambridge University Press; 2009. [Google Scholar]
- 4.Bozell JJ, Petersen GR. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “top 10” revisited. Green Chemistry. 2010;12:539–554. [Google Scholar]
- 5.Bornscheuer U, Huisman G, Kazlauskas R, Lutz S, et al. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 6.Quin MB, Schmidt-Dannert C. Engineering of biocatalysts: from evolution to creation. ACS catalysis. 2011;1:1017–1021. doi: 10.1021/cs200217t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JW, Na D, Park JM, Lee J, et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nature Chemical Biology. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 8.Curran KA, Alper HS. Expanding the chemical palate of cells by combining systems biology and metabolic engineering. Metabolic Engineering. 2012;14:289–297. doi: 10.1016/j.ymben.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen J, Larsson C, van Maris A, Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Current opinion in biotechnology. 2013 doi: 10.1016/j.copbio.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Keasling JD. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 11.Cherubini F. The biorefinery concept: using biomass instead of oil for producing energy and chemicals. Energy Conversion and Management. 2010;51:1412–1421. [Google Scholar]
- 12.Nakamura CE, Whited GM. Metabolic engineering for the microbial production of 1,3-propanediol. Curr Opin Biotechnol. 2003;14:454–459. doi: 10.1016/j.copbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen J, Fussenegger M, Keasling J, Lee SY, et al. Engineering synergy in biotechnology. Nature Chemical Biology. 2014;10:319–322. doi: 10.1038/nchembio.1519. [DOI] [PubMed] [Google Scholar]
- 15.Kwok R. Five hard truths for synthetic biology. Nature. 2010;463:288–290. doi: 10.1038/463288a. [DOI] [PubMed] [Google Scholar]
- 16.Keasling JD. Synthetic biology and the development of tools for metabolic engineering. Metabolic engineering. 2012;14:189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metabolic engineering. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green EM. Fermentative production of butanol—the industrial perspective. Current opinion in biotechnology. 2011;22:337–343. doi: 10.1016/j.copbio.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Takors R. Scale-up of microbial processes: Impacts, tools and open questions. Journal of biotechnology. 2012;160:3–9. doi: 10.1016/j.jbiotec.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Yadav VG, De Mey M, Giaw Lim C, Kumaran Ajikumar P, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metabolic engineering. 2012;14:233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galanie S, Siddiqui MS, Smolke CD. Molecular tools for chemical biotechnology. Current opinion in biotechnology. 2013 doi: 10.1016/j.copbio.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bujara M, Panke S. Engineering in complex systems. Curr Opin Biotechnol. 2010;21:586–591. doi: 10.1016/j.copbio.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Swartz JR. Transforming biochemical engineering with cell-free biology. AIChE Journal. 2012;58:5–13. [Google Scholar]
- 24.Zhang YHP. What is vital (and not vital) to advance economically-competitive biofuels production. Process Biochemistry. 2011;46:2091–2110. [Google Scholar]
- 25.Rollin JA, Tam TK, Zhang YHP. New biotechnology paradigm: cell-free biosystems for biomanufacturing. Green Chemistry. 2013;15:1708–1719. [Google Scholar]
- 26.Ardao I, Hwang ET, Zeng A-P. Fundamentals and Application of New Bioproduction Systems. Springer; 2013. In Vitro Multienzymatic Reaction Systems for Biosynthesis; pp. 153–184. [DOI] [PubMed] [Google Scholar]
- 27.Ye X, Honda K, Morimoto Y, Okano K, Ohtake H. Direct conversion of glucose to malate by synthetic metabolic engineering. Journal of biotechnology. 2013;164:34–40. doi: 10.1016/j.jbiotec.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Guterl JK, Sieber V. Biosynthesis “debugged”: novel bioproduction strategies. Engineering in Life Sciences. 2013;13:4–18. [Google Scholar]
- 29.Billerbeck S, Härle J, Panke S. The good of two worlds: increasing complexity in cell-free systems. Current opinion in biotechnology. 2013;24:1037–1043. doi: 10.1016/j.copbio.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Buchner E, Rapp R. Alkoholische gährung ohne hefezellen. Berichte der deutschen chemischen Gesellschaft. 1897;30:2668–2678. [Google Scholar]
- 31.Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 1961;47:1588. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez S, Demain AL. Enzymes and bioconversions of industrial, pharmaceutical, and biotechnological significance. Organic Process Research & Development. 2010;15:224–230. [Google Scholar]
- 33.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Molecular systems biology. 2008;4 doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DM, Swartz JR. Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnology and bioengineering. 2001;74:309–316. [PubMed] [Google Scholar]
- 35.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnology and bioengineering. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Zhang YP. Cell-free protein synthesis energized by slowly-metabolized maltodextrin. BMC biotechnology. 2009;9:58. doi: 10.1186/1472-6750-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caschera F, Noireaux V. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription–translation system. Biochimie. 2013 doi: 10.1016/j.biochi.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Kim HC, Kim TW, Kim DM. Prolonged production of proteins in a cell-free protein synthesis system using polymeric carbohydrates as an energy source. Process Biochemistry. 2011;46:1366–1369. [Google Scholar]
- 39.Shimizu Y, Inoue A, Tomari Y, Suzuki T, et al. Cell-free translation reconstituted with purified components. Nature biotechnology. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 40.Zawada JF, Yin G, Steiner AR, Yang J, et al. Microscale to manufacturing scale-up of cell-free cytokine production—a new approach for shortening protein production development timelines. Biotechnology and bioengineering. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin G, Garces ED, Yang J, Zhang J, et al. MAbs. 2012:217–225. doi: 10.4161/mabs.4.2.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: applications come of age. Biotechnology advances. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittaker JW. Cell-free protein synthesis: the state of the art. Biotechnology letters. 2013;35:143–152. doi: 10.1007/s10529-012-1075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray CJ, Baliga R. Cell-free translation of peptides and proteins: from high throughput screening to clinical production. Current opinion in chemical biology. 2013;17:420–426. doi: 10.1016/j.cbpa.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Rieckenberg F, Ardao I, Rujananon R, Zeng AP. Cell-free synthesis of 1: 3-propanediol from glycerol with a high yield. Engineering in Life Sciences. 2014 [Google Scholar]
- 46.Siegal-Gaskins D, Tuza ZA, Kim J, Noireaux V, Murray RM. Gene Circuit Performance Characterization and Resource Usage in a Cell-Free “Breadboard”. ACS Synthetic Biology. 2014 doi: 10.1021/sb400203p. [DOI] [PubMed] [Google Scholar]
- 47.Welch P, Scopes RK. Studies on cell-free metabolism: Ethanol production by a yeast glycolytic system reconstituted from purified enzymes. Journal of biotechnology. 1985;2:257–273. [Google Scholar]
- 48.Allain EJ. Cell-free ethanol production: the future of fuel ethanol? Journal of Chemical Technology and Biotechnology. 2007;82:117–120. [Google Scholar]
- 49.Guterl JK, Garbe D, Carsten J, Steffler F, et al. Cell-Free Metabolic Engineering: Production of Chemicals by Minimized Reaction Cascades. ChemSusChem. 2012;5:2165–2172. doi: 10.1002/cssc.201200365. [DOI] [PubMed] [Google Scholar]
- 50.Jung GY, Stephanopoulos G. A functional protein chip for pathway optimization and in vitro metabolic engineering. Science. 2004;304:428–431. doi: 10.1126/science.1096920. [DOI] [PubMed] [Google Scholar]
- 51.Shin J, Noireaux V. An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS synthetic biology. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 52.Bogorad IW, Lin TS, Liao JC. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature. 2013;502:693–697. doi: 10.1038/nature12575. [DOI] [PubMed] [Google Scholar]
- 53.Zhu F, Zhong X, Hu M, Lu L, et al. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnology and bioengineering. 2014 doi: 10.1002/bit.25198. [DOI] [PubMed] [Google Scholar]
- 54.Yu X, Liu T, Zhu F, Khosla C. In vitro reconstitution and steady-state analysis of the fatty acid synthase from Escherichia coli. Proceedings of the National Academy of Sciences. 2011;108:18643–18648. doi: 10.1073/pnas.1110852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Zhang C, Zou R, Zhou K, et al. Statistical Experimental Design Guided Optimization of a One-Pot Biphasic Multienzyme Total Synthesis of Amorpha-4, 11-diene. PloS one. 2013;8:e79650. doi: 10.1371/journal.pone.0079650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korman TP, Sahachartsiri B, Li D, Vinokur JM, et al. A synthetic biochemistry system for the in vitro production of isoprene from glycolysis intermediates. Protein Science. 2014 doi: 10.1002/pro.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Pathway engineered enzymatic de novo purine nucleotide synthesis. ACS chemical biology. 2008;3:499–511. doi: 10.1021/cb800066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Enzymatic De Novo pyrimidine nucleotide synthesis. Journal of the American Chemical Society. 2010;133:297–304. doi: 10.1021/ja1059685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Babich L, van Hemert LJ, Bury A, Hartog AF, et al. Synthesis of non-natural carbohydrates from glycerol and aldehydes in a one-pot four-enzyme cascade reaction. Green Chemistry. 2011;13:2895–2900. [Google Scholar]
- 60.Bechtold M, Brenna E, Femmer C, Gatti FG, et al. Biotechnological development of a practical synthesis of ethyl (S)-2-ethoxy-3-(p-methoxyphenyl) propanoate (EEHP): over 100-fold productivity increase from yeast whole cells to recombinant isolated enzymes. Organic Process Research & Development. 2011;16:269–276. [Google Scholar]
- 61.Zhang YHP, Evans BR, Mielenz JR, Hopkins RC, Adams MW. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS One. 2007;2:e456. doi: 10.1371/journal.pone.0000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye X, Wang Y, Hopkins RC, Adams MW, et al. Spontaneous high-yield production of hydrogen from cellulosic materials and water catalyzed by enzyme cocktails. ChemSusChem. 2009;2:149–152. doi: 10.1002/cssc.200900017. [DOI] [PubMed] [Google Scholar]
- 63.Martín del Campo JS, Rollin J, Myung S, Chun Y, et al. High-Yield Production of Dihydrogen from Xylose by Using a Synthetic Enzyme Cascade in a Cell-Free System. Angewandte Chemie International Edition. 2013;52:4587–4590. doi: 10.1002/anie.201300766. [DOI] [PubMed] [Google Scholar]
- 64.Myung S, Rollin J, You C, Sun F, et al. In vitro metabolic engineering of Hydrogen production at theoretical yield from sucrose. Metabolic Engineering. 2014 doi: 10.1016/j.ymben.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Rollin JA, Ye X, del Campo JM, Adams MW, Zhang Y-HP. Novel Hydrogen Bioreactor and Detection Apparatus. 2014 doi: 10.1007/10_2014_274. [DOI] [PubMed] [Google Scholar]
- 66.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriological reviews. 1977;41:100. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Huang W, Sathitsuksanoh N, Zhu Z, Zhang YHP. Biohydrogenation from biomass sugar mediated by in vitro synthetic enzymatic pathways. Chemistry & biology. 2011;18:372–380. doi: 10.1016/j.chembiol.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 68.You C, Chen H, Myung S, Sathitsuksanoh N, et al. Enzymatic transformation of nonfood biomass to starch. Proceedings of the National Academy of Sciences. 2013;110:7182–7187. doi: 10.1073/pnas.1302420110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu Z, Tam TK, Sun F, You C, Zhang Y-HP. A high-energy-density sugar biobattery based on a synthetic enzymatic pathway. Nature communications. 2014;5 doi: 10.1038/ncomms4026. [DOI] [PubMed] [Google Scholar]
- 70.Minteer SD, Liaw BY, Cooney MJ. Enzyme-based biofuel cells. Current opinion in biotechnology. 2007;18:228–234. doi: 10.1016/j.copbio.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 71.Bujara M, Schümperli M, Billerbeck S, Heinemann M, Panke S. Exploiting cell-free systems: Implementation and debugging of a system of biotransformations. Biotechnology and bioengineering. 2010;106:376–389. doi: 10.1002/bit.22666. [DOI] [PubMed] [Google Scholar]
- 72.Bujara M, Schümperli M, Pellaux R, Heinemann M, Panke S. Optimization of a blueprint for in vitro glycolysis by metabolic real-time analysis. Nature chemical biology. 2011;7:271–277. doi: 10.1038/nchembio.541. [DOI] [PubMed] [Google Scholar]
- 73.Khattak WA, Ul-Islam M, Ullah MW, Yu B, et al. Yeast cell-free enzyme system for bio-ethanol production at elevated temperatures. Process Biochemistry. 2014 [Google Scholar]
- 74.Keller MW, Schut GJ, Lipscomb GL, Menon AL, et al. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proceedings of the National Academy of Sciences. 2013;110:5840–5845. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harper AD, Bailey CB, Edwards AD, Detelich JF, Keatinge-Clay AT. Preparative, in vitro biocatalysis of triketide lactone chiral building blocks. Chem Bio Chem. 2012;13:2200–2203. doi: 10.1002/cbic.201200378. [DOI] [PubMed] [Google Scholar]
- 76.Kawai T, Kubota T, Hiraki J, Izumi Y. Biosynthesis of ε-poly-l-lysine in a cell-free system ofStreptomyces albulus. Biochemical and biophysical research communications. 2003;311:635–640. doi: 10.1016/j.bbrc.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 77.Kim JK, Kang Hi, Chae JS, Park YH, Choi YJ. Synthesis of cefminox by cell-free extracts of Streptomyces clavuligerus. FEMS microbiology letters. 2000;182:313–317. doi: 10.1111/j.1574-6968.2000.tb08914.x. [DOI] [PubMed] [Google Scholar]
- 78.Liu C, Kelly GT, Watanabe CM. In vitro biosynthesis of the antitumor agent azinomycin B. Organic letters. 2006;8:1065–1068. doi: 10.1021/ol052987l. [DOI] [PubMed] [Google Scholar]
- 79.Honda K, Maya S, Omasa T, Hirota R, et al. Production of 2-deoxyribose 5-phosphate from fructose to demonstrate a potential of artificial bio-synthetic pathway using thermophilic enzymes. Journal of biotechnology. 2010;148:204–207. doi: 10.1016/j.jbiotec.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 80.Ye X, Honda K, Sakai T, Okano K, et al. Synthetic metabolic engineering-a novel, simple technology for designing a chimeric metabolic pathway. Microbial cell factories. 2012;11:120. doi: 10.1186/1475-2859-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krutsakorn B, Honda K, Ye X, Imagawa T, et al. In vitro production of n-butanol from glucose. Metabolic engineering. 2013;20:84–91. doi: 10.1016/j.ymben.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 82.Krutsakorn B, Imagawa T, Honda K, Okano K, Ohtake H. Construction of an in vitro bypassed pyruvate decarboxylation pathway using thermostable enzyme modules and its application to N-acetylglutamate production. Microbial cell factories. 2013;12:91. doi: 10.1186/1475-2859-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morimoto Y, Honda K, Ye X, Okano K, Ohtake H. Directed evolution of thermotolerant malic enzyme for improved malate production. Journal of bioscience and bioengineering. 2013 doi: 10.1016/j.jbiosc.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 84.Steffler F, Sieber V. Refolding of a Thermostable Glyceraldehyde Dehydrogenase for Application in Synthetic Cascade Biomanufacturing. PloS one. 2013;8:e70592. doi: 10.1371/journal.pone.0070592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Steffler F, Guterl JK, Sieber V. Improvement of thermostable aldehyde dehydrogenase by directed evolution for application in Synthetic Cascade Biomanufacturing. Enzyme and microbial technology. 2013;53:307–314. doi: 10.1016/j.enzmictec.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 86.Myung S, Zhang YP. Non-complexed four cascade enzyme mixture: simple purification and synergetic co-stabilization. PloS one. 2013;8:e61500. doi: 10.1371/journal.pone.0061500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ninh PH, Honda K, Yokohigashi Y, Okano K, et al. Development of a continuous bioconversion system using a thermophilic whole-cell biocatalyst. Applied and environmental microbiology. 2013;79:1996–2001. doi: 10.1128/AEM.03752-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krauser S, Weyler C, Blaβ LK, Heinzle E. Fundamentals and Application of New Bioproduction Systems. Springer; 2013. Directed Multistep Biocatalysis Using Tailored Permeabilized Cells; pp. 185–234. [DOI] [PubMed] [Google Scholar]
- 89.Krauser S, Kiefer P, Heinzle E. Multienzyme Whole-Cell In Situ Biocatalysis for the Production of Flaviolin in Permeabilized Cells of Escherichia coli. Chem Cat Chem. 2012;4:786–788. [Google Scholar]
- 90.Myung S, You C, Zhang YHP. Recyclable cellulose-containing magnetic nanoparticles: immobilization of cellulose-binding module-tagged proteins and a synthetic metabolon featuring substrate channeling. Journal of Materials Chemistry B. 2013;1:4419–4427. doi: 10.1039/c3tb20482k. [DOI] [PubMed] [Google Scholar]
- 91.You C, Zhang YHP. Self-assembly of synthetic metabolons through synthetic protein scaffolds: one-step purification, co-immobilization, and substrate channeling. ACS synthetic biology. 2012;2:102–110. doi: 10.1021/sb300068g. [DOI] [PubMed] [Google Scholar]
- 92.Jandt U, You C, Zhang Y-P, Zeng A-P. Fundamentals and Application of New Bioproduction Systems. Springer; 2013. Compartmentalization and metabolic channeling for multienzymatic biosynthesis: practical strategies and modeling approaches; pp. 41–65. [DOI] [PubMed] [Google Scholar]
- 93.Zhang YHP. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnology advances. 2011;29:715–725. doi: 10.1016/j.biotechadv.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 94.Peters RJ, Louzao I, van Hest JC. From polymeric nanoreactors to artificial organelles. Chemical Science. 2012;3:335–342. [Google Scholar]
- 95.Agapakis CM, Boyle PM, Silver PA. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nature chemical biology. 2012;8:527–535. doi: 10.1038/nchembio.975. [DOI] [PubMed] [Google Scholar]
- 96.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nature biotechnology. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 97.Conrado RJ, Wu GC, Boock JT, Xu H, et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic acids research. 2012;40:1879–1889. doi: 10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proceedings of the national academy of sciences of the United States of America. 2004;101:17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grimaldi J, Collins C, Belfort G. Toward cell-free biofuel production: Stable immobilization of oligomeric enzymes. Biotechnology progress. 2014 doi: 10.1002/btpr.1876. [DOI] [PubMed]
- 100.Lewandowski AT, Yi H, Luo X, Payne GF, et al. Protein assembly onto patterned microfabricated devices through enzymatic activation of fusion pro-tag. Biotechnology and bioengineering. 2008;99:499–507. doi: 10.1002/bit.21580. [DOI] [PubMed] [Google Scholar]
- 101.Bastian S, Liu X, Meyerowitz JT, Snow CD, et al. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metabolic engineering. 2011;13:345–352. doi: 10.1016/j.ymben.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 102.Campbell E, Wheeldon IR, Banta S. Broadening the cofactor specificity of a thermostable alcohol dehydrogenase using rational protein design introduces novel kinetic transient behavior. Biotechnology and bioengineering. 2010;107:763–774. doi: 10.1002/bit.22869. [DOI] [PubMed] [Google Scholar]
- 103.Katzberg M, Skorupa-Parachin N, Gorwa-Grauslund MF, Bertau M. Engineering cofactor preference of ketone reducing biocatalysts: a mutagenesis study on a γ-diketone reductase from the yeast Saccharomyces cerevisiae serving as an example. International journal of molecular sciences. 2010;11:1735–1758. doi: 10.3390/ijms11041735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lo HC, Leiva C, Buriez O, Kerr JB, et al. Bioorganometallic Chemistry. 13. Regioselective Reduction of NAD+ Models, 1-Benzylnicotinamde Triflate and β-Nicotinamide Ribose-5′-methyl Phosphate, with in Situ Generated [Cp* Rh (Bpy) H]+: Structure-Activity Relationships, Kinetics, and Mechanistic Aspects in the Formation of the 1, 4-NADH Derivatives. Inorganic chemistry. 2001;40:6705–6716. doi: 10.1021/ic010562z. [DOI] [PubMed] [Google Scholar]
- 105.Li Y, Cirino PC. Recent advances in engineering proteins for biocatalysis. Biotechnology and Bioengineering. 2014 doi: 10.1002/bit.25240. [DOI] [PubMed] [Google Scholar]
- 106.Rabaey K, Rozendal RA. Microbial electrosynthesis—revisiting the electrical route for microbial production. Nature Reviews Microbiology. 2010;8:706–716. doi: 10.1038/nrmicro2422. [DOI] [PubMed] [Google Scholar]
- 107.Dominguez-Benetton X, Srikanth S, Satyawali Y, Vanbroekhoven K, Pant D. Enzymatic Electrosynthesis: An Overview on the Progress in Enzyme-Electrodes for the Production of Electricity, Fuels and Chemicals. J Microb Biochem Technol S. 2013;6:2. [Google Scholar]
- 108.Ardao I, Zeng A-P. In silico evaluation of a complex multi-enzymatic system using one-pot and modular approaches: Application to the high-yield production of hydrogen from a synthetic metabolic pathway. Chemical Engineering Science. 2012 [Google Scholar]
- 109.Toya Y, Shimizu H. Flux analysis and metabolomics for systematic metabolic engineering of microorganisms. Biotechnology advances. 2013;31:818–826. doi: 10.1016/j.biotechadv.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 110.Copeland WB, Bartley BA, Chandran D, Galdzicki M, et al. Computational tools for metabolic engineering. Metabolic engineering. 2012;14:270–280. doi: 10.1016/j.ymben.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bujara M, Panke S. In silico assessment of cell-free systems. Biotechnology and Bioengineering. 2012;109:2620–2629. doi: 10.1002/bit.24534. [DOI] [PubMed] [Google Scholar]
- 112.González-Lergier J, Broadbelt LJ, Hatzimanikatis V. Theoretical considerations and computational analysis of the complexity in polyketide synthesis pathways. Journal of the American Chemical Society. 2005;127:9930–9938. doi: 10.1021/ja051586y. [DOI] [PubMed] [Google Scholar]
- 113.Henry CS, Broadbelt LJ, Hatzimanikatis V. Discovery and analysis of novel metabolic pathways for the biosynthesis of industrial chemicals: 3-hydroxypropanoate. Biotechnology and bioengineering. 2010;106:462–473. doi: 10.1002/bit.22673. [DOI] [PubMed] [Google Scholar]
- 114.Moriya Y, Shigemizu D, Hattori M, Tokimatsu T, et al. PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic acids research. 2010;38:W138–W143. doi: 10.1093/nar/gkq318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gao J, Ellis LB, Wackett LP. The University of Minnesota Pathway Prediction System: multi-level prediction and visualization. Nucleic acids research. 2011;39:W406–W411. doi: 10.1093/nar/gkr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carbonell P, Parutto P, Baudier C, Junot C, Faulon J-L. Retropath: Automated Pipeline for Embedded Metabolic Circuits. ACS synthetic biology. 2013 doi: 10.1021/sb4001273. [DOI] [PubMed] [Google Scholar]
- 117.McClymont K, Soyer OS. Metabolic tinker: an online tool for guiding the design of synthetic metabolic pathways. Nucleic acids research. 2013;41:e113–e113. doi: 10.1093/nar/gkt234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Srivastava S, Kotker J, Hamilton S, Ruan P, et al. Proceedings of the 4th International Workshop on Bio-Design Automation (IWBDA); 2012. [Google Scholar]
- 119.Du J, Yuan Y, Si T, Lian J, Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic acids research. 2012;40:e142–e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blazeck J, Liu L, Redden H, Alper H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Applied and environmental microbiology. 2011;77:7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang HH, Huang PY, Xu G, Haas W, et al. Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS synthetic biology. 2012;1:43–52. doi: 10.1021/sb3000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Atsumi S, Wu TY, Machado IM, Huang WC, et al. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Molecular systems biology. 2010;6 doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]