Abstract
Babesia spp. are obligate protozoan parasites of red blood cells. Transmission to humans occurs through bites from infected ticks or blood transfusion. Infections with B. microti account for the majority of the reported cases of human babesiosis in the USA. A lower incidence is caused by the more recently described species B. duncani. The current gold standard for detection of Babesia is microscopic examination of blood smears. Recent PCR-based assays, including real-time PCR, have been developed for B. microti. On the other hand, molecular assays that detect and distinguish between B. microti and B. duncani infections are lacking. Closely related species of Babesia can be differentiated due to sequence variation within the internal transcribed spacer (ITS) regions of nuclear ribosomal RNAs. In the present study, we targeted the ITS regions of B. microti and B. duncani to develop sensitive and species-specific droplet digital PCR (ddPCR) assays. The assays were shown to discriminate B. microti from B. duncani and resulted in limits of detection of ~10 gene copies. Moreover, ddPCR for these species were useful in DNA extracted from blood of experimentally infected hamsters, detecting infections of low parasitemia that were negative by microscopic examination. In summary, we have developed sensitive and specific quantitative ddPCR assays for the detection of B. microti and B. duncani in blood. Our methods could be used as sensitive approaches to monitor the progression of parasitemia in rodent models of infection as well as serve as suitable molecular tests in blood screening.
Keywords: Babesia, Babesiosis, Digital PCR, Real time PCR, Blood infection, Molecular method, Detection, Quantitation
1. Introduction
Babesiosis, caused by intraerythrocytic protozoan parasites of the genus Babesia, has been recognized in recent years as an emerging infectious disease in humans. Transmission occurs primarily by ixodid ticks (Hunfeld et al., 2008). Although uncommon, other routes of transmission include pregnancy and blood transfusion (Joseph et al., 2012; Lobo et al., 2013). The latter is of particular concern since potential blood donors may harbor asymptomatic infections. A 2008 workshop sponsored by the Food and Drug Administration (FDA) discussed various aspects of transfusion-transmitted babesiosis in the United States, including strategies to identify infected blood donors, epidemiology of the disease, and biology and pathogenesis of Babesia spp. (Gubernot et al., 2009). Despite recommendations for the development of tests for babesiosis that met blood donor screening requirements, to this date there is a lack of FDA-approved assays for blood testing against Babesia and other vector-transmitted protozoan parasites.
B. microti is the causative agent for the majority of human cases of babesiosis in the U.S., with a higher prevalence occurring in the Northeastern region of the country (Johnson et al., 2009; Kogut et al., 2005; Leiby, 2011). Infections by B. microti have also been reported in the Upper Midwest, particularly Minnesota and Wisconsin (Centers for Disease Control and Prevention (CDC), 2012; Herwaldt et al., 1995; Setty et al., 2003). A lower incidence of babesiosis is observed in western regions of the U.S. where it is usually caused by infection with B. duncani (Conrad et al., 2006; Herwaldt et al., 1997; Persing et al., 1995; Quick et al., 1993).
Infection with Babesia is usually asymptomatic or results in mild symptoms that resolve within a few days. Infected individuals may display flu-like symptoms within 1–9 weeks which include high fever, headaches, chills, fatigue, and anemia (Hunfeld et al., 2008; Leiby, 2011). Severe cases featuring acute anemia, thrombocytopenia, organ failure, or even death may occur among the elderly, splenectomized, and immunocompromised individuals (Krause et al., 2008). Limited studies in animal models suggest a higher virulence of B. duncani compared with B. microti (Wozniak et al., 1996); however, a correlation between the experimental animal data and human clinical cases remains unclear.
Microscopic examination of blood smears stained with Giemsa has been the gold standard test for the detection of Babesia infection for many years. This method has limitations since it requires well trained personnel due to the fact that Babesia displays morphological similarities with malaria parasites (Leiby, 2011). In addition, microscopic diagnosis is difficult in patients with very low parasitemia during early or chronic stages of infection. Serological methods such as immunofluorescent antibody test and enzymelinked immunosorbent assay have been developed for detection of B. microti infection (Homer et al., 2003; Krause et al., 1994; Luo et al., 2011). As molecular assays have become more commonplace in the diagnosis of parasitic infections, both conventional and real-time polymerase chain reaction (PCR) have been developed for detection of B. microti in human samples (Bloch et al., 2013; Chan et al., 2013; Persing et al., 1992; Rollend et al., 2013; Teal et al., 2012). Of note, highly specific and sensitive molecular assays that detect and distinguish B. microti from B. duncani infections are lacking.
Droplet digital PCR (ddPCR) is a growing technology of nucleic acid detection and quantitation. This method facilitates absolute quantitation of DNA targets without the requirement of standard curves commonly used in real-time PCR (Vogelstein and Kinzler, 1999). In ddPCR, the amplification reaction containing the DNA sample, fluorescently-labeled probe, and components is divided into thousands of microscopic reaction droplets, with each containing one or less copies of the target DNA (Hindson et al., 2011; Nakano et al., 2003; Pinheiro et al., 2012). Following amplification, the measurement of both fluorescent (i.e., positive) and non-fluorescent (i.e., negative) droplets is performed. The number of target DNA molecules present in the sample can be calculated from the fraction of positive reactions using Poisson statistics (Hindson et al., 2011). Uses of ddPCR include measurement of germline DNA copy number variation (Pinheiro et al., 2012), gene expression in single cells (Heredia et al., 2013), detection of genetically modified organisms in food (Morisset et al., 2013), and, more recently, quantitation of bacterial (Roberts et al., 2013) and viral (Hayden et al., 2013; Henrich et al., 2012; White et al., 2012) pathogens in human samples. Applications of the technology in parasitic diseases are lacking to the best of our knowledge.
We report the development of ddPCR assays for the detection and discrimination of B. microti and B. duncani in experimentally infected hamster blood samples. We targeted the ITS regions of nuclear ribosomal RNA given the divergence in sequence variation among closely related Babesia species (Blaschitz et al., 2008; Bostrom et al., 2008; Liu et al., 2008; Niu et al., 2009). Quantitative data of these assays were compared with real-time PCR assays using identical primers and probes. The assays could be used as highly specific and sensitive molecular tests for blood screening of human samples.
2. Materials and methods
2.1. Babesia isolates
Babesia microti GI (BEI Resources NR-44070; ATCC® PRA-398™) and B. duncani WA-1 (BEI Resources NR-12311; ATCC® PRA-302™) were used in this study. Isolates were propagated in Golden Syrian hamsters (Harlan Laboratories, stock: HsdHan:AURA) according to published protocols (Cullen and Levine, 1987; Oz and Hughes, 1996; Wozniak et al., 1996). Blood was drawn at days 3, 7, and 9 or 10 of infection by peri-orbital route. Parasitemia was determined by microscopic examination of blood films stained with Giemsa. On each sample, a minimum of 500 red blood cells (RBCs) were counted to calculate the percent parasitemia. This included all parasitized RBCs regardless of intraerythrocytic stage or number of parasites per cell. Animals were maintained according to protocols approved by the ATCC® IACUC.
2.2. DNA extraction
For DNA extraction, 250 µl of blood were collected in heparin and treated with 750 µl of TSE/Triton X-100 buffer (20 mM Tris– HCl pH 7.5, 48mMEDTA, 100mMNaCl, 0.2% Triton X-100) for 5 min to lyse the erythrocytes. Subsequent steps were followed according to the Gentra® Puregene® Blood Kit (Qiagen) with the addition of 20 µl of proteinase K to the cell lysis buffer and incubation for 2 h. DNA was dissolved in 100 µl of rehydration buffer in the last step of the protocol and frozen at −20 °C.
2.3. PCR and sequencing
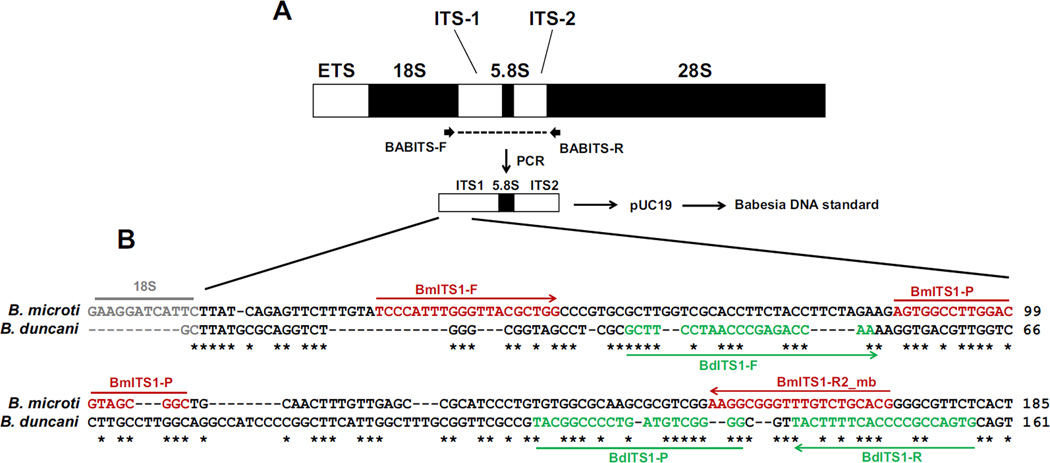
The entire ITS1, 5.8S rRNA, and ITS2 regions of B. microti GI and B. duncani WA-1 were amplified as 930–950 bp PCR products using forward primer BABITS-F that targeted the 3′ region of the 18S rRNA gene and reverse primer BABITS-R that targeted the 5′ region of the 28S rRNA (Table 1 and Fig. 1). These primers were also used to sequence the ITS amplicons using the BigDye® Terminator v3.1 Cycle Sequencing Kit and the ABI 3500×L Genetic Analyzer (Applied Biosystems). ITS1, 5.8S rRNA, and ITS2 sequences from B. microti GI and B. duncani WA-1 were aligned using ClustalW. Sequences in the ITS1 regions that were distinct between each species (Fig. 1B) were selected as targets for real-time PCR and ddPCR assays using the primers and probes listed in Table 1. As positive controls, plasmids were constructed that contained the 930–950 bp ITS1/5.8SrRNA/ITS2 amplicons of B. microti GI and B. duncani WA-1 inserted in pUC19. Plasmid concentrations were determined by the Quant-iT™ PicoGreen® assay (Life Technologies) and their equivalences to copies per µl were calculated assuming a molar mass of dsDNA per base pair equal to 660 g per mole per base pair.
Table 1.
Primers and probes used in this study.
| Designation | Target | Sequence (5′–3′) |
|---|---|---|
| Primera | ||
| BABITS-F | 3′ region of the B. microti or B. duncani 18S rRNA gene | GGTGAACCTGCRGAAGGATC |
| BABITS-R | 5′ region of the B. microti or B. duncani 28S rRNA gene | TCTKCCGCTTARTTATATGC |
| BmITS1-F | B. microti ITS1 | TCCCATTTGGGTTACGCTGG |
| BmITS1-R2_mb | B. microti ITS1 | CGTGCAGACAAACCCGCCTT |
| BdITS1-F | B. duncani ITS1 | GCTTCCTAACCCGAGACCAA |
| BdITS1-R | B. duncani ITS1 | CACTGGCGGGGTGAAAAGTA |
| Probeb | ||
| BmITS1-P | B. microti ITS1 | AGTGGCCTTGGACGTAGCGGC |
| BdITS1-P | B. duncani ITS1 | TACGGCCCCTGATGTCGGGG |
Expected amplicon sizes are as follows: BABITS-F/BABITS-R: 930–950 bp; BmITS1-F/BmITS1-R2_mb: 142 bp; BdITS1-F/BdITS1-R: 125 bp.
Probes were dye-labeled with FAM™. PCR Primers were purchased from IDT. Probes were purchased from Biosearch Technologies.
Fig. 1.
Diagnostic regions targeted by real-time and ddPCR assays for B. microti and B. duncani. (A) Representation of eukaryotic nuclear ribosomal RNA. The entire ITS1, 5.8S rRNA, and ITS2 regions of B. microti GI and B. duncani WA-1 were amplified using primers BABITS-F and BABITS-R. (B) Amplicons were sequenced and aligned using ClustalW. Regions in ITS1 that were unique for each species were selected as targets for real-time PCR and ddPCR assays. Regions shown in red in the B. microti sequence indicate binding sites for forward primer BmITS1-F, reverse primer BmITS1-R2_mb, and FAM-labeled probe BmITS1-P. Regions shown in green in the B. duncani sequence indicate binding sites for forward primer BdITS1-F, reverse primer BdITS1-R, and FAM-labeled probe BdITS1-P. Asterisks indicate identical nucleotides between sequences. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. Real-time PCR
Real-time PCR assays were performed in a Bio-Rad CFX96™ System using 25 µl reaction mixtures containing 1 µMof each primer, 250 nM of probe, 1× Bio-Rad SsoFast™ Probe Supermix, and 5 µl of DNA. Cycling conditions for B. microti were 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s and 60 °C for 10 s. Cycling conditions for B. duncani were 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s and 68 °C for 10 s. Standard curves were generated by using serial dilutions of linearized plasmids containing theITS1/5.8S rRNA/ITS2 regions of B. microti and B. duncani. DNA samples and standards were tested in triplicate. The relative fluorescence unit (RFU) baseline threshold was auto-calculated using the Bio- Rad CFX Manager™3.0 Software. Gene copy numbers of Babesia DNA samples were determined by extrapolating the quantification cycle (Cq) values with those from the standard curves.
2.5. ddPCR
Detection and quantitation of Babesia DNA by ddPCR was performed using the same primers and probes used in real-time PCR assays. ddPCR assays were carried out in 20 µl reaction mixtures containing Bio-Rad 1× ddPCR Master Mix, 1.25 µM of each primer, 312 nM of probe, and 5 µl of DNA. Reactions were loaded into eight-well cartridges and droplets were generated using the Bio-Rad QX100™ Droplet Generator. Droplets containing the PCR mixtures were transferred to a single PCR plate for amplification according to recommendations from Bio-Rad but using annealing temperatures of 60 °C for B. microti DNA and 63 °C for B. duncani DNA. Samples were subsequently analyzed with the Bio-Rad QX100™ Droplet Reader. The absolute limit of quantification and absolute limit of detection were determined for each molecular standard on five replicates. DNA samples from Babesia-infected hamsters were tested in triplicate. Samples measuring above the upper limit of detection by ddPCR (>105 copies per reaction)were diluted and retested. Specificities of real-time and ddPCR assays were examined using template DNA from other vector-borne pathogens (Table 4).
Table 4.
Exclusivity panel organisms tested against Babesia real-time PCR and ddPCR assays.
| Organism/straina | Catalog no. | B. microti | B. duncani | ||
|---|---|---|---|---|---|
| RT-PCR | ddPCR | RT-PCR | ddPCR | ||
| Plasmodium knowlesi H strain | BEI MRA-456G | Neg | Neg | Neg | Neg |
| Plasmodium falciparum 7C46 | BEI MRA-172G | Neg | Neg | Neg | Neg |
| Rickettsia prowazekii BREINL | BEI DD-248 | Neg | Neg | Neg | Neg |
| Borrelia burgdorferi B31 | ATCC 35210D-5 | Neg | Neg | Neg | Neg |
| Trypanosoma cruzi SYLVIO X-10 | ATCC 50823D | Neg | Neg | Neg | Neg |
| Babesia microti GI | BEI NR-44070 | ND | ND | Neg | Neg |
| Babesia duncani WA-1 | BEI NR-12311 | Neg | Neg | ND | ND |
B. microti and B. duncani were used at 1 × 107 copies of standard per reaction. The remaining DNA samples were used at 15 ng per reaction. Neg, negative; ND, not done.
2.6. Statistical analysis
Linear regression analyses of standard curves from real-time PCR were performed with the GraphPad Prism 6.0 software. For ddPCR, positive droplets were discriminated from negative droplets by applying a fluorescence amplitude threshold with the QuantaSoft™ analysis software 1.3.2.0 (Bio-Rad). After being exported, the ddPCR data were further analyzed in Microsoft Excel to calculate means and standard deviation measurements.
3. Results
3.1. Sensitivities of Babesia real-time PCR and ddPCR assays
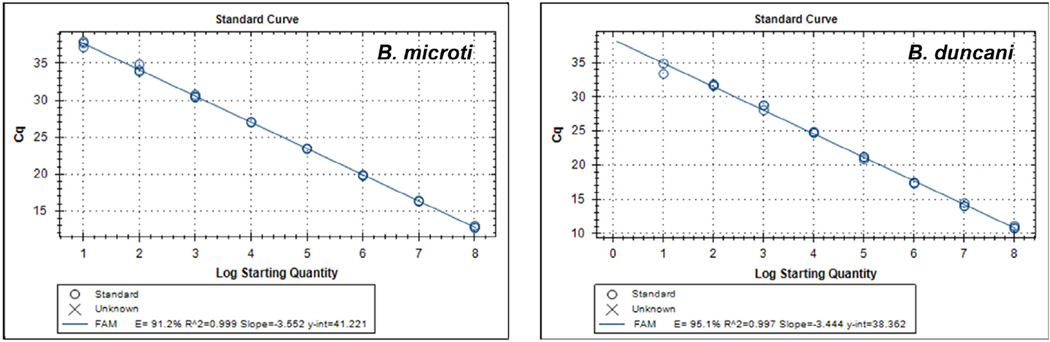
Sensitivities of real-time PCR and ddPCR assays were determined by performing dilution series of positive control plasmids, hereafter referred to as standards, which contained the 930–950 bp ITS1/5.8S rRNA/ITS2 amplicons from B. microti and B. duncani. For real-time PCR, cycle thresholds were calculated for each dilution in triplicate. As shown in Table 2, the sensitivities of detection of Babesia ITS1 DNA by real-time PCR were approximately 10 copies for both B. microti and B. duncani. Plotting the calibration curves based on dilution series of the standards and Cq values resulted in slopes ≥3.44 and R2 values of ≥0.997 for both assays (Fig. 2). Calculated efficiencies were 91.2% and 95.1% for B. microti and B. duncani, respectively.
Table 2.
Real-time PCR detection and quantitation of Babesia DNA standards.
|
B. microti standarda |
Cq |
B. duncani standarda |
Cq | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| 1 × 108 | 12.85 | 0.155 | 1 × 108 | 10.90 | 0.194 |
| 1 × 107 | 16.36 | 0.059 | 1 × 107 | 14.15 | 0.257 |
| 1 × 106 | 19.82 | 0.075 | 1 × 106 | 17.39 | 0.058 |
| 1 × 105 | 23.44 | 0.042 | 1 × 105 | 21.06 | 0.202 |
| 1 × 104 | 27.03 | 0.049 | 1 × 104 | 24.78 | 0.112 |
| 1 × 103 | 30.54 | 0.184 | 1 × 103 | 28.53 | 0.414 |
| 100 | 34.24 | 0.529 | 100 | 31.71 | 0.127 |
| 10 | 37.60 | 0.408 | 10 | 34.13 | 1.057 |
| 1 | 0.0 | 0.0 | 1 | 0.0 | 0.0 |
| H2O | 0.0 | 0.0 | H2O | 0.0 | 0.0 |
Values reflect copy numbers per reaction based on serial dilutions of plasmid DNA standard by DNA concentration and known molecular mass. Data represent the means ± standard deviations of each dilution tested in triplicate from a representative experiment of three performed.
Fig. 2.
Calibration curves of real-time PCR assays run with standards harboring ITS1/5.8S rRNA/ITS2 regions of B. microti (left panel) and B. duncani (right panel). The data correspond to a representative experiment of three performed.
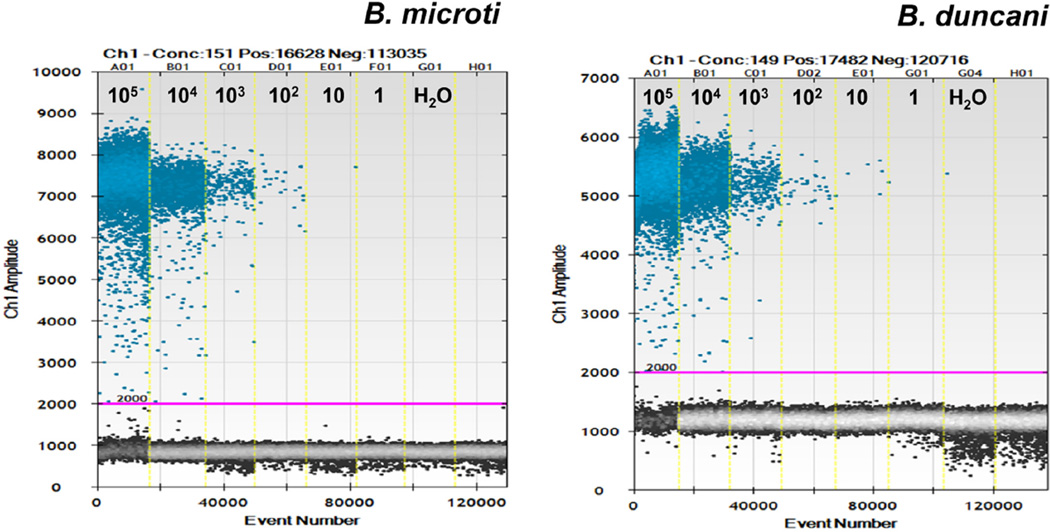
Copy numbers detected by ddPCR of serial dilutions of standards are shown in Table 3. ddPCR detected fewer B. microti and B. duncani ITS1 DNA copies than expected by the calculated standard copy number. However, the sensitivity of ddPCR to detect B. microti or B. duncani ITS1 DNA was similar to that of real time-PCR as both assays obtained positive signals down to 10 copies of diluted standard. A visual representation of positive and negative ddPCR droplet reactions for diluted Babesia DNA standards with minimum threshold for droplet positivity is shown in Fig. 3.
Table 3.
ddPCR detection and quantitation of Babesia DNA standards.
|
B. microti standarda |
ddPCR valueb |
B. duncani standarda |
ddPCR valueb | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| 1 × 105 | 38,200 | 721 | 1 × 105 | 51,733 | 1,858 |
| 1 × 104 | 3,393 | 170 | 1 × 104 | 4,827 | 50 |
| 1 × 103 | 344 | 26.5 | 1 × 103 | 414 | 31.4 |
| 100 | 38.5 | 3.8 | 100 | 44.3 | 3.4 |
| 10 | 1.2 | 0 | 10 | 7.5 | 1.1 |
| 1 | 0 | 0 | 1 | 0 | 0 |
| H2O | 0 | 0 | H2O | 0 | 0 |
Values reflect copy numbers added per reaction based on serial dilutions of plasmid DNA standard by DNA concentration and known molecular mass.
Copies per reaction detected. Data represent the means ± standard deviations of each dilution tested as five replicates from a representative experiment of three performed.
Fig. 3.
A visual representation of positive and negative ddPCR droplet reactions for diluted Babesia DNA standards of B. microti (left panel) and B. duncani (right panel). Minimum threshold for droplet positivity is shown with a pink line. Positive droplets in blue and negative droplets in black are shown for the six diluted linear DNA standards (105 to 1) and water as a negative control (H2O).
3.2. Comparison of real-time PCR and ddPCR using experimentally-infected hamsters
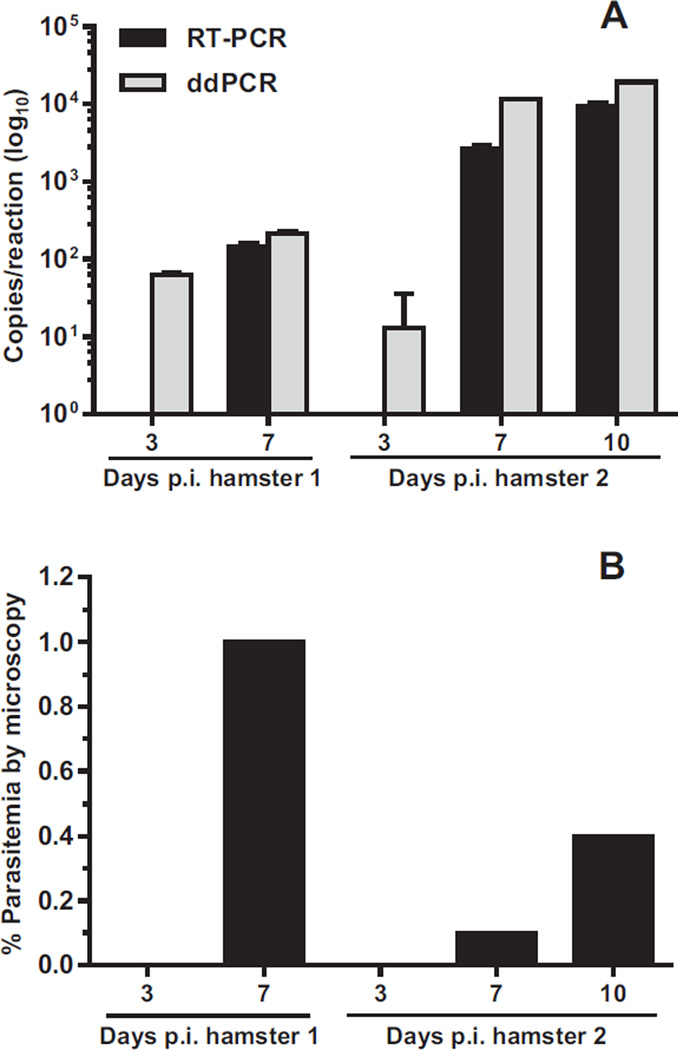
We expanded the utility of these assays to estimate gene copy number in blood samples from experimentally-infected hamsters. Figures 4 and 5, and Appendix: Supplementary Table S1 show the correlations between copy numbers determined by real-time PCR and ddPCR from two hamsters infected with B. microti (Fig. 4A) and two hamsters infected with B. duncani (Fig. 5A). As shown in Fig. 4A, ddPCR was found to be more sensitive than real-time PCR in the detection of B. microti ITS1 DNA at 3 days p.i. in hamsters 1 and 2. Both hamsters were negative for parasitemia by microscopy at this time point (Fig. 4B, 3 days p.i.). At 7 days p.i., increases in copy numbers of B. microti ITS1 DNAwere detected by both assays in hamsters 1 and 2 (Fig. 4A) as parasitemia became detectable by microscopy (Fig. 4B). Levels of B. microti ITS1 DNA at 10 days p.i. were only examined in hamster 2 and showed a continued increase to approximately 9 × 103 and 1.9 × 104 copies per reaction as measured by real-time PCR and ddPCR, respectively (Fig. 4A and Appendix: Supplementary Table S1). In general, measurements of copy numbers at the later time points were two- to fourfold higher by ddPCR compared with real-time PCR (Appendix: Supplementary Table S1). Notably, a correlation between parasite burden determined by microscopy (Fig. 4B) and elevated DNA copy numbers measured by the PCR assays (Fig. 4A) was not observed.
Fig. 4.
Quantification of Babesia microti ITS1 DNA in infected hamsters by real-time PCR (RT-PCR) and ddPCR. (A) DNA copy numbers measured by RT-PCR (black bars) and ddPCR (gray bars) were transformed to log10 values. Non-transformed copy numbers are shown in Appendix: Supplementary Table S1. Data represent the means ± standard deviations of each sample tested in triplicate. Samples that were undetectable by RT-PCR at 3 days p.i. are not shown. (B) Parasitemia of B. microti infected hamsters determined by microscopy.
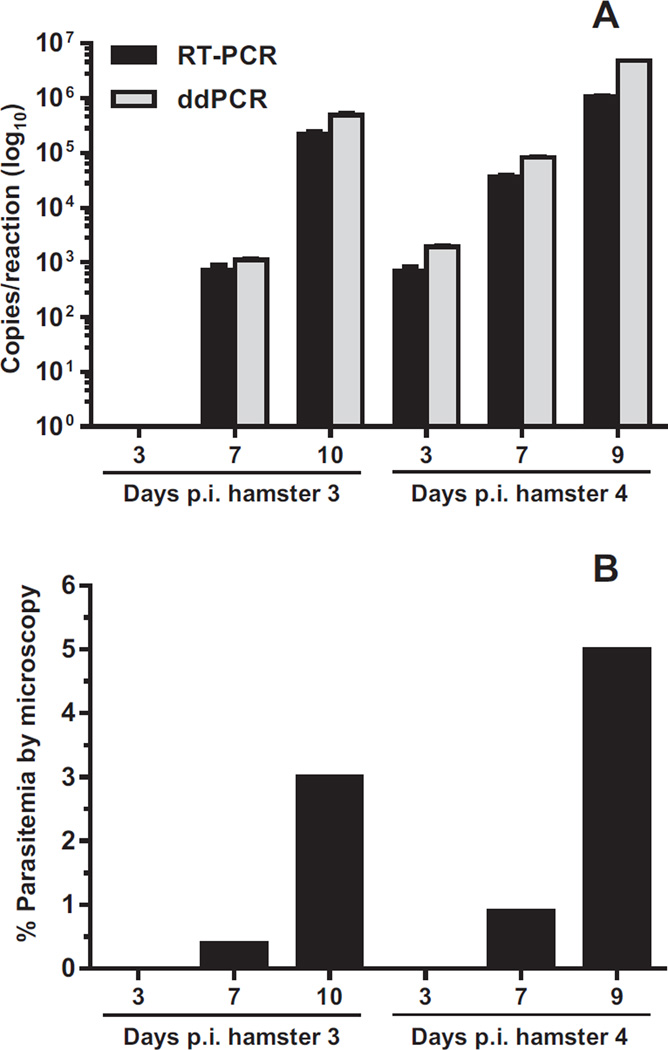
Fig. 5.
Quantification of Babesia duncani ITS1 DNA in infected hamsters by real-time PCR (RT-PCR) and ddPCR. (A) DNA copy numbers measured by RT-PCR (black bars) and ddPCR (gray bars) were transformed to log10 values. Non-transformed copy numbers are shown in Appendix: Supplementary Table S1. Data represent the means ± standard deviations of each sample tested in triplicate. Samples that were undetectable by both assays at 3 days p.i. in hamster 3 are not shown. (B) Parasitemia of B. duncani -infected hamsters determined by microscopy.
Infection of hamsters with B. duncani also resulted in undetectable parasitemia by microscopy at 3 days p.i. (Fig. 5B and Appendix: Supplementary Table S1). However, B. duncani ITS1 DNA at this time point was detected by both real-time PCR and ddPCR in hamster 4 (Fig. 5A, 3 days p.i.). As parasitemia became detectable in both hamsters at 7 days p.i. (Fig. 5B), an increase in B. duncani ITS1 DNA levels measured by real-time PCR and ddPCR was also observed (Fig. 5A). Subsequent blood samples were examined at day 10 p.i. for hamster 3 and day 9 p.i. for hamster 4. As parasitemia by microscopy reached 3% in hamster 3 (Fig. 5B), levels of B. duncani ITS1 DNA measured by real-time PCR and ddPCR continued to increase to approximately 2.2 × 105 and 4.8 × 105 copies per reaction, respectively (Fig. 5A, 10 days p.i. and Appendix: Supplementary Table S1). At 9 days p.i. and a parasitemia of 5%, B. duncani DNA levels in hamster 4 were approximately 1 × 106 copies per reaction by real-time PCR and 4.8 × 106 copies per reaction by ddPCR (Fig. 5A and Appendix: Supplementary Table S1). Similar to the B. microti assays, ddPCR detected two- to fourfold higher copy numbers of B. duncani ITS1 DNA compared with real-time PCR in infected animals. In addition, the extent of parasitemia of B. duncani determined by microscopy correlated minimally with DNA copy number measurements obtained with the PCR assays.
3.3. Specificities of Babesia real-time PCR and ddPCR assays
Specificities of the real-time and ddPCR assays were tested with DNA from five additional vector-borne pathogens: Plasmodium knowlesi, Plasmodium falciparum, Rickettsia prowazekii, Borrelia burgdorferi, and Trypanosoma cruzi (Table 4). Fifteen nanograms of DNA from each organism were used for specificity testing. None of the organisms examined showed cross-reactivity with the Babesia assays. In addition, DNA from B. microti (1 × 107 copies of standard) did not cross-react with the B. duncani assays and vice versa. DNA samples from B. microti strains Peabody (ATCC® PRA-99™) and Nan-Hs-2011 (BEI NR-44071; ATCC® PRA-399™) tested positive in our B. microti ITS1 real-time and ddPCR assays (data not shown). Samples from other Babesia spp. were not available to us at the time of this study.
4. Discussion
Babesiosis was first recognized as a cattle disease of significant economic impact with initial reports of human infections originating in the 1950s. Human babesiosis has been considered an emerging disease in the U.S. in the last 20 years (Hunfeld et al., 2008). A number of factors, such as increased medical awareness, changes in the parasite’s vector ecology, housing development in the countryside, and a growing population of immunocompromised individuals have likely contributed to this emergence. Of note, there has been an increase in Babesia infections transmitted by blood transfusion concurrent with the emergence of human babesiosis in recent years (Bloch et al., 2012; Johnson et al., 2009; Leiby, 2011; Lobo et al., 2013). Current practices used in the prevention of transfusion-transmitted babesiosis include completing a donor questionnaire to identify potential infected individuals (Leiby, 2011; Lobo et al., 2013). Such an approach has many disadvantages as infected blood donors may be asymptomatic or harbor the parasite after symptoms have resolved. The practice may also inadvertently exclude suitable blood donors. Thus, an assay based on serology and/or nucleic acid testing and approved by the FDA will be of great use in blood screening of Babesia.
In the present study, we developed ddPCR assays based on the ITS regions of B. microti and B. duncani that were both sensitive and species-specific. Sensitivities of the assays were comparable with real-time PCR when using the same primers and probes in the amplification of plasmid controls harboring the Babesia ITS sequences. Of note, ddPCR detected fewer DNA copies than the expected standards (Table 3). This may be explained by the fact that the input copy numbers of the standards were low overall. Copy numbers of the standards are calculated prior to amplification and are derived from measuring the number of grams per molecule of the plasmid DNA with a known molecular mass. Discrepancies between the calculated copy numbers of standards and actual detection by ddPCR could be dependent on variations in the measurement of DNA concentration and dilutions of the standards. In ddPCR, the amplification reaction containing the DNA sample, fluorescently-labeled probe, and components is divided into thousands of microscopic reaction droplets, with each containing one or less copies of the target DNA. A limitation of ddPCR as compared with RTPCR is the need to dilute samples with more than 100,000 copies of the target DNA. Overloading the assaymay affect the efficient partitioning of standards into reaction droplets whichmay in turn result in a loss of linearity at high copy numbers.
The ddPCR assays were useful in the detection of parasitemia in hamsters infected with B. microti and B. duncani. Contrary to real-time PCR, the ddPCR assay for B. microti detected parasite infection in 3 day-infected hamsters which were negative by microscopy. Both assays improved detection of parasitemia after 3 days of infection in one of the hamsters infected with B. duncani (Hamster 4 in Fig. 5 and Appendix: Supplementary Table S1). As levels of parasitemia observed by microscopy increased over time in response to Babesia infection, a concomitant increase in copy numbers was detected by the real-time and ddPCR assays. Overall, ddPCR detected two- to fourfold more copy numbers of B. microti and B. duncani ITS DNA compared with real-time PCR. In accordance to previous reports (Wozniak et al., 1996), infection of hamsters with B. duncani resulted in higher levels of parasitemia compared with B. microti, suggestive of a higher virulence potential of the former compared with the latter in animal models. Lastly, the real-time and ddPCR assays were highly specific to B. microti and B. duncani when compared with five additional vector-borne pathogens. Specificity testing against other members of the genus Babesia is pending availability of DNA from these strains.
Importantly, a clear correlation between parasite burden determined by microscopy and DNA copy numbers measured by both PCR assays was not observed in this report. A limitation of our study is that microscopic counts do not take into account RBCs infected with multiple parasites or cells harboring dividing merozoites which may affect the overall DNA copy number detection by the PCR assays. In addition, real-time PCR and ddPCR may detect DNA from extracellular parasites or even free DNA circulating in the blood. These scenarios and the possibility of nuclear-encoded ribosomal RNA genes occurring as multiple copies in the genomes of B. microti and B. duncani could explain the absence of correlation with the microscopy data and even the differences in DNA copy numbers observed between animals. Future research using a larger number of animal subjects accompanied by a more accurate determination of absolute parasitemia expressed as parasites/µl of blood may help determine better trends between parasite burden and copy numbers detected by the molecular PCR assays. Moreover, the addition of an internal amplification control to the reaction mixture targeted by primers unrelated to Babesia may be required to rule out the presence of PCR inhibitors in the DNA samples.
Recently developed quantitative PCR-based methods have shown promise in the diagnosis of Babesia infections in humans (Bloch et al., 2013; Chan et al., 2013; Rollend et al., 2013; Teal et al., 2012). However, many of these methods require further validation prior to becoming standardized assays for blood screening. As opposed to microscopy which can be time consuming and requires proficiency in the detection of parasite morphology, quantitative PCR assays can be useful in high throughput screening, particularly in acute cases of infection when levels of parasitemia are high (Leiby, 2011). On the other hand, such assays including ddPCR may be limited in detecting babesiosis during chronic stages of infection as levels of parasitemia become negligible over time (Krause et al., 1998). The addition of serological tests at this stage to detect increased IgG antibody titers may be necessary to rule out false negative PCR results in chronically infected individuals.
In summary, droplet digital PCR is a novel technology that offers increased sensitivity in the detection and quantitation of nucleic acids from different infectious agents (Hayden et al., 2013; Henrich et al., 2012; Roberts et al., 2013; White et al., 2012). As opposed to other quantitative nucleic acid detection tests, this method does not require the use of molecular standards and provides the absolute quantitation of DNA targets (Hindson et al., 2011; Vogelstein and Kinzler, 1999). We determined the application of ddPCR for protozoan parasites by developing specific tests that are applicable for monitoring parasitemia of B. microti and B. duncani in rodent models of infection. The assays were generally comparable with traditional real-time PCR methods for the detection of Babesia ITS DNA. As digital PCR is adaptable to multiplexing (Zhong et al., 2011), a duplex ddPCR assay is foreseeable to simultaneously detect B. microti and B. duncani using different fluorophores combined with the probes and primers described in this study. The ability to differentiate between both Babesia species may be useful in epidemiological studies in the USA, particularly for B. duncani, to determine the extent of the parasite’s geographic distribution outside the Pacific Coast region (Prince et al., 2010). Pending further validation, the ddPCR methods described herein could be readily adapted for use in the diagnosis of Babesia infections in human specimens and screening of blood bank supplies.
Supplementary Material
HIGHLIGHTS.
This is the first report of ddPCR assays for Babesia microti and B. duncani .
The B. microti assay detected parasitemia as early as 3 days of hamster infection.
The assays were 100% specific when compared with other blood-borne pathogens.
ddPCR may become a useful tool in the diagnosis of Babesia in human blood.
Acknowledgments
This work was supported by the BEI Resources Program (www.beiresources.org) from the National Institute of Allergy and Infectious Diseases (NIH contract # HHSN272201000027C). The views expressed in this publication neither imply review nor endorsement by the Department of Health and Human Services, nor does mention of trade names, commercial practices, or organizations imply endorsement by the U.S. Government. We thank the ATCC Molecular Laboratory Testing Service for assistance with DNA sequencing. We also thank Dr. Andrew Cawthon, Dr. Kurt Langenbach, and Dr. Timothy Stedman for their critical review of the manuscript.
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.exppara.2014.12.003.
References
- Blaschitz M, Narodoslavsky-Gfoller M, Kanzler M, Stanek G, Walochnik J. Babesia species occurring in Austrian Ixodes ricinus ticks. Appl. Environ. Microbiol. 2008;74:4841–4846. doi: 10.1128/AEM.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch EM, Herwaldt BL, Leiby DA, Shaieb A, Herron RM, Chervenak M, et al. The third described case of transfusion-transmitted Babesia duncani. Transfusion. 2012;52:1517–1522. doi: 10.1111/j.1537-2995.2011.03467.x. [DOI] [PubMed] [Google Scholar]
- Bloch EM, Lee TH, Krause PJ, Telford SR, Montalvo L, Chafets D, et al. Development of a real-time polymerase chain reaction assay for sensitive detection and quantitation of Babesia microti infection. Transfusion. 2013;53:2299–2306. doi: 10.1111/trf.12098. [DOI] [PubMed] [Google Scholar]
- Bostrom B, Wolf C, Greene C, Peterson DS. Sequence conservation in the rRNA first internal transcribed spacer region of Babesia gibsoni genotype Asia isolates. Vet. Parasitol. 2008;152:152–157. doi: 10.1016/j.vetpar.2007.11.024. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Babesiosis surveillance – 18 States, 2011. MMWR Morb. Mortal. Wkly Rep. 2012;61:505–509. [PubMed] [Google Scholar]
- Chan K, Marras SA, Parveen N. Sensitive multiplex PCR assay to differentiate Lyme spirochetes and emerging pathogens Anaplasma phagocytophilum and Babesia microti. BMC Microbiol. 2013;13:295–309. doi: 10.1186/1471-2180-13-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad PA, Kjemtrup AM, Carreno RA, Thomford J, Wainwright K, Eberhard M, et al. Description of Babesia duncani n.sp. (Apicomplexa: Babesiidae) from humans and its differentiation from other piroplasms. Int. J. Parasitol. 2006;36:779–789. doi: 10.1016/j.ijpara.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Cullen JM, Levine JF. Pathology of experimental Babesia microti infection in the Syrian hamster. Lab. Anim. Sci. 1987;37:640–643. [PubMed] [Google Scholar]
- Gubernot DM, Nakhasi HL, Mied PA, Asher DM, Epstein JS, Kumar S. Transfusion-transmitted babesiosis in the United States: summary of a workshop. Transfusion. 2009;49:2759–2771. doi: 10.1111/j.1537-2995.2009.02429.x. [DOI] [PubMed] [Google Scholar]
- Hayden RT, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, et al. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 2013;51:540–546. doi: 10.1128/JCM.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J. Virol. Methods. 2012;186:68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia NJ, Belgrader P, Wang S, Koehler R, Regan J, Cosman AM, et al. Droplet Digital PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods. 2013;59:S20–S23. doi: 10.1016/j.ymeth.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Herwaldt BL, Springs FE, Roberts PP, Eberhard ML, Case K, Persing DH, et al. Babesiosis in Wisconsin: a potentially fatal disease. Am. J. Trop. Med. Hyg. 1995;53:146–151. doi: 10.4269/ajtmh.1995.53.146. [DOI] [PubMed] [Google Scholar]
- Herwaldt BL, Kjemtrup AM, Conrad PA, Barnes RC, Wilson M, McCarthy MG, et al. Transfusion-transmitted babesiosis in Washington State: first reported case caused by aWA1-type parasite. J. Infect. Dis. 1997;175:1259–1262. doi: 10.1086/593812. [DOI] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer MJ, Lodes MJ, Reynolds LD, Zhang Y, Douglass JF, McNeill PD, et al. Identification and characterization of putative secreted antigens from Babesia microti. J. Clin. Microbiol. 2003;41:723–729. doi: 10.1128/JCM.41.2.723-729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunfeld KP, Hildebrandt A, Gray JS. Babesiosis: recent insights into an ancient disease. Int. J. Parasitol. 2008;38:1219–1237. doi: 10.1016/j.ijpara.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Johnson ST, Cable RG, Tonnetti L, Spencer B, Rios J, Leiby DA. Seroprevalence of Babesia microti in blood donors from Babesia-endemic areas of the northeastern United States: 2000 through 2007. Transfusion. 2009;49:2574–2582. doi: 10.1111/j.1537-2995.2009.02430.x. [DOI] [PubMed] [Google Scholar]
- Joseph JT, Purtill K, Wong SJ, Munoz J, Teal A, Madison-Antenucci S, et al. Vertical transmission of Babesia microti, United States. Emerg. Infect. Dis. 2012;18:1318–1321. doi: 10.3201/eid1808.110988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogut SJ, Thill CD, Prusinski MA, Lee JH, Backerson PB, Coleman JL, et al. Babesia microti, upstate New York. Emerg. Infect. Dis. 2005;11:476–478. doi: 10.3201/eid1103.040599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause PJ, Telford SR, Ryan R, Conrad PA, Wilson M, Thomford JW, et al. Diagnosis of babesiosis: evaluation of a serologic test for the detection of Babesia microti antibody. J. Infect. Dis. 1994;169:923–926. doi: 10.1093/infdis/169.4.923. [DOI] [PubMed] [Google Scholar]
- Krause PJ, Spielman A, Telford SR, Sikand VK, McKay K, Christianson D, et al. Persistent parasitemia after acute babesiosis. N. Engl. J. Med. 1998;339:160–165. doi: 10.1056/NEJM199807163390304. [DOI] [PubMed] [Google Scholar]
- Krause PJ, Gewurz BE, Hill D, Marty FM, Vannier E, Foppa IM, et al. Persistent and relapsing babesiosis in immunocompromised patients. Clin. Infect. Dis. 2008;46:370–376. doi: 10.1086/525852. [DOI] [PubMed] [Google Scholar]
- Leiby DA. Transfusion-transmitted Babesia spp.: bull’s-eye on Babesia microti. Clin. Microbiol. Rev. 2011;24:14–28. doi: 10.1128/CMR.00022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yin H, Liu G, Guan G, Ma M, Liu A, et al. Discrimination of Babesia major and Babesia ovata based on ITS1-5.8S-ITS2 region sequences of rRNA gene. Parasitol. Res. 2008;102:709–713. doi: 10.1007/s00436-007-0818-y. [DOI] [PubMed] [Google Scholar]
- Lobo CA, Cursino-Santos JR, Alhassan A, Rodrigues M. Babesia : an emerging infectious threat in transfusion medicine. PLoS Pathog. 2013;9:e1003387. doi: 10.1371/journal.ppat.1003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Terkawi MA, Jia H, Aboge GO, Goo YK, Cao S, et al. A double antibody sandwich enzyme-linked immunosorbent assay for detection of secreted antigen 1 of Babesia microti using hamster model. Exp. Parasitol. 2011;130:178–182. doi: 10.1016/j.exppara.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Morisset D, Stebih D, Milavec M, Gruden K, Zel J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS ONE. 2013;8:e62583. doi: 10.1371/journal.pone.0062583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Komatsu J, Matsuura S, Takashima K, Katsura S, Mizuno A. Single-molecule PCR using water-in-oil emulsion. J. Biotechnol. 2003;102:117–124. doi: 10.1016/s0168-1656(03)00023-3. [DOI] [PubMed] [Google Scholar]
- Niu Q, Luo J, Guan G, Liu Z, Ma M, Liu A, et al. Differentiation of two ovine Babesia based on the ribosomal DNA internal transcribed spacer (ITS) sequences. Exp. Parasitol. 2009;121:64–68. doi: 10.1016/j.exppara.2008.09.021. [DOI] [PubMed] [Google Scholar]
- Oz HS, Hughes WT. Acute fulminating babesiosis in hamsters infected with Babesia microti. Int. J. Parasitol. 1996;26:667–670. doi: 10.1016/0020-7519(96)00022-7. [DOI] [PubMed] [Google Scholar]
- Persing DH, Mathiesen D, Marshall WF, Telford SR, Spielman A, Thomford JW, et al. Detection of Babesia microti by polymerase chain reaction. J. Clin. Microbiol. 1992;30:2097–2103. doi: 10.1128/jcm.30.8.2097-2103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persing DH, Herwaldt BL, Glaser C, Lane RS, Thomford JW, Mathiesen D, et al. Infection with a babesia-like organism in northern California. N. Engl. J. Med. 1995;332:298–303. doi: 10.1056/NEJM199502023320504. [DOI] [PubMed] [Google Scholar]
- Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012;84:1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince HE, Lape-Nixon M, Patel H, Yeh C. Comparison of the Babesia duncani (WA1) IgG detection rates among clinical sera submitted to a reference laboratory for WA1 IgG testing and blood donor specimens from diverse geographic areas of the United States. Clin. Vaccine Immunol. 2010;17:1729–1733. doi: 10.1128/CVI.00256-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick RE, Herwaldt BL, Thomford JW, Garnett ME, Eberhard ML, Wilson M, et al. Babesiosis in Washington State: a new species of Babesia. Ann. Intern. Med. 1993;119:284–290. doi: 10.7326/0003-4819-119-4-199308150-00006. [DOI] [PubMed] [Google Scholar]
- Roberts CH, Last A, Molina-Gonzalez S, Cassama E, Butcher R, Nabicassa M, et al. Development and evaluation of a next-generation digital PCR diagnostic assay for ocular Chlamydia trachomatis infections. J. Clin. Microbiol. 2013;51:2195–2203. doi: 10.1128/JCM.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollend L, Bent SJ, Krause PJ, Usmani-Brown S, Steeves TK, States SL, et al. Quantitative PCR for detection of Babesia microti in Ixodes scapularis ticks and in human blood. Vector Borne Zoonotic Dis. 2013;13:784–790. doi: 10.1089/vbz.2011.0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty S, Khalil Z, Schori P, Azar M, Ferrieri P. Babesiosis. Two atypical cases from Minnesota and a review. Am. J. Clin. Pathol. 2003;120:554–559. doi: 10.1309/N3DP-9MFP-NUJD-4XJY. [DOI] [PubMed] [Google Scholar]
- Teal AE, Habura A, Ennis J, Keithly JS, Madison-Antenucci S. A new real-time PCR assay for improved detection of the parasite Babesia microti. J. Clin. Microbiol. 2012;50:903–908. doi: 10.1128/JCM.05848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Digital PCR. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, Quake SR, Curr K. Digital PCR provides absolute quantitation of viral load for an occult RNA virus. J. Virol. Methods. 2012;179:45–50. doi: 10.1016/j.jviromet.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Wozniak EJ, Lowenstine LJ, Hemmer R, Robinson T, Conrad PA. Comparative pathogenesis of humanWA1 and Babesia microti isolates in a Syrian hamster model. Lab. Anim. Sci. 1996;46:507–515. [PubMed] [Google Scholar]
- Zhong Q, Bhattacharya S, Kotsopoulos S, Olson J, Taly V, Griffiths AD, et al. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip. 2011;11:2167–2174. doi: 10.1039/c1lc20126c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.