Abstract
Purpose
To develop a technique for rapid collection of CEST images with the saturation varied to modulate signal loss transfer and enhance contrast.
Theory and Methods
MeLOVARS divides the saturation pulse of length Tsat into N = 3-8 submodules, each consisting of a saturation pulse with length of Tsat/N (~0.3-1 s), one or more low flip-angle gradient-echo readout(s) and a flip back pulse. This results in N readouts with increasing saturation time from Tsat/N to Tsat without extra scan time.
Results
For phantoms, 8 images with Tsat incremented every 0.5s from 0.5-4 s were collected simultaneously using MeLOVARS, which allows rapid determination of exchange rates for agent protons. For live mice bearing glioblastomas, the Z-spectra for 5 different Tsat values from 0.5-2.5 s were acquired in a time normally used for one Tsat. With the additional Tsat-dependence information, LOVARS phase maps were produced with a more clearly defined tumor boundary and an estimated 4.3-fold enhanced CNR. We also show that enhancing CNR is achievable by simply averaging the collected images or transforming them using the Principal Component Analysis (PCA).
Conclusion
MeLOVARS enables collection of multiple saturation-time-weighted images without extra time, producing a LOVARS phase map with increased CNR.
Introduction
Chemical Exchange Saturation Transfer (CEST) imaging is an emerging technology based on the following unique characteristics: 1) the ability to detect signals from low concentration diamagnetic compounds based on selective saturation of rapidly exchanging spins, and 2) the capability of detecting changes in environmental parameters in vivo, including: pH, temperature and metal ion concentration (1-5). There have been a number of preclinical (3,6-13) and clinical applications (12,14) involving the detection of either administered CEST agents or endogenous molecules and metabolites. Upon injection of glucose, glutamate, CT agents and nanocarriers (15-18), CEST imaging has been applied for characterizing tumor vasculature, metabolism, extracellular pH, and nanocarrier uptake.
Tumors also display contrast without administering agents, an effect that has been attributed predominantly to the amide protons of extra soluble peptides/proteins found in brain tumors which resonate ~3.5 ppm from water, known as amide proton transfer (APT) imaging (8,13,14,19). The APT signal has been shown to correlate with the histopathological grade for brain tumors in patients on clinical 3T scanners (14,20,21), and to be a marker for differentiating tumor recurrence from radiation necrosis (8). Other applications include lung (22), breast (23,24) and prostate cancer (25) imaging. Furthermore, APT contrast has been exploited to determine tumor response to various therapeutic methods such as chemotherapy (23,26), radiation (8) and HIFU (27).
Despite the potential for tumor imaging, there are obstacles towards widespread application of CEST, including the low effective contrast (a few percent of the water signal), sensitivity to B0 field inhomogeneities, and susceptibility to interference from other sources of contrast (28). A typical scheme for a CEST pulse sequence is shown in Fig. 1a. Before the water signal readout, a long frequency-selective continuous wave (CW) pulse or pulse train is applied at the resonance frequency of the agent to prepare the magnetization. The saturation preparation is usually on the order of seconds to obtain sufficient amplification of the solute signal.
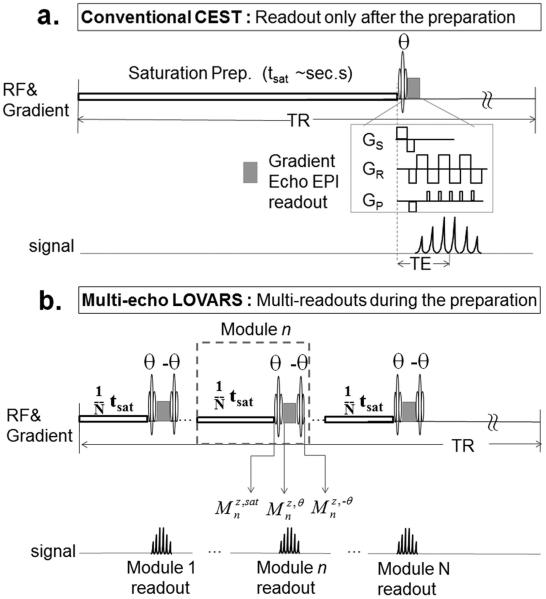
Figure 1.
Acquisition schemes for a. conventional CW CEST with single readout after the saturation preparation and b. MeLOVARS consisting of N saturation modules with readouts
The most common method to detect and quantify CEST contrast is by calculating the asymmetry in the magnetization transfer ratio (MTRasym) at the frequency of the exchangeable protons (Δω): MTRasym=[S(−Δω)–S(+Δω)]/S0 which is the subtraction of the two water signal intensities with saturation pulses at +Δω and −Δω with respect to water, S(+Δω) and S(−Δω), normalized by the signal without saturation (S0). However, for most in vivo data, the MTRasym value is not purely CEST contrast, but also includes interference from other sources of water signal loss generated by the saturation pulse, including conventional magnetization transfer contrast (MTC), direct saturation (DS) and relayed nuclear Overhauser enhancement (NOE) transfer (29,30). In addition, most endogenous CEST agents resonate very close to water (1-4 ppm) (29), resulting in low contrast-to-noise ratio (CNR) and specificity.
Because of these challenges, new CEST methods are needed for improving CNR and specificity or reducing image acquisition times. We previously proposed a strategy based on acquiring multiple saturation transfer weighted (STw) images with different saturation lengths (Tsat), termed Length and Offset VARied Saturation (LOVARS) (31). We propose here a new Multi-echo LOVARS (MeLOVARS) method, which demonstrates the feasibility of collecting all of the saturation length images within one TR through placement of multiple water signal readouts during the saturation preparation instead of at the end as in conventional CEST imaging. This scheme is based on the idea of Look-Locker fast T1 mapping (32,33), with the concept demonstrated using CW saturation, but also readily applicable to the pulse-train saturation modules typically used on clinical scanners by employing the same readout strategy. In addition, we show this multi-echo strategy can be used in vitro to speed up measurements of the exchange properties of CEST compounds.
Methods
MeLOVARS Design
Instead of employing a single long saturation module of length tsat before readout (Fig. 1a), the MeLOVARS method divides the saturation preparation into N = 3-8 sub-modules, each with a length of tsat/N (~0.3 s - 1 s) and interleaves a low flip-angle (FA = θ) fast gradient echo read-out sequence followed by a flip back pulse (FA = −θ) for retaining longitudinal magnetization (Fig. 1b). Thus, multiple (N) readouts are achieved during the preparation, each with an increased effective saturation time. For gradient echo sequences, after the excitation pulse the longitudinal and transverse magnetizations for the nth module become:
| [1] |
where is the FID signal for reconstructing the nth image. After the readout module, the transverse magnetization decays to . Upon application of a flip-back pulse (FA = −θ), the longitudinal magnetization is:
| [2] |
Simulations
To determine initial estimates on the imaging parameters, we numerically solved the 2-pool Bloch equations for N iterations of the MeLOVARS module (Fig. 1b). A decay factor (DF) for the signal was calculated by subtracting MTRasym for MeLOVARS from MTRasym using a single readout with the same Tsat, and normalizing by MTRasym for single readout. The parameters for the water pool were: T1w = 3.55 s, T2*w = 1.11 s, concentration = 111.2 M; for the solute pool: T1S = 1.41 s, T2*S= 0.025 s, ksw = 660 s−1, Δω = 9.3 ppm, concentration = 25 mM. These parameters were also used to perform the QUEST fits for the phantoms.
To simulate the CEST contrast produced by MeLOVARS on mice bearing glioblastomas and to optimize θ and N, we numerically solved the 4-pool Bloch equations including a semi-solid pool (ss), amide pool (am), aliphatic pool (al) and water pool with Δω = 0 ppm, 3.5 ppm, −3.7 ppm and 0 ppm, respectively (28,34). Each module of MeLOVARS was simulated as the pulse-sequence described in Fig. 1b, including a Sat. pulse, an excitation pulse with flip angle = θ, a decay factor (DF) during readout, and a flip-back pulse of angle = −θ. The initial guess of parameters were set as reported previously (28,34-36) e.g. T1w between 1.8 s to 2.5 s and T1 for the remaining 3 pools set to be = T1w, and T2*w between 20 ms to 50 ms, T2*am and T2*al between 1 ms to 10 ms, and T2*ss between 1 μs and 10 μs with taking account of in vivo line-broadening. The simulations used the Levenberg-Marquardt algorithm to fit the experimental MeLOVARS Z-spectra for ROI’s enclosing tumor and contralateral white matter (WM), with the assumption that only the concentration changes for these pools between tumor and WM. Noted that for consistency, we use T2* instead of T2 for the Bloch simulations, without including the B0-inhomogeneity and susceptibility effects. With these fit parameters, we then simulated how the 5 MTRasym values change as a function of N and θ for both tumor and contralateral tissue to maximize the difference for in vivo experiments. All simulations were performed using python scripts written in-house.
In vitro Phantom Experiments
To evaluate the MeLOVARS sequence, a phantom was prepared consisting of four 5mm NMR tubes, with one filled with 0.01 M phosphate-buffered saline (PBS) as the negative control, and the other three each filled with a CEST agent at a concentration of 25 mM in PBS. The three agents were: 1) D-Glucose (Δω = 0.9-1.5 ppm, pH 7.4) (16), 2) Salicylic Acid (Δω = 9.3 ppm, pH 7.1) (37), 3) 5-Chloro-2-(methyl-sulfonamido) benzoic acid (Δω = 7.2 ppm, pH 7.1) (38), with all agents titrated using NaOH and HCl. All in vitro MR scans were acquired on a Bruker vertical 750MHz scanner at a temperature of 310K.
A 2-shot EPI readout scheme was used with TR/TE = 8 s / 5.25 ms, EPI module time = 7.5ms and Matrix Size = 32×32. Z-spectra were acquired using a CW saturation pulse with B1 = 2.4, 3.6, and 4.8 μT and the saturation offset incremented 0.3 ppm for −9.9 ppm −> −6.9 ppm, −2.7 ppm −> 2.7 ppm, and 6.9 ppm −> 9.9 ppm and 0.6 ppm increment for −6.9 −> −2.7 ppm and 2.7 ppm −> 6.9 ppm.
In vivo Animal Studies
SCID/NCR mice (n=5) were xenografted intracranially with 100,000 human glioblastoma stem-like neurosphere cells derived from patients (HSR-GBM1A) (39,40) with MR imaging performed 6 weeks post-injection. Immediately following the final MRI, mice were perfused with 4% paraformaldehyde (PFA). Their brains were removed, cryosectioned (25 μm thick) and stained using hematoxylin and eosin (H&E). MR images were acquired on a Bruker Biospec 11.7T scanner, using a 72mm body coil for transmission and a 4-channel phase-array surface coil for reception. The MeLOVARS parameters were: N=5 for 3 mice with each saturation pulse length of 0.5 s (1/5Tsat) and 6 segment EPI (6.4 ms per segment). The other parameters were: θ = 25°, TR/TE = 4s/4.3ms, FOV = 16.5x15.8x1mm, matrix size = 96×64, saturation offsets = [±4.8, ±4.2, ±3.9, ±3.6, ±3.3, ±3, ±2.4, ±1.5, ±0.6, ±0.3, 0] ppm and B1 was set to 1.2 μT, 1.9 μT and 3 μT. For comparison, conventional CEST images were also acquired using the same EPI readout as MeLOVARS with Tsat = 2.4 s. For MeLOVARS, the Z-spectra acquisition time is 8 min 48 sec, plus an additional 80 sec for the WASSR image-set for B0 mapping and corrections, resulting in ~ 10 min of scanning. Apart from CEST, diffusion-weighted images of the same slice were acquired with three b values of 500, 1000, 1500 for generating apparent diffusion coefficient (ADC) map. Multi-slice STw images (-6ppm) with a matrix size of 128×96 and half slice thickness of 0.5mm were also collected, for checking the partial volume effects of CEST images.
Post-processing
All data were processed using custom-written MATLAB scripts. For both phantom and in vivo studies, a voxel-by-voxel Z-spectra B0 correction was performed through interpolating the original data to every 0.1ppm using a piecewise polynomial fitting, with B0 values from WASSR. CEST contrast was quantified using MTRasym for module n in MeLOVARS, , with being the image with same FA readout without saturation pulse. For in vitro measurements of glucose with ksw > 1,000 s−1 and three different proton types, we calculated an average MTRasym over three frequencies [0.9ppm, 1.2ppm, 1.5ppm], similar to previous studies (15,16). For other agents with a single type, MTRasym is calculated at the peak frequency of the CEST contrast. For in vivo measurements, the contrast maps for amide (-NH, APT weighted) were obtained by averaging MTRasym from 3.3 to 3.9ppm.
The LOVARS phase map was generated using 2 LOVARS units (4 min 48 s), based on averaging the Module 1 and Module 2 images to produce the Tsat,1 images, and averaging the Module 4 and Module 5 images to produce the Tsat,2 images, for both +Δω and −Δω. All other processing was as described previously (31).
The Principal Component Analysis (PC1) map was generated from the 5 MTRasym images with different Tsat using the Matlab function “pca”. The resulting PC1 map preserved the most significant contrast, and was displayed with a scale factor of ½.
RESULTS
To elucidate how the MeLOVARS signal using multiple readouts compares to the CEST signal with the same Tsat and a single readout, we plotted a contour surface displaying a decay factor (DF) of 10% (Fig. 2a), based on 2-pool Bloch simulations. This can be used to select the appropriate θ and n based on T2* for the volume of interest and the TE’s attainable on the scanner. Fig. 2b shows the DF and as a function of TE/T2* and θ, with N = 8, 5 and 3, which we used in the in vitro and in vivo experiments. As seen, to ensure a DF < 10% with θ ~35°, TE/T2* < 0.07 is required for N = 8, TE/T2* < 0.125 for N = 5, TE/T2* < 0.295 for N = 3.
Figure 2.
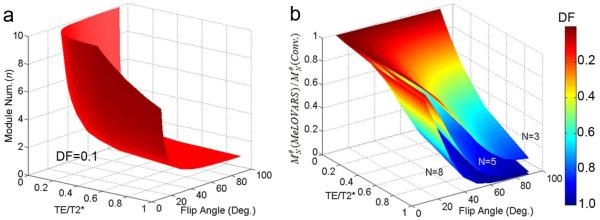
a. Contour surface for the nth MeLOVARS Module readout with a Decay Factor (DF) = 10% comparing the signal using a single readout of same Tsat, to guide the choice of measurement parameters: Flip Angle, Num. of Modules based on TE/T2*. b. Simulations of DF and as a function of TE/T2*, θ with N equal to 3, 5, 8, which we used in the phantom and in vivo experiments.
Phantom experiments
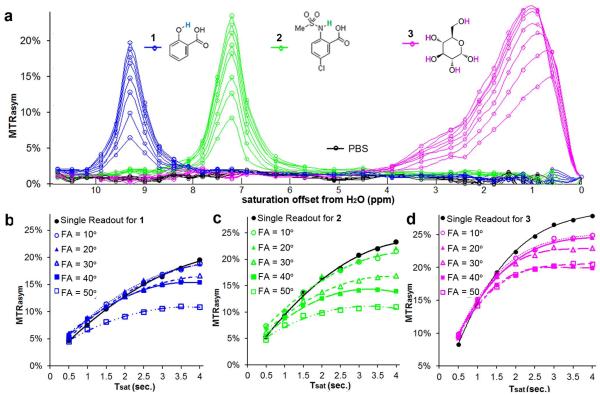
We performed a phantom study to determine whether the MeLOVARS acquisition scheme enables more rapid quantification of exchange rates (ksw) using the QUEST method, and how the data compares with a single module. Z-spectra were collected with number of modules (N) = 8 (from 0.5s to 4s with every 0.5s increment) and fit allowing 8X acceleration over the conventional 8 single readouts. Fig. 3a shows the build-up of MTRasym for three representative diaCEST agents: glucose, 5-Chloro-2-(methyl-sulfonamido) benzoic acid, and salicylic acid (SA) which are spaced over the range of ?ω values currently accessible for diaCEST agents. A negative control (PBS) is also shown. We further compared the conventional method (θ = 20°) build-up curves with MeLOVARS using θ = 10° to 50°. For SA and 5-Chloro-2-(methyl-sulfonamido) benzoic acid, when θ < 30° the QUEST curves are comparable to using a single readout (Fig. 3b&c). For glucose, even for θ = 10 ° and 20°, the MeLOVARS curve deviates substantially from the conventional method for N > 4 (Fig. 3d). This is presumably due to a ~50% reduction of T2 for glucose (T2 < 200 ms, estimated by the Swift-Connick equation (41)), leading to a DF > 10% at N = 8, with T2* dropping to ~100 ms for this phantom of multiple quartz tubes at 17.6 T. There is also a more pronounced spill-over for glucose with its smaller Δω, which reduces MTRasym. For the other two contrast agents, at 25 mM the T2 values are similar to that of PBS and as a result the T2 decay is not as prominent as glucose. It’s reasonable to assume that there is a large error for θ ≥ 40°, as indicated by Fig. 2, keeping DF < 10% requires TE/T2* < 0.03, i.e. T2* > 240 ms which is difficult to guarantee due to the ultra-high B0 field and resulting field inhomogeneity (36).
Figure 3.
Phantom Experiments for 3 CEST agents with different Δω and exchange rates. a. Z-spectra acquired only using 1/8 time of that for conventional method. b. QUEST dataset for SA (1) with different FA comparing with a single readout. For compound 1, the fits were performed assuming a single readout with QUEST determined rates: ksw_single = 620 s−1, ksw_θ=10° = 630 s−1, ksw_ θ= 20° = 660 s−1, ksw_θ=30° = 520 s−1 (above 15% error). c. QUEST dataset for 5-Chloro-2-(methyl-sulfonamido) benzoic acid (2) with different FA compared with a single readout. For compound 2, QUEST determined rates: ksw_single = 940 s−1, ksw_θ=10° = 980 s−1, ksw _θ= 20° = 940 s−1, ksw _θ= 30° = 1800 s−1 (above 15% error). d. QUEST dataset for glucose (3) with different FA compared with a single readout, showing a pronounced contrast decay when n>5, even for FA = 10°.
In vivo Imaging of mice brain
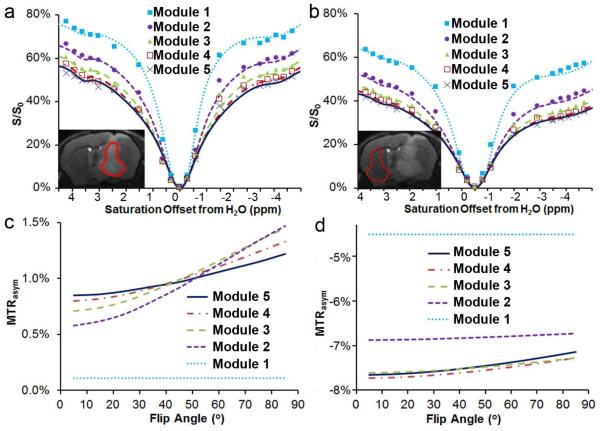
We then acquired MeLOVARS data on mice bearing glioblastomas, with 5 modules each 0.5 s long (Fig. 4). The saturation pulse perturbs the magnetization of solute protons, semi-solid protons and water protons in vivo, and as a result the Z-spectra are influenced by CEST, conventional MT, and also NOE-relayed transfers from aliphatic protons. We were able to fit the 5 Z-spectra collected on both tumor (Fig. 4a) and contralateral WM (Fig. 4b) tissue to a 4-pool Bloch equation model. Based on these parameters, we simulated how the multiple MTRasym data collected in MeLOVARS changes as a function of flip angle (τ). As is shown in Fig. 4c, for the tumor tissue which has a longer water T2, higher concentration of exchangeable protons and smaller concentration of semi-solid and aliphatic protons, the MTRasym value increases from Module 1 to Module 5 when θ < 35°. However, for WM, the MTRasym value is largest for Module 1 and drops for the remaining Modules. Also, for both tumor and WM, Module 1 is constant as a function of θ because there is no extra T2* decay.
Figure 4.
In vivo experimental Z-spectra (symbols) and 4-pool Bloch equation simulations using MeLOVARS (lines) of tumor (a) and contralateral WM (b), with the experimental DF reaching ~10 % for θ = 25°, N = 5 compared to the conventional acquisition method with the same tsat ; Plot of simulations using the same parameters to determine how MTRasym changes as a function of FA (θ) for tumor (c) and contraleral WM (d). The fit parameters were: bulk water pool: T1_tumor = 2.35 s, T1_WM = 2.0 s, T2_tumor = 35 ms, T2_WM = 26 ms; macromolecular pool: T2 = 0.0078 ms, ksw = 159 s−1, conc._tumor = 4598 mM, conc._WM = 9800 mM; exchangeable amide pool: T2 = 6 ms, ksw = 20 s−1, conc._tumor = 629 mM, conc._WM = 290 mM; and aliphatic pool: T2 = 2.7 ms, ksw=10.5 s−1, conc._tumor = 1100 mM, conc._WM = 2900 mM. The T1 values for the other 3 pools were set to be the same as bulk water.
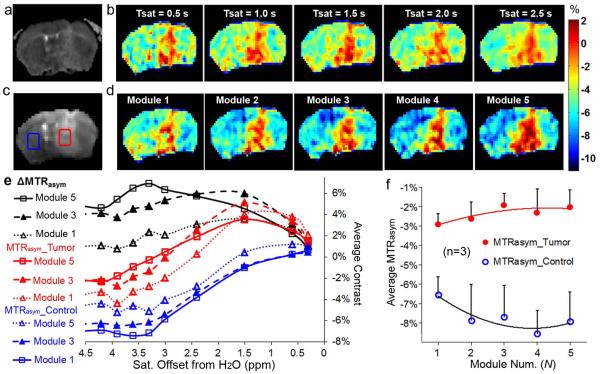
Fig. 5 further illustrates the performance of MeLOVARS with N=5, and 0.5s long modules. The ADC map created using an EPI readout (Fig. 5a) shows the tumor as hyperintense as previously reported for GBM (42). For comparison, Fig. 5b displays the 5 MTRasym maps acquired using the conventional CEST method, for Tsat = 0.5 s, 1 s, 1.5 s, 2 s and 2.5 s respectively. The STw image at +3.6 ppm from the 5th Module readout using MeLOVARS is shown in Fig. 5c, with the same B1 as that in Fig. 5b. Fig. 5d displays the 5 MTRasym maps from MeLOVARS (n =1,2,3,4,5), which only used 1/5 of acquisition time of the conventional method. Note that the tumor rim is highlighted in Module 1 as was seen previously for other tumors (31). In addition, the contrast between tumor and contralateral tissue increases for MeLOVARS when N > 1 because of the differences in T2* for these tissues. As MeLOVARS enables acquisition of 5 MTRasym spectra simultaneously, we further display the build-up of the MTRasym for both tumor and contralateral WM and the metric ΔMTRasym obtained by taking the subtraction of MTRasym_tumor and MTRasym_WM (Fig. 5e&f). As can be seen, in the MTRasym_tumor increases in the region from 3 – 4 ppm from Module 1 to Module 5 while MTRasym_WM decreases, resulting in a more significant increase in ΔMTRasym for MeLOVARS. Fig. 5f further highlights the MTRasym dependence on Tsat at frequencies between 3.3 – 3.9 ppm for n = 3 mice.
Figure 5.
MeLOVARS performance in a mouse bearing glioblastoma. a. ADC map of the diffusion-weighted image based on EPI readout; b. the conventional MTRasym maps (B1 = 1.9 uT) with Tsat = 0.5s, 1s, 1.5s, 2s and 2.5s respectively, which requires 5×’s the scanner time. c. STw image at +Δω from the 5th Module readout in MeLOVARS scheme; d. MeLOVARS MTRasym maps (B1 = 1.9 μT) for 5 Modules respectively; Note that in both MeLOVARS echo1 in b and Tsat = 0.5s in d) only the rim of the tumor is enhanced, but not in a. e. The MTRasym build-up for tumor core and for contralateral control region, ΔMTRasym were obtained by taking the subtraction of MTRasym for Tumor core (red ROI in c) and MTRasym for the contralateral WM (blue ROI in c). f. MTRasym changes as a function of Module Num. for tumor and contralateral tissue is different, based on the data for n = 3 mice.
We also quantitatively compared the MTRasym and the CNR values for the CEST images collected using conventional or MeLOVARS methods on n = 3 mice (Table 1). The first 3 rows compare the averaged MTRasym_Tumor, MTRasym_Ctrl and ΔMTRasym of the 3 mice and their standard deviations. As is seen, the averaged values of MTRasym for tumor tissue are very similar between the two methods for Tsat = 1 s or larger, whereas these values determined using MeLOVARS in WM are consistently lower than measured using conventional methods. This is presumably due to the shorter T2* of WM and larger decay factor. This leads to an increase in contrast from 3.6% for Module 1 to ~6% by Modules 4&5 for MeLOVARS. The same saturation times show a more constant ΔMTRasym using conventional methods. Furthermore, we evaluated how the SNR of the MeLOVARS images changes compared to conventional images. There is a slight decrease of SNR for the MeLOVARS Module 2 and higher images due to T2* decay from the readouts, however because the contrast between tumor and control tissue (ΔMTRasym) increases the resulting CNR is nearly the same.
Table 1.
MTRasym Contrast and CNR comparison for Conventional and MeLOVARS at 11.7 T
| (n=3) | Tsat = 0.5s (Module 1) |
Tsat = 1s (Module 2) |
Tsat = 1.5s (Module 3) |
Tsat = 2s (Module 4) |
Tsat = 2.5s (Module 5) |
|
|---|---|---|---|---|---|---|
| MTRasym_
Tumor (%) |
Conventional MeLOVARS |
−0.9 ± 0.3 −2.4 ± 0.6 |
−1.8 ± 1.0 −2.2 ± 0.9 |
−1.4 ± 0.4 −1.4 ± 0.6 |
−2.0 ± 0.7 −1.8 ± 1.2 |
−2.1 ± 0.5 −1.6 ± 0.9 |
| MTRasym_
Ctrl. (%) |
Conventional MeLOVARS |
−4.9 ± 0.2 −6.0 ± 0.9 |
−6.0 ± 1.2 −7.4 ± 1.8 |
−6.0 ± 1.0 −7.2 ± 1.6 |
−5.9 ± 1.1 −8.1 ± 1.2 |
−5.8 ± 0.6 −7.4 ± 1.5 |
| ΔMTRasym
(%) |
Conventional MeLOVARS |
4.0 ± 0.1 3.6 ±0.8 |
4.2 ± 0.2 5.2 ± 1.6 |
4.5 ± 0.6 5.8 ± 1.5 |
3.9 ± 0.5 6.2 ± 1.2 |
3.6 ± 0.0 5.9 ± 1.8 |
| aSNR_S0 | Conventional MeLOVARS |
74 ± 2.5 79 ± 1.0 |
74 ± 2.5 73 ± 7.6 |
74 ± 2.5 62 ± 8.6 |
74 ± 2.5 58 ± 10 |
74 ± 2.5 55 ± 8.4 |
|
bCNR_ ΔMTRasym |
Conventional MeLOVARS |
3.6 ± 0.2 3.5 ± 0.7 |
3.7 ± 0.0 4.7 ± 1.4 |
4.1 ± 0.4 4.3 ± 0.7 |
3.5 ± 0.3 4.4 ± 1.0 |
3.3 ± 0.1 4.0 ± 1.3 |
The SNR for S0 in nth module in MeLOVARS was calculated using , with the noise level σn calculated by times of standard deviation for a control ROI on the subtraction of two consecutively-acquired images.
The CNR of the MTRasym maps for the nth module was calculated using
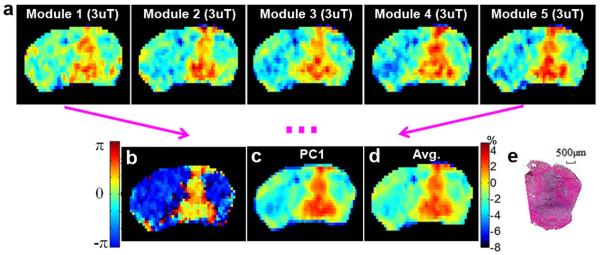
The images from the multiple modules of MeLOVARS can be combined and analyzed in different ways for improving image CNR (Fig. 6). Apart from directly generating 5 MTRasym maps from each module (Fig. 6a, B1 = 3 uT), a LOVARS phase map (Fig.6b) was produced from the two repetitions of the MeLOVARS acquisition at +Δω and −Δω, showing a more clearly defined tumor boundary and enhanced CNR. Based on Table 1 which shows that the images for modules 2-5 have comparable CNR with conventionally acquired images, the displayed phase map from 2 LOVARS units with each image averaging 2 modules achieves a 4.3-fold CNR enhancement (Table 2 in (31)) compared with conventionally acquired MTRasym maps with the same readout. Alternatively, a PCA contrast map can be generated (Fig. 6c) which retains the same MTRasym contrast scale instead of transforming to phase, and displays a higher CNR than the individual images. Also, simple averaging of all 5 MTRasym maps can be performed (Fig. 6d), with ~√5 times increase in CNR over the single module image. The tumor shape was further confirmed through H&E stain (Fig. 6e).
Figure 6.
a. MeLOVARS MTRasym maps with B1 = 3μT for 5 Modules respectively; b. LOVARS phase map for defining the tumor territories (2 LOVARS unit, with the average of Module 1 and Module 2 as Tsat,1 and the average of Module 4 and Module 5 as Tsat,2. ). c. ½ PC1 map of the 5 maps in a using PCA; d. the MTRasym map by averaging five images in a; e. H&E staining for one frozen slice
DISCUSSION
We propose an efficient method for acquisition of multiple saturation length images that can be readily applied to both in vitro and in vivo CEST imaging. Compared to the conventional CW saturation method with a single module, there is an additional dephasing process caused by the multiple readouts. We have introduced simple guidelines based on the two pool model for choosing the N, θ and TE/T2* for the sequence to limit the T2* decay. For in vivo APT imaging, we determined an optimal θ = 25° based on setting DF to ~10% and using a 4-pool model with the parameters fitted to the experimental data. Several acquisition methods have been developed to measure exchange rates or isolate protons with a certain rate e.g. CERT (43), Spin-Lock (44), SAFARI (45), Two-frequency (46), VDMP (47)and FLEX (48,49). As shown in Fig. 3, our method in phantoms enables fast acquisition of QUEST data which can be fit to determine ksw for θ < 30° at high magnetic fields.
Acceleration of CEST data acquisition can also be accomplished using CEST-FISP (50) or other steady-state sequences (51-53) or through application of a gradient during the saturation pulses (54,55). One major advantage of MeLOVARS is that it can be readily implemented in live animal and patient studies as it is based on a gradient-echo readout (GE or GRE) scheme (52,56,57). As proof-of-principle, we applied MeLOVARS for imaging endogenous APT contrast in mice bearing glioblastoma at 11.7 T, and demonstrate this sequence produces Z-spectra and MTRasym spectra with an additional Tsat dimension. As is shown in Fig. 5&6 and also quantitatively in Table 1, each module of MeLOVARS has either a higher or comparable ΔMTRasym and CNR than the conventional method using the same readout sequence and parameters (i.e. GE-EPI). The N-fold image-increase allows an increase of CNR by √N. The N groups of experimental Z-spectra with different Tsat values allows a more stable fitting to 4-pool Block equation models because of the additional measured points. We demonstrate that the 5 MTRasym images can be combined to create LOVARS phase maps, PCA maps or average MTRasym maps as well. There could be other methods for analyzing MeLOVARS data, such as the QUESTRA method (58). Also, while the T2* is between 7–28 ms for mouse gray matter at 11.7 T allowing acquisition of 5 modules, T2* ~50 ms for human frontal gray matter at 3 T (59) which might allow acquisition of 10 modules if the same θ and TE are used. Although we have only focused on the endogeneous APT contrast of brain tumor in the manuscript, this method is applicable to many other applications either for detecting endogenous molecules such as glutamate (60), creatine, and glycosaminoglycans or exogenous compounds. Although the concept of MeLOVARS is demonstrated on a high field small animal scanner, this method can be readily translated to clinical scanners, either using a multi-channel parallel transmission coil for generating the CW saturation pulse (21), or through substitution of a train of saturation pulses for the CW pulse in between the multiple “Look-Locker” readouts (61).
Regarding the gradient-echo readout used in MeLOVARS, there could be practical concerns, especially at high magnetic field due to imperfect shimming, air-tissue interfaces and the distribution of magnetic susceptibility. Fortunately on lower-field clinical scanners (e.g. 1.5 T and 3 T) with longer and more homogeneous T2*, gradient-echo readout sequences are used very frequently esp. in brain such as in fast T1w imaging, Dynamic Contrast Enhanced (DCE) imaging, perfusion and BOLD functional imaging. Although the small θ sacrifices signal, it also allows shorter recovery times which increase efficiency as has been discussed previously (62).
CONCLUSION
We developed MeLOVARS as a new CEST acquisition method, which enables rapid acquisition of multiple STw images with different effective Tsat values. For phantoms, MeLOVARS collects images with 8 Tsat’s from 0.5 s to 4 s simultaneously, enabling the measurement of the exchange rates for three CEST agents. For detecting glioblastoma in live mice, MeLOVARS enable acquisition of 5 Z-spectra, MTRasym spectra and contrast maps in 8.5 min, with 5 Tsat’s from 0.5 s to 2.5 s which can be employed to generate LOVARS phase maps and increase the CNR.
ACKNOWLEDGEMENTS
This work was supported by NIH Grants R01EB012590, R01EB015031, R01EB015032, S10RR028955, S10RR025118 and the Pearl and Yueh-Heng Yang Foundation. The simulation scripts employed in the manuscript can be accessed by emailing the corresponding author. .
References
- 1.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 2.Trokowski R, Ren JM, Kalman FK, Sherry AD. Selective sensing of zinc ions with a PARACEST contrast agent. Angew Chem-Int Edit. 2005;44(42):6920–6923. doi: 10.1002/anie.200502173. [DOI] [PubMed] [Google Scholar]
- 3.Longo DL, Dastru W, Digilio G, Keupp J, Langereis S, Lanzardo S, Prestigio S, Steinbach O, Terreno E, Uggeri F, Aime S. Iopamidol as a responsive MRI-chemical exchange saturation transfer contrast agent for pH mapping of kidneys: In vivo studies in mice at 7 T. Magn Reson Med. 2010;65(1):202–211. doi: 10.1002/mrm.22608. [DOI] [PubMed] [Google Scholar]
- 4.Schilling F, Schröder L, Palaniappan KK, Zapf S, Wemmer DE, Pines A. MRI Thermometry Based on Encapsulated Hyperpolarized Xenon. Chemphyschem : a European journal of chemical physics and physical chemistry. 2010;11(16):3529–3533. doi: 10.1002/cphc.201000507. [DOI] [PubMed] [Google Scholar]
- 5.McMahon MT, Gilad AA, Zhou J, Sun PZ, Bulte JW, van Zijl PC. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): Ph calibration for poly-L-lysine and a starburst dendrimer. Magn Reson Med. 2006;55(4):836–847. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilad AA, McMahon MT, Walczak P, Winnard PT, Jr., Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25(2):217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 7.Sun PZ, Zhou J, Sun W, Huang J, van Zijl PC. Detection of the ischemic penumbra using pH-weighted MRI. J Cereb Blood Flow Metab. 2007;27(6):1129–1136. doi: 10.1038/sj.jcbfm.9600424. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, Wang S, Yan K, Fu DX, Ford E, Tyler B, Blakeley J, Laterra J, van Zijl PC. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med. 17(1):130–134. doi: 10.1038/nm.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasalatiy O, Gerard RD, Zhao P, Sun X, Sherry AD. Labeling of adenovirus particles with PARACEST agents. Bioconjug Chem. 2008;19(3):598–606. doi: 10.1021/bc7002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinogradov E, He H, Lubag A, Balschi JA, Sherry AD, Lenkinski RE. MRI detection of paramagnetic chemical exchange effects in mice kidneys in vivo. Magn Reson Med. 2007;58(4):650–655. doi: 10.1002/mrm.21393. [DOI] [PubMed] [Google Scholar]
- 11.Liu G, Li Y, Pagel MD. Design and characterization of a new irreversible responsive PARACEST MRI contrast agent that detects nitric oxide. Magn Reson Med. 2007;58(6):1249–1256. doi: 10.1002/mrm.21428. [DOI] [PubMed] [Google Scholar]
- 12.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST) Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2266–2270. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PC. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9(8):1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 14.Wen Z, Hu S, Huang F, Wang X, Guo L, Quan X, Wang S, Zhou J. MR imaging of high-grade brain tumors using endogenous protein and peptide-based contrast. Neuroimage. 51(2):616–622. doi: 10.1016/j.neuroimage.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker-Samuel S, Ramasawmy R, Torrealdea F, Rega M, Rajkumar V, Johnson SP, Richardson S, Gonçalves M, Parkes HG, Arstad E, Thomas DL, Pedley RB, Lythgoe MF, Golay X. In vivo imaging of glucose uptake and metabolism in tumors. Nat Med. 2013;19(8):1067–1072. doi: 10.1038/nm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan KW, McMahon MT, Kato Y, Liu G, Bulte JW, Bhujwalla ZM, Artemov D, van Zijl PC. Natural D-glucose as a biodegradable MRI contrast agent for detecting cancer. Magn Reson Med. 2012;68(6):1764–1773. doi: 10.1002/mrm.24520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flament J, Geffroy F, Medina C, Robic C, Mayer JF, Mériaux S, Valette J, Robert P, Port M, Le Bihan D, Lethimonnier F, Boumezbeur F. In vivo CEST MR imaging of U87 mice brain tumor angiogenesis using targeted LipoCEST contrast agent at 7 T. Magn Reson Med. 2013;69(1):179–187. doi: 10.1002/mrm.24217. [DOI] [PubMed] [Google Scholar]
- 18.Chen LQ, Howison CM, Jeffery JJ, Robey IF, Kuo PH, Pagel MD. Evaluations of extracellular pH within in vivo tumors using acidoCEST MRI. Magn Reson Med. 2013 doi: 10.1002/mrm.25053. 10.1002/mrm.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salhotra A, Lal B, Laterra J, Sun PZ, van Zijl PC, Zhou J. Amide proton transfer imaging of 9L gliosarcoma and human glioblastoma xenografts. NMR Biomed. 2008;21(5):489–497. doi: 10.1002/nbm.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Wen Z, Huang F, Lu S, Wang X, Hu S, Zu D, Zhou J. Saturation power dependence of amide proton transfer image contrasts in human brain tumors and strokes at 3 T. Magn Reson Med. 2011;66(4):1033–1041. doi: 10.1002/mrm.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Togao O, Yoshiura T, Keupp J, Hiwatashi A, Yamashita K, Kikuchi K, Suzuki Y, Suzuki SO, Iwaki T, Hata N, Mizoguchi M, Yoshimoto K, Sagiyama K, Takahashi M, Honda H. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro Oncol. 2014;16(3):441–448. doi: 10.1093/neuonc/not158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Togao O, Kessinger CW, Huang G, Soesbe TC, Sagiyama K, Dimitrov I, Sherry AD, Gao J, Takahashi M. Characterization of lung cancer by amide proton transfer (APT) imaging: an in-vivo study in an orthotopic mouse model. PLoS One. 2013;8(10):e77019. doi: 10.1371/journal.pone.0077019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dula AN, Arlinghaus LR, Dortch RD, Dewey BE, Whisenant JG, Ayers GD, Yankeelov TE, Smith SA. Amide proton transfer imaging of the breast at 3 T: establishing reproducibility and possible feasibility assessing chemotherapy response. Magn Reson Med. 2013;70(1):216–224. doi: 10.1002/mrm.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klomp DW, Dula AN, Arlinghaus LR, Italiaander M, Dortch RD, Zu Z, Williams JM, Gochberg DF, Luijten PR, Gore JC, Yankeelov TE, Smith SA. Amide proton transfer imaging of the human breast at 7T: development and reproducibility. NMR Biomed. 2013;26(10):1271–1277. doi: 10.1002/nbm.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia G, Abaza R, Williams JD, Zynger DL, Zhou J, Shah ZK, Patel M, Sammet S, Wei L, Bahnson RR, Knopp MV. Amide proton transfer MR imaging of prostate cancer: a preliminary study. J Magn Reson Imaging. 2011;33(3):647–654. doi: 10.1002/jmri.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagiyama K, Mashimo T, Togao O, Vemireddy V, Hatanpaa KJ, Maher EA, Mickey BE, Pan E, Sherry AD, Bachoo RM, Takahashi M. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proc Natl Acad Sci U S A. 2014;111(12):4542–4547. doi: 10.1073/pnas.1323855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hectors SJ, Jacobs I, Strijkers GJ, Nicolay K. Amide proton transfer imaging of high intensity focused ultrasound-treated tumor tissue. Magn Reson Med. 2014;72(4):1113–1122. doi: 10.1002/mrm.25000. [DOI] [PubMed] [Google Scholar]
- 28.Liu G, Song X, Chan KW, McMahon MT. Nuts and bolts of chemical exchange saturation transfer MRI. NMR Biomed. 2013;26(7):810–828. doi: 10.1002/nbm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): What is in a name and what isn't? Magn Reson Med. 2011;65(4):927–948. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9(8):1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 31.Song X, Gilad AA, Joel S, Liu G, Bar-Shir A, Liang Y, Gorelik M, Pekar JJ, van Zijl PC, Bulte JW, McMahon MT. CEST phase mapping using a length and offset varied saturation (LOVARS) scheme. Magn Reson Med. 2012;68(4):1074–1086. doi: 10.1002/mrm.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Look DC. Time Saving in Measurement of NMR and EPR Relaxation Times. RevSciInstrum. 1970;41:250. LD. [Google Scholar]
- 33.Gowland P, Mansfield P. Accurate measurement of T1 in vivo in less than 3 seconds using echo-planar imaging. Magn Reson Med. 1993;30(3):351–354. doi: 10.1002/mrm.1910300312. [DOI] [PubMed] [Google Scholar]
- 34.Li AX, Hudson RH, Barrett JW, Jones CK, Pasternak SH, Bartha R. Four-pool modeling of proton exchange processes in biological systems in the presence of MRI-paramagnetic chemical exchange saturation transfer (PARACEST) agents. Magn Reson Med. 2008;60(5):1197–1206. doi: 10.1002/mrm.21752. [DOI] [PubMed] [Google Scholar]
- 35.de Graaf RA, Brown PB, McIntyre S, Nixon TW, Behar KL, Rothman DL. High magnetic field water and metabolite proton T1 and T2 relaxation in rat brain in vivo. Magn Reson Med. 2006;56(2):386–394. doi: 10.1002/mrm.20946. [DOI] [PubMed] [Google Scholar]
- 36.Seehafer JU, Kalthoff D, Farr TD, Wiedermann D, Hoehn M. No increase of the blood oxygenation level-dependent functional magnetic resonance imaging signal with higher field strength: implications for brain activation studies. J Neurosci. 2010;30(15):5234–5241. doi: 10.1523/JNEUROSCI.0844-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Song X, Li Y, Liu G, Ray Banerjee S, Pomper MG, McMahon MT. Salicylic Acid and Analogues as diaCEST MRI Contrast Agents with Highly Shifted Exchangeable Proton Frequencies. Angew Chem Int Ed Engl. 52(31):8116–8119. doi: 10.1002/anie.201302764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song X, Yang X, Ray Banerjee S, Pomper MG, McMahon MT. Anthranilic acid analogues as diamagnetic CEST (diaCEST) MRI contrast agents that feature an IntraMolecular-bond Shifted HYdrogen (IM-SHY) Con Med & Mol Imag. 2014 doi: 10.1002/cmmi.1597. doi: 10.1002/cmmi.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun P, Xia S, Lal B, Eberhart CG, Quinones-Hinojosa A, Maciaczyk J, Matsui W, Dimeco F, Piccirillo SM, Vescovi AL, Laterra J. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differentiation and tumor propagation. Stem cells. 2009;27(7):1473–1486. doi: 10.1002/stem.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, Guerrero-Cazares H, Quinones-Hinojosa A, Laterra J, Xia S. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30(31):3454–3467. doi: 10.1038/onc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swift TJ, Connick RE. Nmr-Relaxation Mechanisms of 017 in Aqueous Solutions of Paramagnetic Cations and Lifetime of Water Molecules in First Coordination Sphere. Journal of Chemical Physics. 1962;37(2):307. [Google Scholar]
- 42.Desprechins B, Stadnik T, Koerts G, Shabana W, Breucq C, Osteaux M. Use of diffusion-weighted MR imaging in differential diagnosis between intracerebral necrotic tumors and cerebral abscesses. AJNR Am J Neuroradiol. 1999;20(7):1252–1257. [PMC free article] [PubMed] [Google Scholar]
- 43.Zu Z, Janve V, Li K, Does M, Gore J, Gochberg D. Multi-angle ratiometric approach to measure chemical exchange in amide proton transfer imaging. Magnetic Resonance in Medicine. 2012;68(3):711–719. doi: 10.1002/mrm.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin T, Autio J, Obata T, Kim S. Spin-Locking Versus Chemical Exchange Saturation Transfer MRI for Investigating Chemical Exchange Process Between Water and Labile Metabolite Protons. Magnetic Resonance in Medicine. 2011;65(5):1448–1460. doi: 10.1002/mrm.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheidegger R, Vinogradov E, Alsop DC. Amide proton transfer imaging with improved robustness to magnetic field inhomogeneity and magnetization transfer asymmetry using saturation with frequency alternating RF irradiation. Magn Reson Med. 2011;66(5):1275–1285. doi: 10.1002/mrm.22912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J, Khitrin A, Regatte R, Jerschow A. Uniform saturation of a strongly coupled spin system by two-frequency irradiation. Journal of Chemical Physics. 2011;134(23) doi: 10.1063/1.3600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J, Yadav NN, Bar-Shir A, Jones CK, Chan KW, Zhang J, Walczak P, McMahon MT, van Zijl PC. Variable delay multi-pulse train for fast chemical exchange saturation transfer and relayed-nuclear overhauser enhancement MRI. Magn Reson Med. 2014;71(5):1798–1812. doi: 10.1002/mrm.24850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman JI, McMahon MT, Stivers JT, Van Zijl PC. Indirect detection of labile solute proton spectra via the water signal using frequency-labeled exchange (FLEX) transfer. J Am Chem Soc. 132(6):1813–1815. doi: 10.1021/ja909001q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav NN, Jones CK, Hua J, Xu J, van Zijl PC. Imaging of endogenous exchangeable proton signals in the human brain using frequency labeled exchange transfer imaging. Magn Reson Med. 69(4):966–973. doi: 10.1002/mrm.24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah T, Lu L, Dell KM, Pagel MD, Griswold MA, Flask CA. CEST-FISP: a novel technique for rapid chemical exchange saturation transfer MRI at 7 T. Magn Reson Med. 65(2):432–437. doi: 10.1002/mrm.22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones CK, Huang A, Xu J, Edden RA, Schar M, Hua J, Oskolkov N, Zaca D, Zhou J, McMahon MT, Pillai JJ, van Zijl PC. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7T. Neuroimage. 77:114–124. doi: 10.1016/j.neuroimage.2013.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu H, Jones CK, van Zijl PC, Barker PB, Zhou J. Fast 3D chemical exchange saturation transfer (CEST) imaging of the human brain. Magn Reson Med. 64(3):638–644. doi: 10.1002/mrm.22546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones CK, Polders D, Hua J, Zhu H, Hoogduin HJ, Zhou J, Luijten P, van Zijl PC. In vivo three-dimensional whole-brain pulsed steady-state chemical exchange saturation transfer at 7 T. Magn Reson Med. 2012;67(6):1579–1589. doi: 10.1002/mrm.23141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X, Lee J, Jerschow A. Ultrafast Scanning of Exchangeable Sites by NMR Spectroscopy. Angew Chem-Int Edit. 2013;52(32):8281–8284. doi: 10.1002/anie.201303255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dopfert J, Witte C, Schroder L. Fast Gradient-Encoded CEST Spectroscopy of Hyperpolarized Xenon. Chemphyschem. 2014;15(2):261–264. doi: 10.1002/cphc.201300888. [DOI] [PubMed] [Google Scholar]
- 56.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nat Med. 2012;18(2):302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixon WT, Hancu I, Ratnakar SJ, Sherry AD, Lenkinski RE, Alsop DC. A multislice gradient echo pulse sequence for CEST imaging. Magn Reson Med. 63(1):253–256. doi: 10.1002/mrm.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun PZ. Simplified and scalable numerical solution for describing multi-pool chemical exchange saturation transfer (CEST) MRI contrast. J Magn Reson. 2010;205(2):235–241. doi: 10.1016/j.jmr.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wansapura JP, Holland SK, Dunn RS, Ball WS. NMR relaxation times in the human brain at 3.0 tesla. J Magn Reson Imaging. 1999;9(4):531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 60.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nat Med. 2012;18(2):302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitt B, Zaiss M, Zhou J, Bachert P. Optimization of pulse train presaturation for CEST imaging in clinical scanners. Magn Reson Med. 2011;65(6):1620–1629. doi: 10.1002/mrm.22750. [DOI] [PubMed] [Google Scholar]
- 62.Sun PZ, Lu J, Wu Y, Xiao G, Wu R. Evaluation of the dependence of CEST-EPI measurement on repetition time, RF irradiation duty cycle and imaging flip angle for enhanced pH sensitivity. Phys Med Biol. 2013;58(17):N229–240. doi: 10.1088/0031-9155/58/17/N229. [DOI] [PMC free article] [PubMed] [Google Scholar]