Abstract
C. elegans molting is a process during which the apical extracellular matrix of the epidermis, the cuticle, is remodeled through a process of degradation and re-synthesis. Using a genetic approach, we identified nekl-3 as essential for the completion of molting. NEKL-3 is highly similar to the mammalian NEK kinase family members NEK6 and NEK7. Animals homozygous for a hypomorphic mutation in nekl-3, sv3, had a novel molting defect in which the central body region, but not the head or tail, was unable to shed the old cuticle. In contrast, a null mutation in nekl-3, gk506, led to complete enclosure within the old cuticle. nekl-2, which is most similar to mammalian NEK8, was also essential for molting. Mosaic analyses demonstrated that NEKL-2 and NEKL-3 were specifically required within the large epidermal syncytium, hyp7, to facilitate molting. Consistent with this, NEKL-2 and NEKL-3 were expressed at the apical surface of hyp7 where they localized to small spheres or tubular structures. Inhibition of nekl-2, but not nekl-3, led to the mislocalization of LRP-1/megalin, a cell surface receptor for low-density lipoprotein (LDL)-binding proteins. In addition, nekl-2 inhibition led to the mislocalization of several other endosome-associated proteins. Notably, LRP-1 acts within hyp7 to facilitate completion of molting, suggesting at least one mechanism by which NEKL-2 may influence molting. Notably, our studies failed to reveal a requirement for NEKL-2 or NEKL-3 in cell division, a function reported for several mammalian NEKs including NEK6 and NEK7. Our findings provide the first genetic and in vivo evidence for a role of NEK family members in endocytosis, which may be evolutionarily conserved.
Keywords: C. elegans, molting, kinases, NEK, endocytosis, hyp7, NEKL-2, NEKL-3
INTRODUCTION
Studies on cell cycle regulation in the fungus Aspergillus nidulans identified Never in Mitosis A (NimA), a serine-threonine protein kinase involved in multiple aspects of mitosis (Oakley and Morris, 1983; Osmani et al., 1988). Subsequently, NimA family members, referred to as NEKs, have been identified in a wide range of organisms (supplementary material Fig. S1) (Fry et al., 2012; Moniz et al., 2011). The nematode Caenorhabditis elegans has four NIMA-related kinase genes. The product of nekl-1 (NEver in mitosis Kinase Like) resembles mammalian NEK9 and, to a lesser extent, NEK8, both of which have regulator of chromosome condensation (RCC1) repeats carboxyl-terminal to their kinase domain. NEKL-4 is an ortholog of Nek10, which has Armadillo/β-catenin–like repeats amino-terminal to the kinase domain. nekl-2 and nekl-3 both encode small proteins that consist mainly of a kinase domain.
NEKL-3 is a compelling ortholog of mammalian NEK6 and NEK7, which are 82% identical to one another and, like NEKL-3, contain little more than a protein kinase domain. Studies in tissue culture systems have implicated NEK6 and NEK7 in mitotic spindle formation, cytokinesis, centrosome separation, centriole duplication, and the control of microtubule dynamics (Bertran et al., 2011; Kim and Rhee, 2011; Lee et al., 2008; Motose et al., 2011; Motose et al., 2012; O'Regan et al., 2007; O'Regan and Fry, 2009; Rapley et al., 2008; Sdelci et al., 2011; Yin et al., 2003; Yissachar et al., 2006). In addition, overexpression of NEK6 and NEK7 are associated with cancer progression (Nassirpour et al., 2010; Wang et al., 2013) and NEK6 may inhibit cellular senescence (Jee et al., 2010; Jee et al., 2013). Mice homozygous for a deletion mutation in NEK7 typically arrest in late embryogenesis or within a month after birth, and mouse embryonic fibroblasts derived from NEK7 mutants show increased aneuploidy (Salem et al., 2010). NEK6 and NEK7 are stimulated by NEK9-mediated phosphorylation (Belham et al., 2003; Bertran et al., 2011; O'Regan et al., 2007; O'Regan and Fry, 2009; Richards et al., 2009), but the substrates of Nek6 and Nek7, with the possible exception of the kinesin Eg5 (Bertran et al., 2011; Rapley et al., 2008), have not been clearly defined (Belham et al., 2001; Lizcano et al., 2002). Interestingly, NEK6 and NEK7 were identified in a non-biased high-throughput screen as positive regulators of clathrin-mediated endocytosis, suggesting that these NEK family members have functions unrelated to cell division (Pelkmans et al., 2005).
NEKL-2, although also composed almost exclusively of a kinase domain, does not closely resemble NEKL-3 (supplementary material Figs S1 and S2). Its closest mammalian relative is NEK8, but NEK8 has RCC1 repeats, which are lacking in NEKL-2. In vertebrates, NEK8/NPHP9 has been implicated primarily in ciliogenesis (Fry et al., 2012; Mahjoub et al., 2004; Mahjoub et al., 2005) and kidney function (Liu et al., 2002; Mahjoub et al., 2005; Otto et al., 2008; Smith et al., 2006; Trapp et al., 2008). More recent studies have implicated NEK8 in the response to DNA-replication stress and maintenance of genomic stability (Choi et al., 2013; Jackson, 2013). NEK8 may negatively regulate caveolae/raft-mediated endocytosis but function as a positive regulator of clathrin-mediated endocytosis (Pelkmans et al., 2005), suggesting, as for NEK6 and NEK7, that individual NEKs may carry out a broad range of biological functions, including roles in intracellular trafficking.
As described below, we have shown that C. elegans nekl-2 and nekl-3 are required for the completion of molting. Nematodes typically undergo four juvenile or larval molts of their exoskeleton, termed the cuticle. The cuticle is composed of cross-linked collagen and other components and is largely derived from the apical surface of hyp7, a large syncytium that makes up most of the epidermis (termed hypodermis). As one of the larval molts commences and a new cuticle is synthesized under the old, a series of movements ensue, followed by rupture and escape from the old cuticle (Page and Johnstone, 2007; Singh and Sulston, 1978). In C. elegans, inactivation of many individual genes by mutation or by RNA interference (RNAi) results in a frequent failure to shed the old cuticle completely (Frand et al., 2005; Kang et al., 2013). Gene products required for proper molting include nuclear hormone receptors (Gissendanner and Sluder, 2000; Hayes et al., 2006; Kostrouchova et al., 1998, 2001; Monsalve and Frand, 2012), matrix metalloproteases (Altincicek et al., 2010; Davis et al., 2004; Hashmi et al., 2004; Kim et al., 2011; Stepek et al., 2011; Suzuki et al., 2004), selenoproteins (Stenvall et al., 2011), enzymes controlling sterol and fatty acid synthesis (Entchev and Kurzchalia, 2005; Jia et al., 2002; Kuervers et al., 2003; Li and Paik, 2011), hedgehog-related proteins (Hao et al., 2006; Zugasti et al., 2005), and LRP-1, an ortholog of megalin, a large member of the LDL receptor family (Yochem et al., 1999). A number of molting genes are associated with secretion, endocytosis, and vesicle trafficking (Frand et al., 2005; Kang et al., 2013; Liegeois et al., 2007), and several of these, including the Disabled adaptor ortholog dab-1, the Saccharomyces cerevisiae Vps27p ortholog hgrs-1, and the S. cerevisiae Sec23p ortholog sec-23, affect trafficking and endocytosis of LRP-1 from the apical membrane of hyp7 (Holmes et al., 2007; Kamikura and Cooper, 2006; Kang et al., 2013; Roberts et al., 2003; Roudier et al., 2005).
The genetic studies described herein strengthen the notion that NEK kinases have functions not linked to cell division or to ciliogenesis. The requirement of nekl-2 and nekl-3 for proper completion of molting, instead, suggests their involvement in extracellular matrix remodeling and vesicular trafficking. Data consistent with this possibility are presented.
MATERIALS AND METHODS
Genetic strains and maintenance
All strains were cultured at 20°C (unless otherwise stated) on nematode growth medium (NGM) supplemented with Escherichia coli OP50 as a food source according to standard protocols (Stiernagle, 2006). Preexisting strains used in theses studies are as follows: N2, var. Bristol, designated the wild-type strain (Brenner, 1974); LH191 (eqIs1[lrp-1::gfp]; rrf-3(pk1426)II) (Kang et al., 2013); VC1733 (nekl-2(gk839) l/hT2[bli-4(e937) let-?(q782) qls48](I;III)); VC1774 (nekl-2(gk841) l/hT2[bli-4(e937) let-?(q782) qls48](I;III)); and VC2632 (nekl-2(ok3240) l/hT2[bli-4(e937) let-?(q782) qls48](I;III)). LH243 (nekl-3(sv3)X; eqIs3 [nekl-3::gfp; pRF4]), LH373 (nekl-3(gk506)X; mnEx174[F19H6; pTG96]), SP2735 (nekl-3(sv3)/lin-2(e1309) unc-9(e101) X), SP2736 (nekl-3(sv3)X; mnEx174[F19H6; pTG96]), WY967 (eqIs1[lrp-1::gfp]; rrf-3(pk1426)II; nekl-3(sv3)X; mnEx174[F19H6; pTG96]), WY977 (eqIs1[lrp-1::gfp]; rrf-3(pk1426)II; nekl-3(gk506)X; mnEx174[F19H6; pTG96]), WY1061 (nekl-2(gk839); fdEx257), WY1070 (fbn-1(tm290), fdEx250, eqIs1[lrp-1::gfp]; rrf-3(pK1426)II), WY1073 (nekl-2(gk839); fdEx261), WY1077 (nekl-2(gk839); fdEx261; nekl-3(sv3)X; eqIs3 [nekl-3::gfp; pRF4]); WY976 (eqIs1[lrp-1::gfp]; dab-1(gk291)II). RT424 (pwIs126; EEA-1::GFP), RT1113 (pwIs439; RAB- 5::GFP), RT1378 (pwIs528; CHC-1::GFP), RT225 (pwIs94; SNX-1::GFP), WY1095 (lin-35(n745); pwIs94). The gk506 mutation was outcrossed eight times prior to strain construction.
Isolation of sv3
The sv3 allele was identified in a screen for recessive mutations that affect the completion of molting. L4 larvae of the N2 strain were treated with 50 mM ethyl methanesulfonate (Brenner, 1974). F1 animals from the mutagenesis were placed on many NGM plates (three animals per plate). After growth at 25°C, the plates were examined for the presence of F2 progeny exhibiting defects in molting, including a failure to shed all of the old cuticle. Because mutants exhibiting defective molting are often incapable of reaching adulthood, non-mutant siblings were individually isolated from candidate plates for the identification of heterozygotes that could be used to maintain the mutations until they were mapped and balanced. Because none of the mutations proved to be temperature sensitive, a temperature of 20° was used for their maintenance and analysis. In all, 3,600 haploid genomes were examined, and 12 mutations identified. Consistent with its large size (Yochem et al., 1999), lrp-1 proved a large target, accounting for six of the mutations. Standard genetic mapping placed sv3 ~0.04 cM to the left of lin-2 on LGX. After the sv3 isolate was outcrossed eight times, the stock SP2735 [sv3/lin- 2(e1309) unc-9(e101) X] was established. Stocks cannot be maintained if sv3 is homozygous.
Plasmid construction and site-directed mutagenesis
A functional fusion between nekl-3 coding sequences and GFP sequences was constructed from DNA generated by PCR (Pfu Turbo, Stratagene) using a rescuing cosmid clone, F19H6, as template. A DNA fragment for the 5' portion of the gene, comprising ~5 kb from the BglII through the EcoRI sites of the genomic sequence, was generated using LRB-504 and LRB-505 (supplementary material Table S1) and cloned into BlueScript. The 3' portion of the gene was generated in two fragments, to add a unique PstI site immediately before the stop codon of the open reading frame. Three alanine codons were also added immediately preceding the PstI site. First, LRB-506 and LRB-507 were used to generate a fragment containing an EcoRI site from within the gene and the new PstI site. Then, LRB-508 and LRB-509 were used to generate a fragment containing the new PstI site and a genomic XhoI site and including the stop codon and 3'UTR. These two PCR products were combined in the proper order into Bluescript. The EcoRI to XhoI region was then excised and subsequently ligated into the plasmid containing the 5' BglII to EcoRI fragment, thereby creating a complete gene with three alanine codons and a PstI site immediately before the stop codon. Successful rescue of sv3 following microinjection demonstrated that this altered plasmid retained nekl-3 activity. Finally, a fragment encoding GFP, taken from the plasmid pPD103.87 as a PstI fragment, was cloned in the proper orientation into the PstI site of the modified nekl-3 plasmid to generate LH#pLRB59-6.
A functional fusion between the nekl-2 gene and mCherry sequences was generated using a modified pPD95.77 vector in which GFP sequences have been replaced by mCherry (Kuzmanov et al., 2014). A 1,888-kb nekl-2 genomic fragment containing 891 bp of 5' flanking DNA was obtained following PCR amplification using a rescuing fosmid clone, WRM0639aE11, as template using primers NEKL2Ch1 and NEKL2Ch2. The fragment was cloned into the mCherry vector using KpnI and BamHI restriction enzymes. The final construct contained the full-length nekl-2 coding sequence with a spacer of three glycines between nekl-2 and mCherry. Microinjection of balanced nekl-2(gk839) animals proved that this translational fusion is functional.
The proline codon at position 194 of nekl-3 was changed to a leucine codon by means of the QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) and primers P194Lm1 and P194Lm2, which incorporate a change from C to T at nucleotide position 1,209 following the A of the ATG start codon. The template for the mutagenesis was LH#pLRB59-6. The alteration was verified by DNA sequencing of plasmids prior to microinjection of balanced nekl-3(gk506) animals. The K52M K53M nekl-3 variant was generated in the same manner using primers K5253Mm1 and K5253Mm2. Sequences for all primers used in this study can be found in Supplementary Table 1.
For the nekl-1 RNAi-feeding construct, the entire thirteenth exon was cloned. For the nekl-4 RNAi-feeding construct the entire eighth exon was cloned. Both sequences were inserted using XhoI and XbaI restriction sites into pPD129.36 (Timmons et al., 2001).
PCR-based analysis of mRNA
Total RNA was isolated from mixed-staged populations of N2 worms by means of TRIzol (Invitrogen). The true 5-prime ends of mRNA were determined with a FirstChoice RNA ligase– mediated rapid amplification of cDNA ends (RLM-RACE) kit (Ambion). The 3' ends were determined by means of reagents supplied with the kit. For both ends, gene-specific primers were used in combination with primers provided with the kit. Internal splice sites were verified by RT- PCR with a cMaster RTplusPCR System kit (Eppendorf) and gene-specific primers.The DNA sequence of the PCR products generated from cDNA was determined en masse as above.
Microinjection of DNA and dsRNA
Rescue of the sv3 mutation was assessed by the microinjection (Mello and Fire, 1995) of balanced heterozygotes with cosmid DNA or with long-range PCR products based on the genomic DNA sequence of the relevant regions. Transgenic lines were identified on the basis of the co-injection of either of the transformation markers pRF4 (Mello et al., 1991) or pTG96 (Yochem et al., 1998). Long-range PCR products were generated with the Expand Long Template PCR System (Roche Diagnostics). For nekl-3, the primers for the PCR product bearing the number 287 were LRB-124 and LRB-125 and for PCR#289, LRB-128 and LRB-129.
For initial RNAi experiments (Fire et al., 1998), 250 to 500 µg/ml of gene-specific double-stranded RNA (dsRNA) was microinjected into the gonads or intestines of hermaphrodites, which were then periodically transferred to new plates to allow discrete intervals of egg laying. dsRNAs were produced by generating PCR products with oligonucleotide primers that contained the T7 promoter sequence (See supplemental Methods). The primers were LRB-189 and LRB- 190 for nekl-3 and LRB-405 and LRB-406 for nekl-1. RNA was transcribed from both strands of the PCR products by means of a T7 MEGA-Script kit (Ambion). dsRNA was allowed to form from the complementary strands during the transcription.
RNAi feeding studies
For RNAi feeding, L4 animals were placed on lawns of bacteria expressing gene-specific double-stranded RNA (dsRNA) using standard methods (Ahringer, 2005). As a negative control, worms were placed on lawns of the same bacterial host transformed with a plasmid, pDF129.36, that expresses a ~200-bp dsRNA that is not homologous to any C. elegans genes (Timmons et al., 2001). RNAi clones for nekl-2, nekl-3, qua-1 and fbn-1 were obtained from the Ahringer library. The construction of RNAi-feeding clones for nekl-1 and nekl-4 are described in the Supplemental Methods. For an analysis of the ability of RNAi clones to enhance sv3, L4 larvae transgenic for a GFP-marked extrachromosomal array expressing wild-type nekl-3 were placed on the appropriate lawns at 22°C. Non-green eggs were transferred to new plates expressing the appropriate RNAi clones. Body lengths and head widths of larvae were measured 4 days after hatching. For gonad measurements, worms were collected on the second day after hatching. For testing endocytic marker strains (described in Fig. 8) with nekl-2(RNAi), strains were propagated on lin-35(RNAi) plates for at least two generations before transferring to nekl- 2(RNAi) plates.
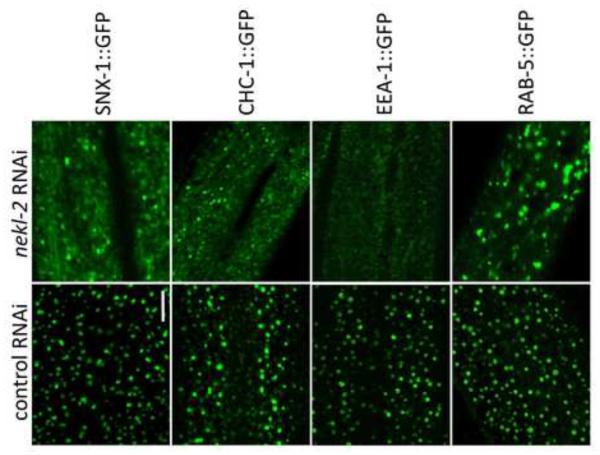
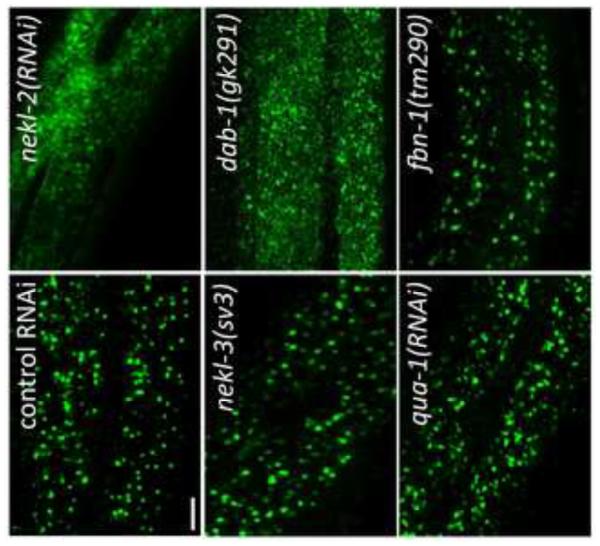
Fig. 8. NEKL-2 is required for correct localization of endosomal markers.
Strains containing integrated arrays expressing N-terminal fusions of GFP to SNX-1, CHC-1, EEA-1, and RAB-5 were assayed following treatment with control RNAi or nekl-2(RNAi) following RNAi hypersensitization using either the lin-35(n745) mutation (SNX-1::GFP) or lin-35(RNAi) (other markers). Representative images are shown for each marker in larval-stage animals. Note the altered expression pattern of all 4 markers following nekl-2(RNAi). Scale bar = 5 µm.
Microscopy and measurements
For body length and head width measurements, an Olympus IX71 inverted epifluorescence microscope with a scientific-grade CMOS camera (Orca Flash 2.8, Hamamatsu) and MetaMorph 7.7 (Molecular Devices) software were used. Body length was measured from the tip of the worm’s head to the end of the tail. At least 30 animals were analyzed for each condition. For gonad area and rectal width measurements, a Nikon Eclipse epifluorescence microscope and OpenLab software were used. The same equipment was used for fluorescence comparisons in supplementary material Fig. S3. At least 10 animals were used for gonad area measurements for each condition. Only worms that were in the dorso-ventral orientation and with a clearly visible gonad were scored. Rectal width was measured in a minimum of 30 animals for each strain with worms in the latero-lateral position. Width was measured in three to five places in the chosen focal plane, and an average value was then calculated.
For fluorescence images (except for Fig. 6E and Fig. S3), an Olympus IX71 inverted microscope equipped with a spinning-disc confocal head (CSU-X1 Yokogawa) was used. Confocal illumination was provided by an LMM5 laser launch (Spectral Applied Research). Image acquisition was performed using MetaMorph 7.7 software. Colocalization analysis was performed with ImageJ software, using the “Colocalization” plugin, with manual determination of threshold level.
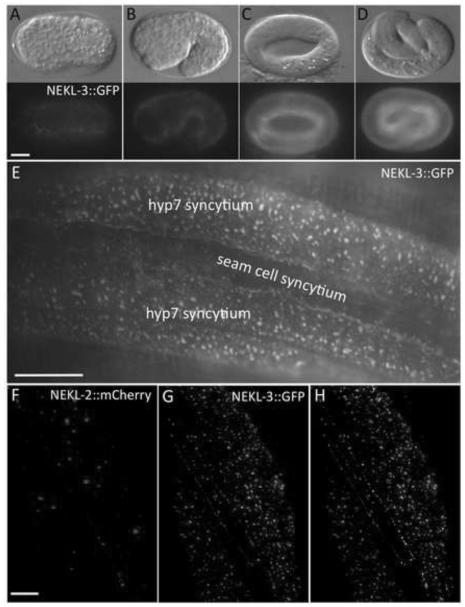
Fig. 6. NEKL-2 and NEKL-3 reporters are expressed in the hypodermis.
(A-D) DIC and corresponding fluorescence images of NEKL-3::GFP expression in embryos. Expression is first detected in hypodermal cells coincident with the initiation of embryonic morphogenesis (A), with expression increasing in the hypodermis during the remainder of embryogenesis (B-D). Embryonic stages depicted are bean (A, ~350 min post fertilization), 1.5-fold (B, ~420 min), 3- fold (C, ~520 min) and pretzel (D, >600 min). In adults (E) and larvae (not shown), NEKL-3::GFP is present in the apical region of the parts of hyp7 that do not overlie body muscles and in the apical region of other hypodermal cells, but expression is not evident in the seam cells. The expression of NEKL-3::GFP can be globular, punctate or slightly tubular. (F-H) NEKL-2 (F) and NEKL-3 (G) reporters show little overlap in subcellular localization (H, white), although NEKL-2::mCherry is expressed in puncta near the apical surface of hyp7. Scale bar in A = 10 µm for A-D; in E = 10 µm; in F = 5 µm for F-H.
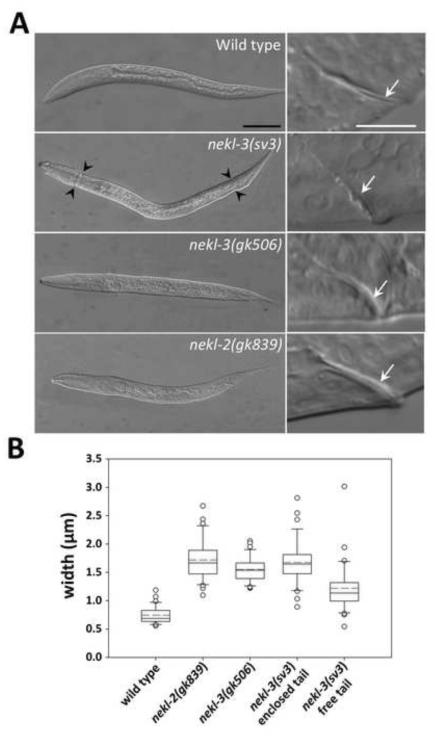
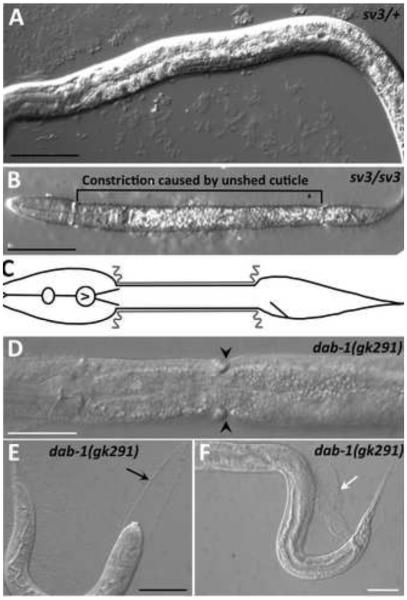
Fig. 3. Unshed cuticles of the nekl-2 and nekl-3 mutants.
(A) DIC images of arrested nekl-3(gk506) and nekl-2(gk893) homozygotes compared with a wild-type L2 larva and a nekl-3(sv3) homozygote arrested at L2. In nekl-3(gk506) and nekl-2(gk893) homozygotes, the cuticle always encloses the entire larva, whereas nekl-3(sv3) mutants typically shed the cuticle in the head and tail regions (black arrowheads). White arrows in right panels indicate corresponding rectums, which are thicker in nekl-3(gk506) and nekl-2(gk893) homozygotes because of a double layer of cuticle. Scale bar for left panels = 50 µm; scale bar for right panels = 10 µm. (B) Box-and-whisker plot showing the rectal width of mutants. The box represents the middle 50% of the data points that are the closest to the median value (black line); the dashed red line represents the mean value. Dots are outliers that differ by ≥1.5-fold from the upper or lower quartiles; whiskers encompass values between the outliers (dots) and the upper and lower quartiles (top and bottom of box). sv3 homozygotes with free tails are somewhat variable in rectal widths but are closer to wild type than are nekl-2(gk506) or nekl-3(gk893) deletion mutants. sv3 homozygotes with enclosed tails have rectal widths that are indistinguishable from deletion mutants. n ≥ 30 for each genotype. For raw data and statistical analyses, see Supplementary File 1.
Genetic mosaic analysis
Genetic mosaic analysis of nekl-2(gk839) and nekl-3(sv3) was carried out using established procedures (Yochem et al., 1998; Yochem et al., 2000; Yochem, 2006). The source of the mosaics was the strain SP2736 (nekl-3(sv3); mnEx174[F19H6; pTG96]) for nekl-3 and WY1061 (nekl-2(gk839); fdEx257[nekl-2(+)(fosmid clones wrm0639aE11 and wrm0636aD02; pTG96]) for nekl-2.
RESULTS
sv3 affects the completion of molting, conferring the Corset or Dumbbell phenotype
We isolated the sv3 mutation on the basis of its effect on the completion of molting, but the phenotype differed from that of previously studied genes that affect molting. At hatching, the homozygotes segregating from heterozygous mothers were indistinguishable from their non-mutant siblings (Fig. 1A and data not shown). The mutant homozygotes, however, were unable to completely shed the old cuticle at the end of the first molt. Although the head became free of the old cuticle, most of the middle part of the body remained encased by it (Fig. 1B,C). The tail was usually also free of the old cuticle, but sometimes it remained encased. Those worms that had only the middle part of the body constricted by the old cuticle had the appearance of dumbbells or of being encased in a long corset, because the head and tail were free to increase in size. In contrast, previously studied molting mutants can be completely encased in the old cuticle, have bits of old cuticle attached to their tails, or have narrow rings of old cuticle that constrict only a small region of the body (Fig. 1D-F).
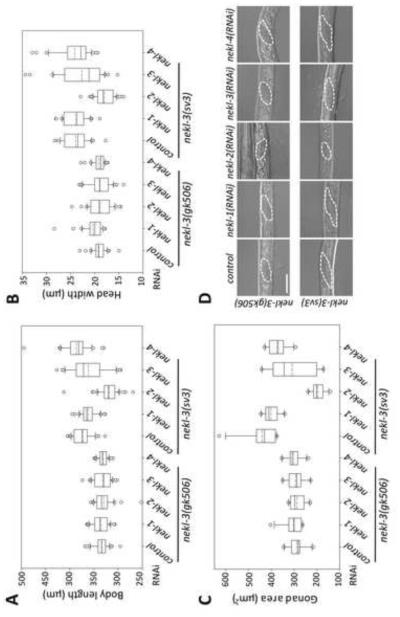
Fig. 4. Enhancement of the nekl-3(sv3) phenotype by nekl-2.
Enhancement of molting defects was assayed in nekl-3(sv3) and nekl-3(gk506) mutant backgrounds. (A-C) Measurements of body length (A), head width (B), and the area of the gonad from a longitudinal section (C) are summarized as box-and-whisker plots (see legend for Fig. 3). A,B, n ≥ 30 for each genotype; C, n ≥ 10. Control RNAi was carried out using strain HT115 carrying vector plasmid pPD129.36. nekl-2(RNAi) enhanced all three aspects of the sv3 mutation but did not affect those of the gk506 mutation. Whereas nekl-1 and nekl-4 RNAi had no effect on either mutation, nekl-3(RNAi) conferred a slight but statistically significant enhancement of gonad defects on sv3 homozygotes. For raw data and statistical analyses (A-C) see Supplementary File 1. (D) Representative images of gonads used for the measurements in C. Gonads of nekl-3(gk506) animals are similar in all RNAi conditions and indicate development to late L1 or early L2. Gonads of nekl-3(sv3) animals typically exhibit more extensive development than that seen for gk506 within the time frame of the experiment and indicate development to mid-to-late L2 stages (sv3 homozygotes can progress to L3 if allowed further growth). RNAi of nekl-2 in the sv3 background leads to L1 or early L2 arrest based on gonad size. Dashed line indicates the gonad periphery. Scale bar = 20 µm.
sv3 homozygotes were for the most part immobile and did not feed well. Postembryonic development did, however, continue, with some of the mutants achieving the third larval stage (L3), based on the morphology of the gonad and incipient vulva. The failure to shed the old cuticle from the middle of the body was repeated for homozygotes that entered the molt from the second larval stage (L2) to L3, and such mutants therefore were encased in two old cuticles (data not shown). The persistence of the constriction of sv3 homozygous larvae by the first larval stage (L1) cuticle resulted in worms that were small for their developmental stage (Fig. 1B and data not shown). A comparison of head widths and body lengths between arrested sv3 homozygotes and wild-type larvae during the L2-L3 molt is consistent with sv3 homozygotes arresting following the second molt and indicated that body length was primarily affected in arrested sv3 mutants (Supplementary File 1).
sv3 is an allele of the NIMA-related kinase gene nekl-3
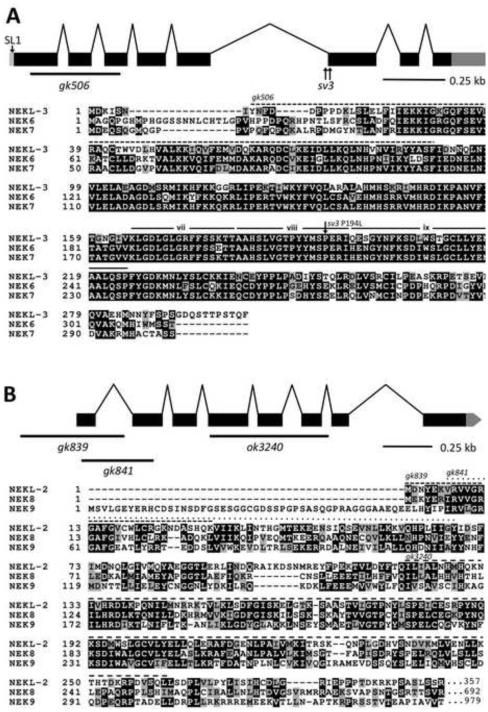
Genetic mapping placed sv3 ~15 kb to the left of lin-2 on the X chromosome. Proof that the affected gene in sv3 homozygotes is mlt-1/nekl-3 (F19H6.1) was obtained as follows. (1) A PCR fragment corresponding specifically to nekl-3 fully rescued the mutant phenotype; (2) RNAi of nekl-3 by injection or feeding methods conferred defective completion of molting, including the corset phenotype; (3) sv3 homozygotes had two closely linked independent mutations in F19H6.1 (Fig. 2A; see below); (4) gk506, a deletion allele of nekl-3, also conferred defective completion of molting. The 302-amino-acid sequence predicted for the nekl-3 product indicated that it belongs to a family of serine/threonine protein kinases and showed striking similarity to NEK6 and NEK7 (Kandli et al., 2000), two NIMA-related kinases that are paralogs of each other in mammals (Fig. 2A; supplementary material Fig. S1). That kinase activity is essential for NEKL-3 function was supported by the observation that an engineered variant of NEKL-3 (K52M / K53M) predicted to lack kinase activity (O'Regan and Fry, 2009) failed to rescue nekl-3 mutants (data not shown).
Fig. 2. The structure of nekl-2 and nekl-3 and alignments of their products with human NEKs.
(A) The structure of the nekl-3 locus with the locations of the sv3 alterations and the gk506 deletion is depicted along with an alignment of its predicted amino-acid sequence with human NEK6 and NEK7. The predicted kinase domain of NEKL-3 spans amino acids 23–281; the region deleted by gk506 is indicated by the dashed line. The P194L mutation conferred by sv3 affects a conserved proline in subdomain viii of the kinase domain (subdomains vii, viii and ix are indicated (Hanks and Hunter, 1995)). (B) The structure of the nekl-2 locus with the locations of the gk839, gk841, and ok3240 deletion alleles along with an alignment of its predicted kinase domain (amino acids 4–267) with the kinase domains of human NEK8 and NEK9. The C-terminal halves of both NEK8 and NEK9 have RCC1 repeats (not shown), whereas NEKL-2 extends only for an additional 90 amino acids after the kinase domain (also see supplementary material Fig. S2). In peptide alignments, black boxes with white lettering indicate identical amino acids; gray boxes with black lettering, similar amino acids; white background with black lettering, dissimilar amino acids.
nekl-3 has eight exons, and its resulting mRNA is 1,097 nucleotides long and has an SL1 splice leader. Although RLM-RACE analysis indicated variation in the start of transcription, alternative forms of the open reading frame of the mRNA were not abundant. A comparison of the nekl-3 gene in sv3 with wild type indicated that the sv3 allele of nekl-3 was associated with two mutations that are separated by 24 bp (Fig. 2A). One is a T-to-A transversion in the fifth intron 20 nucleotides upstream of the start of the sixth exon. The other is a C-to-T transition of the fifth nucleotide of the sixth exon, leading to a proline-to-leucine change in the coding sequence (P194L). The latter mutation is likely to be causative, because a proline is invariant at this relative position, the activation subdomain, in various members of the NIMA family and is usually conserved in other protein kinases as well (Hanks and Hunter, 1995). The importance of this change was further indicated by the failure of a plasmid expressing the P194L form of nekl-3 (but wild type for the intron mutation) to rescue the sv3 mutant phenotype.
The gk506 deletion allele of nekl-3 removes 340 nucleotides of the open reading frame (amino acids Y8–F90) and generates a frame shift (Fig. 2A). The deleted codons include those encoding the conserved ATP-binding domain, a vital feature of protein kinases. The gk506 deletion is therefore likely to be null. Moreover, its phenotype when homozygous was stronger than that conferred by sv3, indicating that the latter allele is hypomorphic. Similar to the sv3 allele, nekl-3(gk506) homozygotes had normal embryonic development and were normal L1 larvae until they entered their first molt. Unlike sv3 larvae, however, nekl-3(gk506) larvae remained completely encased in the old cuticle after they entered the first molt (Fig. 3A). Consistent with nekl-3(gk506) larvae arresting following the first molt, the head width and body length of nekl-3(gk506) larvae corresponded closely with wild-type larvae during the L1-L2 molt (Supplementary File 1).
Analysis of the rectum indicated that nekl-3(gk506) homozygotes contained a double layer of cuticle, leading to visible rectal thickening (Fig. 3). nekl-3(sv3) mutants that had shed the tail portion of their old cuticle did not typically contain a double layer of cuticle within the rectum, whereas sv3 homozygotes with an encased tail had an increased rectal width that was similar to gk506 homozygotes (Fig. 3). Consistent with gk506 homozygotes having a more severe phenotype than sv3 homozygotes, the former animals were slightly smaller and their gonads were less developed (Fig. 4). Although gk506 homozygotes arrested growth at the molt from L1 to L2, they were quite mobile and could live for several days, although they were likely unable to feed because of the unshed L1 cuticle.
nekl-2 is required for molting
Our findings for NEKL-3 prompted us to examine the three other NEK family members in C. elegans for roles in molting. RNAi of nekl-2 in RNAi-hypersensitive backgrounds (Simmer et al., 2002; Wang et al., 2005), but not in wild type, conferred a range of molting defects, including the Corset phenotype, whereas no effects on molting were observed for RNAi feeding or injection of nekl-1 or for RNAi feeding of nekl-4 (data not shown and see below). NEKL-2 resembles NEKL-3 in consisting mostly of a protein kinase domain, but sequence divergence between them (supplementary material Figs S1 and S2) indicated that they are not likely to be functional counterparts. To investigate the phenotype of nekl-2 in more detail, three loss-of- function alleles of nekl-2 were obtained (Fig. 2B). gk839 is a deletion that begins 276 bp upstream of the ATG start codon and includes the first exon and most of the first intron and is therefore likely to be null. Like nekl-3(gk506), gk839 homozygotes also arrested at the L1-to-L2 molt, remaining completely enclosed in the old cuticle, which is clearly evident in the rectum (Fig. 3). The phenotypes of two other alleles, gk841 and ok3240, resembled that of gk839 but were not examined in detail. gk841 deletes a region corresponding to amino acids V9 to Y68 and leads to a frame shift in the second exon; ok3240 deletes a region corresponding to amino acids I212 to L261 and induces a frame shift after exon 6.
nekl-2 enhances the molting defect of nekl-3(sv3)
The hypomorphic nature of nekl-3(sv3) permitted an examination of nekl-2 for genetic interactions with nekl-3. We also further examined nekl-1 and nekl-4, because, although they did not appear to be required for molting, genetic enhancement provides a sensitive test for involvement in a process. Although neither nekl-1(RNAi) nor nekl-4(RNAi) enhanced the phenotype of sv3, nekl-2(RNAi) showed strong effects. Even 4 days after hatching, nekl-3(sv3); nekl-2(RNAi) worms failed to form corsets, remaining instead completely encased in the old cuticle at the L1-to-L2 molt (Fig. 4A,B). A failure to continue to grow and develop was also reflected in the size and morphology of the gonad. Two days after hatching, the gonads of sv3 animals could be clearly seen to have left and right arms, whereas the gonads of nekl-3(sv3); nekl-2(RNAi) failed to develop (Fig. 4C and data not shown). In contrast, nekl-2(RNAi) of wild- type (non-RNAi hypersensitive) worms did not cause any detectable molting defect (data not shown), indicating true enhancement. RNAi feeding of nekl-3 itself also enhanced sv3 defects, but the effects were slight relative to those of nekl-2 feeding (Fig. 4), which may reflect low efficacy of nekl-3 RNAi. The enhancement of nekl-3 by nekl-2 and their similar mutant phenotypes suggest that these genes might act in a common pathway or have complementary functions.
Mosaic analyses indicate that nekl-2 and nekl-3 are required in hyp7
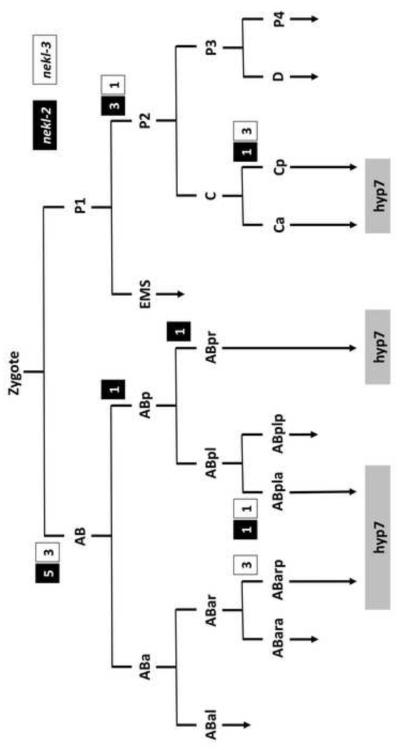
To determine the tissue or cell-types that require the function of nekl-2 and nekl-3 for proper molting, genetic mosaics analyses were carried out for both genes. Our approach made use of extrachromosomal arrays containing wild-type copies of either nekl-2 or nekl-3 together with a cell autonomous GFP reporter (Yochem et al., 1998; Yochem and Herman, 2005; Yochem et al., 2000). Such arrays are capable of rescuing nekl-2 or nekl-3 molting defects provided that the array has been inherited within the cell lineage(s) that requires nekl-2 or nekl-3 function. Although the isolated nekl-2 and nekl-3 mosaics varied greatly with respect to which cell or cells expressed the array, one feature in common to all of them was that hyp7, a large hypodermal syncytium containing 139 nuclei that is formed by cell fusion events (Podbilewicz and White, 1994; Sulston et al., 1983), was positive for the array (Fig. 5). In the case of nekl-3, three mosaics were identified in which the array was present only in cells derived from Cp, which gives rise to hyp11, some of the nuclei of hyp7, and 16 body muscles. In addition, on the basis of three AB(+) P1(–) nekl-3 mosaics, neither hyp11 nor the body muscles are likely to be the foci of nekl-3 activity as only one body muscle (AB.prpppppaa) descends from AB (Sulston et al., 1983) and hyp11 lacked expression in these mosaics (Fig. 5). Furthermore, nekl-3 mosaics were identified in which the array was present only in ABarp, ABarpaa, or ABpla, consistent with a hyp7 focus (Fig. 5). Finally, four mosaics were identified in which the hyp7 syncytium was the only nekl-3(+) cell in the animal, although the precursor cell that contributed the array to hyp7 could not be determined in these mosaics.
Fig. 5. Mosaic analyses implicate hyp7 as the focus of gene activity for nekl-2 and nekl-3.
Results from healthy L4 or adult mosaics in which nekl-2 or nekl-3 were inherited within a restricted portion of the lineage. The number of mosaics in which a particular cell established a positive clone for nekl-2 or nekl-3 are indicated within white or black boxes next to the division (horizontal lines) that produced that cell. Both genes have a similar pattern and have positive clones that in all cases contribute descendants to the large hyp7 hypodermal syncytium. In addition to the mosaics depicted, hyp7 was the only positive cell in four nekl-3 mosaics and one nekl-2 mosaic.
Similarly, inheritance of a nekl-2-rescuing array either by AB only (eight mosaics) or by P1 only (four mosaics) was sufficient to rescue the nekl-2 mutant phenotype (Fig. 5). For all of the healthy mosaics, the only tissue that was always positive for GFP was hyp7. That hyp7 is the focus for nekl-2 was strengthened by some specific patterns of inheritance. For example, in one animal, ABplaa appeared to be the only cell to establish a positive clone, with the array being expressed in hyp3, hyp4, hyp7, and several head neurons. On the P1 side of the lineage, several mosaics were positive for hyp7 and for body muscles that arise from the P2 cell (Fig. 5). Of particular note, establishment of a positive clone only by Cpa, which contributes seven nuclei to hyp7 and one to hyp11, was sufficient for robust rescue, and, in one mosaic, only hyp7 nuclei expressed GFP (data not shown). Thus, the nekl-2 analysis bears a striking resemblance to that of nekl-3, with hyp7 implicated as the focus for both genes.
Notably, the mosaic analyses provided no evidence for a maternal contribution from either nekl-2 or nekl-3 that might mask a requirement for either gene, for example, during embryogenesis: the progeny of germ-line mosaics had the same phenotype as that of homozygotes that segregated from heterozygous mothers. This is in agreement with the RNAi studies mentioned above, as RNAi is likely to deplete or reduce expression of the targeted genes in the germ line. We also note that a genome-wide RNAi screen suggested that nekl-2 affects the development, function, or integrity of the vulva (Parry et al., 2007). Although nekl-2(gk839) normally confers an early larval arrest that precludes an examination of the vulva, an effect of nekl-2 on the vulva could be examined in certain mosaics that were rescued for the early requirement but were array minus for the cells of the vulva. We observed that the vulva was morphologically normal in nine of nine mosaics in which the vulva failed to inherit the array and six animals with array-minus vulvae were observed to lay eggs. Thus, the vulva appeared not to require nekl-2, although it is possible that nekl-2 activity in another tissue may affect vulval integrity.
Fluorescent nekl-2 and nekl-3 constructs are expressed in hyp7
We analyzed nekl-3 expression with strains containing eqIs3, a spontaneous insertion of a transgene expressing a functional C-terminal fusion of GFP to full-length NEKL-3. NEKL-3::GFP expression initiated in the hypodermis of embryos coincident with the onset of morphogenesis (Fig. 6A) and increased during the remainder of embryogenesis (Fig. 6B-D). In larvae and adults, NEKL-3::GFP was observed in hyp7 and other hypodermal cells but not in seam cells. In hyp7, NEKL-3::GFP was present in small spheres or puncta and small tubules that were almost exclusively in the extreme apical region and apparently beneath the plasma membrane (Fig. 6E). Although not particularly mobile, some movement of the puncta was observed. In most worms, GFP was also present in hyp7 at its boundary with the seam cells (Fig. 6E). NEKL-3::GFP was also observed in the vulva and in certain undefined neurons that make contact with body muscles and in the uterus (data not shown).
We also analyzed expression of a functional full-length C-terminal fusion of mCherry to NEKL-2. NEKL-2::mCherry was observed in spheres or puncta near the apical surface of the hypodermis including the hyp7 syncytium (Fig. 6F). Similar to NEKL-3::GFP, NEKL- 2::mCherry was not observed in the seam cells (data not shown). In contrast to NEKL-3::GFP, NEKL-2::mCherry was not observed in the vulva (data not shown). Of particular interest, NEKL-3::GFP puncta only rarely co-localized with NEKL-2::mCherry in hyp7, suggesting that these kinases may function in distinct cellular compartments (Fig. 6G,H).
nekl-2, but not nekl-3, prevents endocytosis of LRP-1::GFP from the apical surface of hyp7
Because of their requirement for proper molting and their expression in hyp7, we examined nekl-2 and nekl-3 for effects on the localization of LRP-1::GFP, a convenient marker for endocytosis from the apical membrane of hyp7 (Kang et al., 2013). lrp-1 is also required for proper molting, and, at steady state, LRP-1::GFP is predominantly associated with endosomes that lie beneath the apical membrane (Kang et al., 2013; Yochem et al., 1999). Notably, in 44/50 rrf-3(pk1426) (RNAi hypersensitive) animals treated with nekl-2 RNAi (nekl-2 deletion mutations were not used in these studies, because the small size of the arrested worms made GFP observations difficult), LRP-1::GFP expression was strikingly altered from the normal endosomal pattern to one indicating dramatic accumulation on the apical membrane (Fig. 7). This pattern resembles that seen when LRP-1::GFP worms are treated with RNAi that targets proteins known to be required for endocytosis, such as CHC-1, the clathrin heavy chain ortholog, or DAB-1, an ortholog of Drosophila Disabled (Kang et al., 2013) (Fig. 7). These results suggest that NEKL-2 is required for the formation or pinching off of clathrin-coated pits in the apical membrane of hyp7.
Fig. 7. NEKL-2 affects endocytosis of LRP-1::GFP from the apical surface of hyp7.
The major localization of LRP-1::GFP in hyp7 of living worms changes from endosomes in the apical region (control RNAi in wild type) to a smeared pattern on the apical surface upon treatment with nekl-2 RNAi. The pattern resembles the apical accumulation of LRP-1::GFP conferred by dab-1(gk291). In contrast, subcellular localization of LRP-1::GFP is not strongly perturbed by nekl-3(sv3), qua-1(RNAi), or fbn-1(tm290), which also affect the completion of molting. For all LRP-1::GFP experiments, the strains contained the rrf-3(pk1426) mutation, which confers RNAi hypersensitivity. Scale bar = 5 µm. For additional information, see Fig. S3.
In contrast to the nekl-2(RNAi) studies, LRP-1::GFP did not accumulate on the apical surface of hyp7 in nekl-3(sv3) or nekl-3(gk506) homozygotes (because nekl-3(RNAi) has low efficacy, the mutations were used; Fig. 7 and data not shown). Instead, approximately half of the worms exhibited the wild-type pattern (Fig. 7). For the other half, the spacing between puncta was variably altered, but the puncta remained apical, suggesting that they were endosomes (supplementary material Fig. S3). Because this alteration of the wild-type pattern could be a mechanical consequence of the constriction caused by the unshed cuticle and by an overall unhealthy state of the mutants, LRP-1::GFP was examined in animals treated with RNAi against a hedgehog-like protein, QUA-1, or against FBN-1, which resembles fibrillin. Inhibition of either of these causes a molting defect (Frand et al., 2005; Hao et al., 2006; Kang et al., 2013), but neither protein is likely to function in membrane trafficking. For both proteins, the localization of LRP-1::GFP was normal in some animals but moderately changed in others, sometimes in a manner similar to that seen with the nekl-3 mutations (Fig 7.; supplementary material Fig. S3), suggesting that constrictions caused by the old cuticle can indeed mechanically alter the spacing between endosomes. In any case, the nekl-2 and nekl-3 mutant patterns differed strongly from each other in terms of appearance and penetrance of the defect. Although a role, if any, for NEKL-3 in trafficking is currently unclear, NEKL-2 appears to have a significant and broad function in endocytosis from the apical surface of a polarized epithelium in C. elegans.
nekl-2 is required for the correct localization of multiple endocytic compartment markers
To determine if NEKL-2 has a more general role in vesicular trafficking at the apical surface we examined the localization of four markers for early endosomes in wild-type and nekl-2(RNAi) larvae. For these studies we used either lin-35 mutations or pretreatment of marker strains with lin-35(RNAi) to increase the sensitivity of strains to nekl-2(RNAi) (Wang et al., 2005), which led to a majority of treated larvae displaying molting defects, although some animals were able to reach adulthood. SNX-1 is a component of the retromer complex, co-localizes with early endosomes and regulates clathrin dynamics (Sato et al., 2014; Shi et al., 2009). Moreover, SNX- 1 expression overlaps extensively with LRP-1 (data not shown). As expected, SNX-1::GFP expression was altered in a manner similar to LRP-1::GFP following nekl-2(RNAi) with expression becoming diffuse across the apical surface (Figs. 7 and 8). We next tested CHC-1, a clathrin heavy chain ortholog that is essential for clathrin-mediated endocytosis (Grant and Hirsh, 1999; Sato et al., 2014; Shen et al., 2013). Again, diffuse apical expression was observed following inhibition of nekl-2 (Fig. 8), indicating that NEKL-2 plays an important role in the early formation of endosomes in the larval hypodermis. Two additional markers, EEA-1::GFP and RAB-5::GFP were also examined. EEA-1 is an effector of RAB-5, a small GTPase required for early endosome movement, docking and fusion (Christoforidis et al., 1999; Nielsen et al., 1999; Sato et al., 2014). Although both markers were altered in nekl-2(RNAi) larvae, they displayed distinct defects. In the case of EEA-1::GFP, nekl-2(RNAi) most typically led to diffuse apical expression. The effect of nekl-2(RNAi) on RAB-5::GFP localization was somewhat weaker and more variable than that observed for the other markers, but included both diffuse expression as well as the formation of larger puncta or blobs (Fig. 8 and data not shown). Taken together, our results demonstrate that NEKL-2 plays a broad role in the regulation of endocytosis in hyp7.
DISCUSSION
Our studies have implicated NEKL-2 and NEKL-3, two conserved members of the NEK family in C. elegans, in the process of larval molting. Both nekl-2 and nekl-3 are predicted to encode serine-threonine kinases that were required in hyp7, the major source of the cuticle of C. elegans. NEKL-2 affected endocytosis of LRP-1::GFP, as well as a number of additional markers for endocytosis, from the apical surface of hyp7, a polarized epithelium. In contrast, NEKL-3 did not have an obvious effect on LRP-1::GFP, but its expression pattern and requirement in hyp7 suggests that it might be involved in a separate aspect of membrane trafficking. That the specific cellular functions and targets of these two kinases might differ from each is also supported by a lack of overlap in their expression patterns at the apical surface of hyp7. The striking similarity in their phenotypes and the enhancement of nekl-3(sv3) by nekl-2 RNAi does, however, suggest that these genes have overlapping or complementary functions. For example, NEKL-3 might be required for the recycling of receptors such as LRP-1 from endosomes to the plasma membrane of hyp7. A potential combination of partially blocking endocytosis with nekl-2(RNAi) and partially blocking recycling with nekl-3(sv3) might therefore prove strongly additive or synergistic. Our hypothesis that NEKL-2 and NEKl-3 function in vesicular trafficking is supported by a high-throughput RNAi study of mammalian protein kinases, which identified NEK2, NEK6, NEK7 and NEK8 as candidate regulators of endocytosis (Pelkmans et al., 2005). Thus, trafficking and endocytosis may represent an evolutionarily conserved function for some NEK family members. Importantly, our studies represent the first genetic demonstration of a role for NEK family members in vesicular trafficking and ECM remodeling within an intact organism.
Most studies of mammalian NEK6, NEK7, and other NIMA-related kinases, some of which have used dominant-negative forms of the genes, suggest an extensive involvement in cell division (Fry et al., 2012; Moniz et al., 2011; O'Regan et al., 2007). In contrast, no association with the cell cycle or cell division was apparent for either NEKL-2 or NEKL-3 during embryogenesis or early larval development. Embryos derived from germ-line mosaics, and therefore completely lacking nekl-2 or nekl-3 function, or derived from mothers subjected to RNAi, appeared completely normal. For each gene under either condition, a defect only became apparent when larvae entered the first molt. In addition, we found that brood sizes in nekl-2 and nekl-3 germ- line mosaic animals were not strongly affected, indicating that these genes are not likely to play a role in germ cell proliferation (Supplementary File 1). Also, expression of fully functional reporter constructs for both genes was largely restricted to the hypodermis. This lack of broad expression suggests that these genes are unlikely to regulate a basic cellular function, such as division, at least in the majority of cell types. Although genetic redundancy between C. elegans NEKs could potentially account for a lack of obvious cell division defects, the worm genes lack paralogs. In addition, embryonic defects were not observed in nekl-3 mutants that had been subjected to nekl-1, nekl-2, or nekl-4 RNAi (Fig. 4 and data not shown). Curiously, deletion of NEK6 in mice does not lead to any phenotype under standard laboratory conditions (Bian et al., 2014). Furthermore, NEK7−/− mice progress to either late embryogenesis or early postnatal stages of development before arresting (Salem et al., 2010). Although isolated mouse embryonic fibroblast cells from NEK7−/− mice do show indications of cell cycle disturbances, the lack of early embryonic arrest or documentation of cell cycle defects in the majority of cell types raises questions about the importance of NEK7 in the global regulation of the cell cycle. Although it could be argued that double mutants of NEK6 and NEK7 may be required to observe a stronger cell cycle phenotype in mice, this argument does not apply to C. elegans, which contains only a single ancestral ortholog of these genes, nekl-3. In addition, it has been reported that murine NEK6 and NEK7 have only a limited overlap in their expression patterns (Feige and Motro, 2002).
Three additional differences between the worm and mammalian genes are suggested by our studies. The first is the failure of RNAi of NEKL-1, the putative ortholog of NEK9, to elicit a molting defect, as it should if its product, by analogy to mammalian studies, activates NEKL-3. This is a preliminary result, however, as not all genes in C. elegans respond to RNAi (Fire et al., 1998). Second, a nekl-3 plasmid was engineered to have mutations (K52M and K53M) that correspond to kinase-dead dominant-negative forms of mammalian NEK6 (K74M and K75M) and NEK7 (K63M and K64M) (O'Regan and Fry, 2009). In contrast to the tissue-culture studies, however, wild-type worms transgenic for multiple copies of the plasmid had no obvious defects (data not shown), although as expected, this construct also failed to rescue the sv3 mutation (data not shown), indicating that these amino acids are critical for function. Three, overexpression of a NEKL-3 variant (Y86A) that corresponds to constitutively active versions of NEK6 and NEK7 (Richards et al., 2009) rescued nekl-3 mutations but otherwise conferred no obvious phenotype (data not shown). In contrast, hyperactivity of NEK6 or NEK7 in tissue culture leads to various defects including cancer progression (Cao et al., 2012; Cohen et al., 2013; Fry et al., 2012; Jee et al., 2010; Jee et al., 2011; Kim and Rhee, 2011; Moniz et al., 2011; Nassirpour et al., 2010). Interestingly, the equivalent tyrosine residues in NEK6 (Y108A) and NEK7 (Y97A) are phosphorylated by NEK9, the mammalian ortholog of NEKL-1, which did not appear to functionally interact with NEKL-2 and NEKL-3.
Questions remain concerning the molecular and developmental basis for the Corset phenotype and the correlation between the Corset phenotype and the severity of knockdown for nekl-2 and nekl-3. The rarity of the Corset phenotype in various molting screens does, however, suggest that these genes represent a subclass of molting genes. Studies are underway with the hope that they will provide insight into the completion of molting with potential relevance to polarized epithelia of worms and mammals. In any case, our studies of nekl-2 and nekl-3 suggest a serious evaluation of members of the NIMA family in processes other than cell division.
Supplementary Material
Fig. S1. A phylogenetic tree of NIMA family kinases generated with MAFTT in combination with ProtTest 2.4 and PhyML 3.0. Red, Aspergillus nidulans (An) NIMA, the founding member of the family; orange, the two Drosophila melanogaster (Dm) members; green, the four C. elegans (Ce) members; blue, the eleven Homo sapiens (Hs) members. Numbers indicate branch lengths. Scale bar = 0.1 branch length.
Fig. S2. Amino acid alignments of C. elegans NEKLs (A,B) Complete peptide alignments of (A) NEKL-2 with human NEK8 and NEK9 and (B) NEKL-3 with NEKL-2. In peptide alignments, black boxes with white lettering indicate identical amino acids; gray boxes with black lettering, similar amino acids; white background with black lettering, dissimilar amino acids.
Fig. S3. Molting defects may non-specifically alter LRP-1::GFP expression in some worms. nekl-3(sv3), qua-1(RNAi), and fbn-1(tm290) can lead to an altered patchy expression of LRP- 1::GFP. Unlike nekl-2(RNAi), which leads to variable defects in LRP-1::GFP expression in 100% of worms, nekl-3(sv3), qua-1(RNAi), and fbn-1(tm290) cause LRP-1::GFP misexpression in only about 50% of worms, despite the fully penetrant molting defects displayed by all three strains (data not shown). Scale bar = 5 µm.
Supplementary Table 1. Table of primer sequences used in this study.
Supplementary File 1. Excel file with data that accompany Figures 3 and 4 along with the results of statistical analyses for comparisons between the various genotypes. In addition, further descriptive phenotypes for RNAi results are provided.
Highlights.
Two NEK kinase family members in C. elegans, NEKL-2 and NEKl-3, control molting.
NEKL-2 and NEKL-3 localize to the apical surface of epidermal cells in puncta.
C. elegans NEKL-2 and NEKL-3 may function as regulators of vesicular trafficking.
NEKL-2 regulates the endocytosis of LRP-1, a megalin ortholog required for molting.
5) In contrast to expectations, NEKL-2 and NEKl-3 do not control cell division.
Fig. 1. The Corset or Dumbbell phenotype.
(A) sv3 heterozygous larvae develop normally and do not show molting defects. (B,C) A similarly staged sv3 homozygote that has failed to shed the old cuticle from the middle section (corset phenotype) is shown (B) along with a schematic representation of a worm with a constricted middle section (C), in which the gray lines indicate unshed old cuticle. (D-F) In contrast, the corset defect is less frequently seen with other mutations that affect the completion of molting such as dab-1(gk291). Pictured are images of dab-1(gk291) mutants in which the larva has a narrow constriction only (D; black arrowheads), is completely enclosed in old cuticle (E; black arrow indicates unshed cuticle in head region), or has failed to completely shed the old cuticle from the tail (F; white arrow). Black scale bars = 50 µm; white scale bars = 100 µm.
ACKNOWLEDGEMENTS
Cosmid clones were generously provided by Audrey Fraser, Ratna Shownkeen, and Alan Coulson of the Wellcome Trust Sanger Institute, Hinxton, UK. We thank Barth Grant for generously providing C. elegans strains. Some strains were provided by the CGC, which is funded by the US National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). We are also greatly indebted to Bob Herman and Simon Tuck for their generous support of this project, to Amy Fluet for editing and to Jay Gatlin for help with microscopy. Support at the University of Minnesota was from NIH (grant GM22387 to LC). Support at the University of Wyoming was from NIH (grant GM066868 to DSF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahringer J. WormBook. The C. elegans Research Community; 2005. Reverse Genetics. [Google Scholar]
- Altincicek B, Fischer M, Luersen K, Boll M, Wenzel U, Vilcinskas A. Role of matrix metalloproteinase ZMP-2 in pathogen resistance and development in Caenorhabditis elegans. Dev Comp Immunol. 2010;34:1160–1169. doi: 10.1016/j.dci.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Belham C, Comb MJ, Avruch J. Identification of the NIMA family kinases NEK6/7 as regulators of the p70 ribosomal S6 kinase. Curr Biol. 2001;11:1155–1167. doi: 10.1016/s0960-9822(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Belham C, Roig J, Caldwell JA, Aoyama Y, Kemp BE, Comb M, Avruch J. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem. 2003;278:34897–34909. doi: 10.1074/jbc.M303663200. [DOI] [PubMed] [Google Scholar]
- Bertran MT, Sdelci S, Regue L, Avruch J, Caelles C, Roig J. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 2011;30:2634–2647. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Liao H, Zhang Y, Wu Q, Zhou H, Yang Z, Fu J, Wang T, Yan L, Shen D, Li H, Tang Q. Never in mitosis gene A related kinase-6 attenuates pressure overload-induced activation of the protein kinase B pathway and cardiac hypertrophy. PLoS One. 2014;9:e96095. doi: 10.1371/journal.pone.0096095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Xia Y, Yang J, Jiang J, Chen L, Ni R, Li L, Gu Z. Clinical and biological significance of never in mitosis gene A-related kinase 6 (NEK6) expression in hepatic cell cancer. Pathol Oncol Res. 2012;18:201–207. doi: 10.1007/s12253-011-9429-0. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Lin JR, Vannier JB, Slaats GG, Kile AC, Paulsen RD, Manning DK, Beier DR, Giles RH, Boulton SJ, Cimprich KA. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol Cell. 2013;51:423–439. doi: 10.1016/j.molcel.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Cohen S, Aizer A, Shav-Tal Y, Yanai A, Motro B. Nek7 kinase accelerates microtubule dynamic instability. Biochim Biophys Acta. 2013;1833:1104–1113. doi: 10.1016/j.bbamcr.2012.12.021. [DOI] [PubMed] [Google Scholar]
- Davis MW, Birnie AJ, Chan AC, Page AP, Jorgensen EM. A conserved metalloprotease mediates ecdysis in Caenorhabditis elegans. Development. 2004;131:6001–6008. doi: 10.1242/dev.01454. [DOI] [PubMed] [Google Scholar]
- Entchev EV, Kurzchalia TV. Requirement of sterols in the life cycle of the nematode Caenorhabditis elegans. Semin Cell Dev Biol. 2005;16:175–182. doi: 10.1016/j.semcdb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Feige E, Motro B. The related murine kinases, Nek6 and Nek7, display distinct patterns of expression. Mech Dev. 2002;110:219–223. doi: 10.1016/s0925-4773(01)00573-1. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, O'Regan L, Sabir SR, Bayliss R. Cell cycle regulation by the NEK family of protein kinases. J Cell Sci. 2012;125:4423–4433. doi: 10.1242/jcs.111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hao L, Mukherjee K, Liegeois S, Baillie D, Labouesse M, Burglin TR. The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev Dyn. 2006;235:1469–1481. doi: 10.1002/dvdy.20721. [DOI] [PubMed] [Google Scholar]
- Hashmi S, Zhang J, Oksov Y, Lustigman S. The Caenorhabditis elegans cathepsin Z-like cysteine protease, Ce-CPZ-1, has a multifunctional role during the worms' development. J Biol Chem. 2004;279:6035–6045. doi: 10.1074/jbc.M312346200. [DOI] [PubMed] [Google Scholar]
- Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- Holmes A, Flett A, Coudreuse D, Korswagen HC, Pettitt J. C. elegans Disabled is required for cell-type specific endocytosis and is essential in animals lacking the AP-3 adaptor complex. J Cell Sci. 2007;120:2741–2751. doi: 10.1242/jcs.03474. [DOI] [PubMed] [Google Scholar]
- Jackson PK. Nek8 couples renal ciliopathies to DNA damage and checkpoint control. Mol Cell. 2013;51:407–408. doi: 10.1016/j.molcel.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Jee HJ, Kim AJ, Song N, Kim HJ, Kim M, Koh H, Yun J. Nek6 overexpression antagonizes p53-induced senescence in human cancer cells. Cell Cycle. 2010;9:4703–4710. doi: 10.4161/cc.9.23.14059. [DOI] [PubMed] [Google Scholar]
- Jee HJ, Kim HJ, Kim AJ, Song N, Kim M, Lee HJ, Yun J. The inhibition of Nek6 function sensitizes human cancer cells to premature senescence upon serum reduction or anticancer drug treatment. Cancer Lett. 2013;335:175–182. doi: 10.1016/j.canlet.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Jee HJ, Kim HJ, Kim AJ, Song N, Kim M, Yun J. Nek6 suppresses the premature senescence of human cancer cells induced by camptothecin and doxorubicin treatment. Biochem Biophys Res Commun. 2011;408:669–673. doi: 10.1016/j.bbrc.2011.04.083. [DOI] [PubMed] [Google Scholar]
- Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- Kamikura DM, Cooper JA. Clathrin interaction and subcellular localization of Ce-DAB-1, an adaptor for protein secretion in Caenorhabditis elegans. Traffic. 2006;7:324–336. doi: 10.1111/j.1600-0854.2006.00386.x. [DOI] [PubMed] [Google Scholar]
- Kandli M, Feige E, Chen A, Kilfin G, Motro B. Isolation and characterization of two evolutionarily conserved murine kinases (Nek6 and nek7) related to the fungal mitotic regulator, NIMA. Genomics. 2000;68:187–196. doi: 10.1006/geno.2000.6293. [DOI] [PubMed] [Google Scholar]
- Kang YL, Yochem J, Bell L, Sorensen EB, Chen L, Conner SD. Caenorhabditis elegans reveals a FxNPxY-independent low-density lipoprotein receptor internalization mechanism mediated by epsin1. Mol Biol Cell. 2013;24:308–318. doi: 10.1091/mbc.E12-02-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Rhee K. NEK7 is essential for centriole duplication and centrosomal accumulation of pericentriolar material proteins in interphase cells. J Cell Sci. 2011;124:3760–3770. doi: 10.1242/jcs.078089. [DOI] [PubMed] [Google Scholar]
- Kim TH, Kim YJ, Cho JW, Shim J. A novel zinc-carboxypeptidase SURO-1 regulates cuticle formation and body morphogenesis in Caenorhabditis elegans. FEBS Lett. 2011;585:121–127. doi: 10.1016/j.febslet.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuervers LM, Jones CL, O'Neil NJ, Baillie DL. The sterol modifying enzyme LET-767 is essential for growth, reproduction and development in Caenorhabditis elegans. Mol Genet Genomics. 2003;270:121–131. doi: 10.1007/s00438-003-0900-9. [DOI] [PubMed] [Google Scholar]
- Kuzmanov A, Karina EI, Kirienko NV, Fay DS. The conserved PBAF nucleosome- remodeling complex mediates the response to stress in Caenorhabditis elegans. Mol Cell Biol. 2014;34:1121–1135. doi: 10.1128/MCB.01502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Kim HJ, Kim MA, Jee HJ, Kim AJ, Bae YS, Park JI, Chung JH, Yun J. Nek6 is involved in G2/M phase cell cycle arrest through DNA damage-induced phosphorylation. Cell Cycle. 2008;7:2705–2709. doi: 10.4161/cc.7.17.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Paik YK. A potential role for fatty acid biosynthesis genes during molting and cuticle formation in Caenorhabditis elegans. BMB Rep. 2011;44:285–290. doi: 10.5483/BMBRep.2011.44.4.285. [DOI] [PubMed] [Google Scholar]
- Liegeois S, Benedetto A, Michaux G, Belliard G, Labouesse M. Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics. 2007;175:709–724. doi: 10.1534/genetics.106.066035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lu W, Obara T, Kuida S, Lehoczky J, Dewar K, Drummond IA, Beier DR. A defect in a novel Nek-family kinase causes cystic kidney disease in the mouse and in zebrafish. Development. 2002;129:5839–5846. doi: 10.1242/dev.00173. [DOI] [PubMed] [Google Scholar]
- Lizcano JM, Deak M, Morrice N, Kieloch A, Hastie CJ, Dong L, Schutkowski M, Reimer U, Alessi DR. Molecular basis for the substrate specificity of NIMA-related kinase-6 (NEK6). Evidence that NEK6 does not phosphorylate the hydrophobic motif of ribosomal S6 protein kinase and serum- and glucocorticoid-induced protein kinase in vivo. J Biol Chem. 2002;277:27839–27849. doi: 10.1074/jbc.M202042200. [DOI] [PubMed] [Google Scholar]
- Mahjoub MR, Qasim Rasi M, Quarmby LM. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol Biol Cell. 2004;15:5172–5186. doi: 10.1091/mbc.E04-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Trapp ML, Quarmby LM. NIMA-related kinases defective in murine models of polycystic kidney diseases localize to primary cilia and centrosomes. J Am Soc Nephrol. 2005;16:3485–3489. doi: 10.1681/ASN.2005080824. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz L, Dutt P, Haider N, Stambolic V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011;6:18. doi: 10.1186/1747-1028-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve GC, Frand AR. Toward a unified model of developmental timing: A "molting" approach. Worm. 2012;1:221–230. doi: 10.4161/worm.20874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motose H, Hamada T, Yoshimoto K, Murata T, Hasebe M, Watanabe Y, Hashimoto T, Sakai T, Takahashi T. NIMA-related kinases 6, 4, and 5 interact with each other to regulate microtubule organization during epidermal cell expansion in Arabidopsis thaliana. Plant J. 2011;67:993–1005. doi: 10.1111/j.1365-313X.2011.04652.x. [DOI] [PubMed] [Google Scholar]
- Motose H, Takatani S, Ikeda T, Takahashi T. NIMA-related kinases regulate directional cell growth and organ development through microtubule function in Arabidopsis thaliana. Plant Signal Behav. 2012;7:1552–1555. doi: 10.4161/psb.22412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassirpour R, Shao L, Flanagan P, Abrams T, Jallal B, Smeal T, Yin MJ. Nek6 mediates human cancer cell transformation and is a potential cancer therapeutic target. Mol Cancer Res. 2010;8:717–728. doi: 10.1158/1541-7786.MCR-09-0291. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- O'Regan L, Blot J, Fry AM. Mitotic regulation by NIMA-related kinases. Cell Div. 2007;2:25. doi: 10.1186/1747-1028-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Regan L, Fry AM. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol Cell Biol. 2009;29:3975–3990. doi: 10.1128/MCB.01867-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J Cell Biol. 1983;96:1155–1158. doi: 10.1083/jcb.96.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Pu RT, Morris NR. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell. 1988;53:237–244. doi: 10.1016/0092-8674(88)90385-6. [DOI] [PubMed] [Google Scholar]
- Otto EA, Trapp ML, Schultheiss UT, Helou J, Quarmby LM, Hildebrandt F. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. Genomewide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B, White JG. Cell fusions in the developing epithelial of C. elegans. Dev Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards MW, O'Regan L, Mas-Droux C, Blot JM, Cheung J, Hoelder S, Fry AM, Bayliss R. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol Cell. 2009;36:560–570. doi: 10.1016/j.molcel.2009.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B, Clucas C, Johnstone IL. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol Biol Cell. 2003;14:4414–4426. doi: 10.1091/mbc.E03-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier N, Lefebvre C, Legouis R. CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic. 2005;6:695–705. doi: 10.1111/j.1600-0854.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- Salem H, Rachmin I, Yissachar N, Cohen S, Amiel A, Haffner R, Lavi L, Motro B. Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene. 2010;29:4046–4057. doi: 10.1038/onc.2010.162. [DOI] [PubMed] [Google Scholar]
- Sato K, Norris A, Sato M, Grant BD. C. elegans as a model for membrane traffic. WormBook. 2014:1–47. doi: 10.1895/wormbook.1.77.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdelci S, Bertran MT, Roig J. Nek9, Nek6, Nek7 and the separation of centrosomes. Cell Cycle. 2011;10:3816–3817. doi: 10.4161/cc.10.22.18226. [DOI] [PubMed] [Google Scholar]
- Shen Q, He B, Lu N, Conradt B, Grant BD, Zhou Z. Phagocytic receptor signaling regulates clathrin and epsin-mediated cytoskeletal remodeling during apoptotic cell engulfment in C. elegans. Development. 2013;140:3230–3243. doi: 10.1242/dev.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Sun L, Banerjee R, Tobin M, Zhang Y, Grant BD. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 2009;28:3290–3302. doi: 10.1038/emboj.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Singh RN, Sulston JE. Some observations on the moulting of Caenorhabditis elegans. Nematologica. 1978;24:63–71. [Google Scholar]
- Smith LA, Bukanov NO, Husson H, Russo RJ, Barry TC, Taylor AL, Beier DR, Ibraghimov-Beskrovnaya O. Development of polycystic kidney disease in juvenile cystic kidney mice: insights into pathogenesis, ciliary abnormalities, and common features with human disease. J Am Soc Nephrol. 2006;17:2821–2831. doi: 10.1681/ASN.2006020136. [DOI] [PubMed] [Google Scholar]
- Stenvall J, Fierro-Gonzalez JC, Swoboda P, Saamarthy K, Cheng Q, Cacho-Valadez B, Arner ES, Persson OP, Miranda-Vizuete A, Tuck S. Selenoprotein TRXR-1 and GSR-1 are essential for removal of old cuticle during molting in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:1064–1069. doi: 10.1073/pnas.1006328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepek G, McCormack G, Birnie AJ, Page AP. The astacin metalloprotease moulting enzyme NAS-36 is required for normal cuticle ecdysis in free-living and parasitic nematodes. Parasitology. 2011;138:237–248. doi: 10.1017/S0031182010001113. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Sagoh N, Iwasaki H, Inoue H, Takahashi K. Metalloproteases with EGF, CUB, and thrombospondin-1 domains function in molting of Caenorhabditis elegans. Biol Chem. 2004;385:565–568. doi: 10.1515/BC.2004.069. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Trapp ML, Galtseva A, Manning DK, Beier DR, Rosenblum ND, Quarmby LM. Defects in ciliary localization of Nek8 is associated with cystogenesis. Pediatr Nephrol. 2008;23:377–387. doi: 10.1007/s00467-007-0692-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D, Jr., Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- Wang R, Song Y, Xu X, Wu Q, Liu C. The expression of Nek7, FoxM1, and Plk1 in gallbladder cancer and their relationships to clinicopathologic features and survival. Clin Transl Oncol. 2013;15:626–632. doi: 10.1007/s12094-012-0978-9. [DOI] [PubMed] [Google Scholar]
- Yin MJ, Shao L, Voehringer D, Smeal T, Jallal B. The serine/threonine kinase Nek6 is required for cell cycle progression through mitosis. J Biol Chem. 2003;278:52454–52460. doi: 10.1074/jbc.M308080200. [DOI] [PubMed] [Google Scholar]
- Yissachar N, Salem H, Tennenbaum T, Motro B. Nek7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett. 2006;580:6489–6495. doi: 10.1016/j.febslet.2006.10.069. [DOI] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Herman RK. Genetic mosaics. WormBook. 2005:1–6. doi: 10.1895/wormbook.1.58.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Sundaram M, Bucher EA. Mosaic analysis in Caenorhabditis elegans. Methods Mol Biol. 2000;135:447–462. doi: 10.1385/1-59259-685-1:447. [DOI] [PubMed] [Google Scholar]
- Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- Yochem JK. Nomarski images for learning the anatomy, with tips for mosaic analysis. WormBook. 2006:1–47. doi: 10.1895/wormbook.1.100.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugasti O, Rajan J, Kuwabara PE. The function and expansion of the Patched- and Hedgehog-related homologs in C. elegans. Genome Res. 2005;15:1402–1410. doi: 10.1101/gr.3935405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A phylogenetic tree of NIMA family kinases generated with MAFTT in combination with ProtTest 2.4 and PhyML 3.0. Red, Aspergillus nidulans (An) NIMA, the founding member of the family; orange, the two Drosophila melanogaster (Dm) members; green, the four C. elegans (Ce) members; blue, the eleven Homo sapiens (Hs) members. Numbers indicate branch lengths. Scale bar = 0.1 branch length.
Fig. S2. Amino acid alignments of C. elegans NEKLs (A,B) Complete peptide alignments of (A) NEKL-2 with human NEK8 and NEK9 and (B) NEKL-3 with NEKL-2. In peptide alignments, black boxes with white lettering indicate identical amino acids; gray boxes with black lettering, similar amino acids; white background with black lettering, dissimilar amino acids.
Fig. S3. Molting defects may non-specifically alter LRP-1::GFP expression in some worms. nekl-3(sv3), qua-1(RNAi), and fbn-1(tm290) can lead to an altered patchy expression of LRP- 1::GFP. Unlike nekl-2(RNAi), which leads to variable defects in LRP-1::GFP expression in 100% of worms, nekl-3(sv3), qua-1(RNAi), and fbn-1(tm290) cause LRP-1::GFP misexpression in only about 50% of worms, despite the fully penetrant molting defects displayed by all three strains (data not shown). Scale bar = 5 µm.
Supplementary Table 1. Table of primer sequences used in this study.
Supplementary File 1. Excel file with data that accompany Figures 3 and 4 along with the results of statistical analyses for comparisons between the various genotypes. In addition, further descriptive phenotypes for RNAi results are provided.