Abstract
Radical prostatectomy is a common treatment option for prostate cancer before it has spread beyond the prostate. Examination for surgical margins is performed post-operatively with positive margins reported to occur in 6.5 – 32% of cases. Rapid identification of cancerous tissue during surgery could improve surgical resection. Desorption electrospray ionization (DESI) is an ambient ionization method which produces mass spectra dominated by lipid signals directly from prostate tissue. With the use of multivariate statistics, these mass spectra can be used to differentiate cancerous and normal tissue. The method was applied to 100 samples from 12 human patients to create a training set of MS data. The quality of the discrimination achieved was evaluated using principal component analysis - linear discriminant analysis (PCA-LDA) and confirmed by histopathology. Cross validation (PCA-LDA) showed >95% accuracy. An even faster and more convenient method, touch spray (TS) mass spectrometry, not previously tested to differentiate diseased tissue, was also evaluated by building a similar MS data base characteristic of tumor and normal tissue. An independent set of 70 non-targeted biopsies from six patients was then used to record lipid profile data resulting in 110 data points for an evaluation dataset for TS-MS. This method gave prediction success rates measured against histopathology of 93%. These results suggest that DESI and TS could be useful in differentiating tumor and normal prostate tissue at surgical margins and that these methods should be evaluated intra-operatively.
Introduction
Prostate cancer is estimated to be the most commonly diagnosed cancer in the United States, representing 14% (233,000 cases) of all newly diagnosed cancer cases in 20141. Treatments for prostate cancer include surgery, radiation therapy, and hormone therapy, the choice between them being primarily dependent on the patient’s health and the stage of the cancer. At the time of diagnosis, over 90% of prostate cancer cases have tumors confined to the prostate gland, representing stage 1 or stage 2. Before cancer has spread to the outer layer of the prostate, the disease is curable through complete surgical resection by radical prostatectomy (RP)2. Standard practice relies on preoperative measurements such as rectal examination and clinical biopsy data to guide surgical resection. Positive surgical margins are only identified after completion of the surgery and their incidence is reported to range from 6.5–32%3.
Needle biopsies are used for early diagnosis of prostate cancer, however cancer detection using such biopsies is prone to false-negatives, reportedly up to 25%4. Difficulty in diagnosis is the result of the limited sample size, the limited number of malignant glands among many benign, and confusion of benign histological features which mimic prostate cancer such as paraganglia or xanthoma. Immunohistochemistry (IHC) has been used to label p63, a marker for basal cells, which is present in benign and absent in cancerous tissue. However, negative staining is not reliably diagnostic given the limited number and sizes of samples. Positive staining for α-methylacyl-CoA racemase (AMACR) can be used since it is greatly up-regulated in cancer, but this method too has pitfalls with false-negative rates up to 20–30% in cases such as pseudohyperplastic, atrophic, and foamy gland adenocarcinoma of the prostate5. Currently unrepresented in prostate molecular diagnostics is the use of the lipid constituents of tissue, either in early diagnosis or in post-surgical tumor margin diagnostics.
Lipid constituents of tissues are readily measured by ambient ionization MS methods such as desorption electrospray ionization MS (DESI-MS). The basis for ambient ionization is sample examination in the ambient environment with little to no prior sample preparation6. The two methods used in this study, DESI and touch spray (TS) ionization7 fall into this category as do others like nanospray desorption electrospray ionization (nanoDESI)8, probe electrospray ionization (PESI)9 and laser ablation electrospray ionization (LAESI)10.
A number of recent studies have reported the application of mass spectrometry (MS) as a molecular diagnostic tool for cancer. Zare and coworkers used DESI imaging with statistical analysis methods to classify gastrointestinal cancer tissues from banked and surgical specimens11. Agar and coworkers showed that it is possible to distinguish diseased from healthy brain tissue using libraries of mass spectra characteristic of particular disease states12. The work on brain cancer has allowed clear differentiation of healthy from diseased tissue as well as glioma subtype, the stage of the disease in particular tumor regions, and the tumor cell concentrations at particular locations13. Another mass spectrometry method, ultraperformance liquid chromatography MS, has been shown to accurately identify prostate cancer from serum using a metabolite-based assay14. It is possible to recognize in these and related studies the beginnings of a significant effort to perform cancer molecular diagnostics through the use of mass spectrometry (MS).
The origins of recent interest in diagnostics by mass spectrometry can be traced to the development of matrix-assisted laser desorption ionization (MALDI) imaging and its application to mapping the distribution of proteins and peptides in tissue sections15, 16. Ambient ionization methods which do not use matrices have also given highly encouraging results, especially for small molecules including drug metabolites, hormones and lipids17. The earliest application of DESI to cancer diagnosis demonstrated changes in lipid profiles in normal human liver tissue compared to metastatic liver adenocarcinoma. This study established the fact that tumor margins can be recognized by mass spectrometry imaging18. Subsequent work using DESI differentiated tumor from normal tissue using lipid profiles in kidney cancer19, bladder cancer20, testicular cancer21, and brain cancer12, 13, 22, 23. The development of ambient ionization methods for cancer diagnostics was accelerated by the rapid evaporative ionization mass spectrometry experiments of Takats and coworkers24. In this approach, the smoke produced during electrosurgical dissection is transferred and analyzed by a mass spectrometer. The smoke contains phospholipids and other biomolecules released from the region being resected and comparison to a library of spectra allows tissue identification. DESI and TS are both spray-based ambient ionization methods. However, their complementary features make them suitable for different tasks. DESI-MS is primarily an imaging technique, allowing collection of mass spectra, pixel by pixel, to create a 2D molecular image. The collected data can be assembled in hyperspectral datacubes25 from which 2D ion images corresponding to specific components can be extracted to show the spatial distribution and relative amounts of analytes. Applications of DESI in distinguishing diseased and non-diseased tissue rely on the use of multivariate statistics25, typically principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA). DESI provides a key connection between the information from histopathology and the characteristic mass spectrum that is produced for each tissue and disease type because the same (or adjacent) tissue sections analyzed by DESI can be evaluated by the pathologist. On the other hand, TS-MS is a user-guided method in which a spot of interest is directly sampled with a probe, e.g. teasing needle, and transferred to the interface of a mass spectrometer, and ionized by the application of solvent and high voltage. TS produces information-rich spectra which are similar to DESI, but in a localized and very rapid process which typically takes a few seconds.
We explore the potential of DESI-MS and TS-MS for prostate cancer differentiation through in vitro analysis of radical prostatectomy specimens. First, DESI-MS was used to establish the relationship of MS features to pathology and then touch spray was used to characterize unknown tissue samples. Training sets for both methods were built using data obtained from 12 radical prostatectomy specimens and evaluated by PCA and linear discriminant analysis (LDA). Both methods resulted in >95% correct sample identification when confirmed against histopathology. A TS-MS test dataset, obtained from another six radical prostatectomy specimens, was used for further evaluation of the method using a non-targeted analysis of the prostate tissue samples.
Results and discussion
DESI and TS lipid profiles and training datasets
Prostate cancer is a heterogeneous disease. It is commonly diagnosed by histopathology which at times can be ambiguous. The morphological variations that are the basis for diagnosis by histology result from underlying biochemical changes. The two main criteria for diagnosis are atypical architectural features (e.g. perineural infiltration) and atypical cytological features (e.g. enlarged nuclei). Prostate cancer diagnosis does not currently use the lipid components of the cancerous cells or tissue. Thus, complementary information provided by direct molecular diagnosis of prostate cancer by mass spectrometry could be useful to improve decision-making strategies. In this study, two different ionization techniques, DESI and TS, were used to acquire lipid profiles and investigate differences between tumor and normal tissue. The biochemical features recovered by DESI and TS were also compared, since each ambient ionization technique provides unique capabilities.
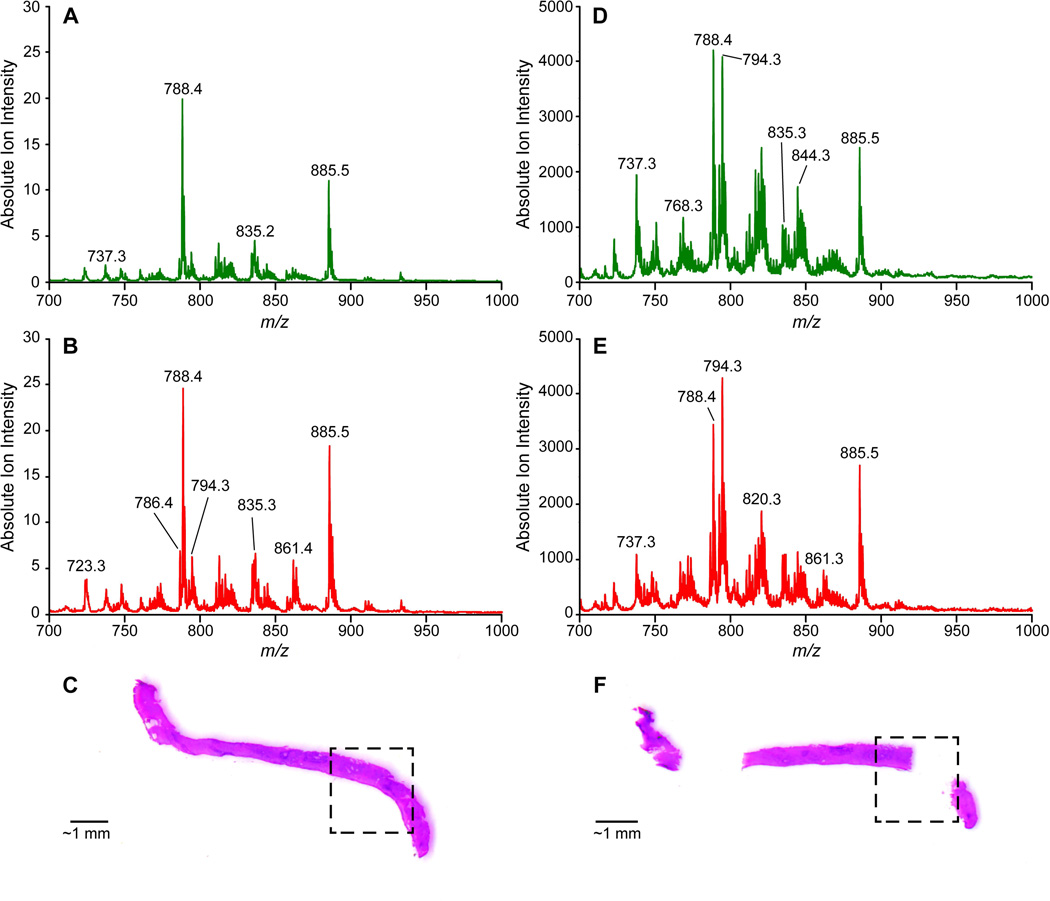
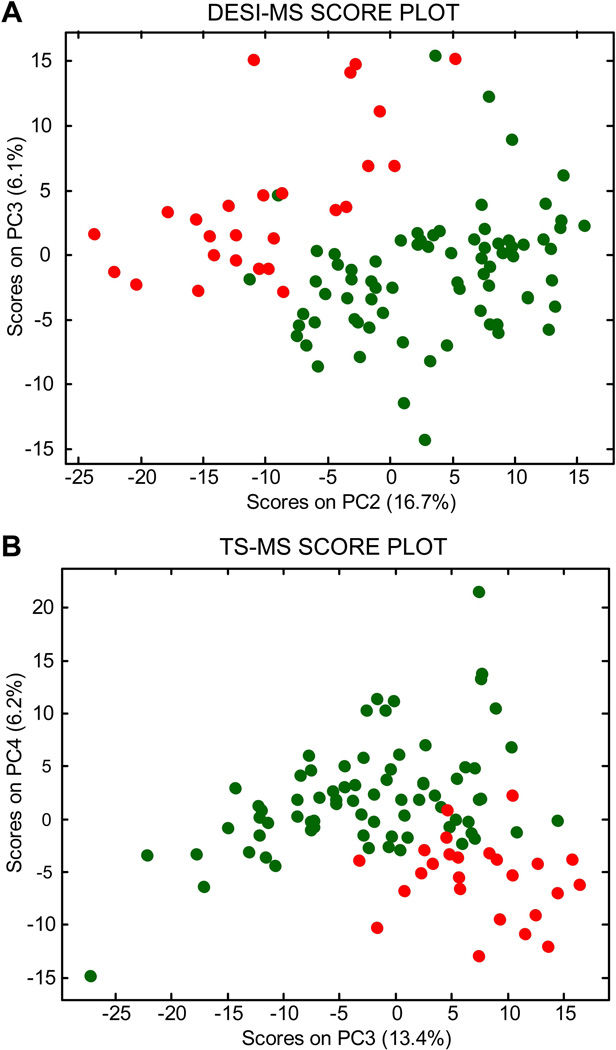
For prostate tissue, the primary components of lipid profiles (detected as negative ions) are glycerophospholipids: phosphotidylethanolamines (PhE), phosphotidylserines (PhS), phosphotidylcholines (PhC), and phosphatidylinositols (PhI). These lipids can be observed in the average normal (74 samples) and tumor (26 samples) mass spectra shown in Figure 1 for DESI (A and B) and TS (C and D). Many of the same differences between normal tissue and tumor are seen in both DESI and TS average spectra. These include ratio changes for ions of m/z 788 and 885, an increase in relative abundance of ions of m/z 786, 835, 861, 863, and a decrease in relative abundance of ions such as m/z 737. However, there are also differences in the mass spectra recorded by DESI and TS (Figure 1 A vs D, Figure 1 B vs. E). For example, chlorinated adducts on phosphatidylcholine species (e.g. m/z 794) are more abundant in the TS than the DESI spectra (as well in the case of m/z 794 being more abundant in tumor than in normal tissue). This example suggests that some differences between the ionization methods can be attributed to different salt content tolerances (TS has a lower salt tolerance than DESI) in the complex matrix when analyzed with no sample pretreatment, as discussed elsewhere8. The same spectra also show that the average ion intensity in TS is two orders of magnitude greater than that in DESI, suggesting that extraction/desorption occurs more rapidly in comparison to DESI. Considering the lipid profiles recorded for the entire dataset, 12 patients and 100 samples (thousands of mass spectra), the differences between disease states are difficult to address by eye therefore a robust statistical strategy is needed to efficiently explore the chemical information contained in the mass spectra. PCA was performed as an exploratory tool to identify chemical features that characterize tumor and normal tissue based on the DESI and TS lipid profiles. The PCA score plots are shown in Figure 2 A and B, respectively for DESI and TS. Both plots show the lowest-order principal components (PCs) for which separation between normal and tumor tissue is apparent. Indeed, distinct groupings of normal (dark green objects) and tumor tissue samples (red objects) are present. The loading plots shown in Supplementary Figure 1 A and B display those lipids that contribute the most to the PC differentiation.
Figure 1.
(A) Average DESI-MS of normal prostate tissue samples (N=74) from training set (B) Average DESI-MS of prostate cancer tissue samples (N=26) from training set (C) Prostate tissue biopsy with outlined region of cancerous tissue which was analyzed by DESI imaging (D) Average TS-MS of normal prostate tissue samples (N=74) from training set (E) Average TS-MS of prostate cancer tissue samples (N=26) from training set (F) Prostate tissue biopsy analyzed by TS-MS, tissue is adjacent to DESI imaged biopsy again the cancerous region outlined, the two missing regions correspond to locations of TS-MS analysis [Updated figure attached separately]
Figure 2.
(A) DESI-MS score plot, PC2 vs. PC3, displaying separation of prostate cancer (red objects) and normal prostate tissue samples (dark green objects) (B) TS-MS score plot, PC3 vs. PC4, displaying separation of prostate cancer (red objects) and normal prostate tissue samples (dark green objects) [Updated figure attached separately]
The first principal component by definition encompasses the largest variation in the multidimensional space of the original variables, PC2 is orthogonal and includes the largest remaining variation, etc. Separation in PC space between normal and tumor tissue is interpreted in terms of differences between their DESI (and TS) mass spectra (i.e. biochemical features). The TS spectra are usually noisier than the DESI spectra, which may be attributed to the smaller number of scans acquired over a shorter time window before signal exhaustion. In this case, the best separation among red and dark green objects is in the PC3 vs. PC4 score plot and is associated with a slightly lower percentage of the total data variation. By contrast, DESI mass spectra result from averaging many pixels in the hyperspectral images, selected as regions of interest (tumor or normal tissue sections) and this helps reduce random noise. In DESI, the best separation is along PC2 vs. PC3.
Independently of the ionization method used for analysis, the loading plots display a similar relationship between the objects and variables. This observation provides initial insight into the consistency between DESI and TS in recovering the biochemical features associated with the tumor/normal condition. In particular, the ions of m/z 835, 861, 885, and 887 are higher in relative abundance in prostate cancer tissue, while m/z 788 is higher in relative abundance in normal prostate tissue. It is noteworthy that the ions observed in higher abundance in prostate cancer tissue correspond to phosphatidylinositol species: PhI(38:4), PhI(38:3), PhI(36:4), and PhI(34:5) respectively. Oncogenes (e.g. PI3K, AKT, PTEN) pertaining to lipid-based signaling are relevant to prostate and other human cancers26–28. Intriguingly, the dynamics in the PhI class of glycerophospholipids could relate to PI3K, an oncogene which utilizes phosphorylated-PhIs (not directly detected here) for cancer cell growth and survival signaling29. The observed increase in non-phosphorylated PhIs might prove to have an unexplored but important role in prostate cancer.
Classification of prostate cancer/normal tissue by molecular profiling
LDA was performed on the DESI and TS target datasets (data that was directly correlated to identified tumor or normal samples by histopathology) to quantify the separation between normal and tumor samples and build a classification model capable of predicting the disease state of unknown samples. The first eight and nine PCs were used for DESI and TS datasets, respectively, which provided the highest prediction rates for both classes (normal and tumor tissue). The average cross-validation (CV) prediction rate was 98% for DESI and 96% for TS. The number of false results per class obtained through the cross-validation process can be seen in Table 1, where the CV confusion matrices are reported. Notably, DESI performs slightly better than TS-MS, although both methods exceed 95% in correct predictions.
Table 1.
Cross validation of training set
| (A) DESI-MS CV prediction rates | (C) TS-MS CV prediction rates | ||||
| Normal | 98.7% | Normal | 96.1% | ||
| Tumor | 96.2% | Tumor | 95.8% | ||
| Average | 97.5% | Average | 96% | ||
| (B) DESI-MS CV confusion matrix | (D) TS-MS CV confusion matrix | ||||
| Species | Normal | Tumor | Species | Normal | Tumor |
| Normal | 73 | 1 | Normal | 73 | 3 |
| Tumor | 1 | 25 | Tumor | 1 | 23 |
To further test the performance of TS-MS, which was used here for the first time to differentiate diseased and healthy tissue, an additional evaluation set was tested. Samples were acquired from patients #13–18 and a total number of 70 biopsies were performed from which 110 samples were analyzed randomly. The LDA model built on the target TS dataset was used to predict the tissue condition (tumor or normal) of these unknown samples. The LDA predictions, based on MS molecular diagnostics, were compared with pathological evaluation of adjacent, unanalysed, tissue sections. Nine samples were diagnosed as prostate cancer by an expert pathologist, while the remaining 101 samples were normal tissue. The LDA results were only moderately discriminatory which we attribute to the low reproducibility of some ions (e.g. chloride adducts) in the TS spectra acquired in full-scan MS mode. We therefore built an LDA model using the top five discriminant ions in the range m/z 700–1000, as opposed to compressing the entire mass spectra by PCA, using a stepwise variable selection strategy to select the most discriminant ions30. This strategy proved more efficient and robust in differentiating tumor and normal tissue for the TS target dataset (see Table 2), no prediction errors in cross validation. Moreover, the average percentage of correct prediction of tumor and normal tissue for the external evaluation set (110 samples) was equal to 93%. It should be noted that the small number of samples with tumor tissue was due to the non-targeted selection of the samples analyzed. These results suggest that further refinements of the strategy used for data analysis could strengthen a decision-making strategy based on molecular diagnostics. A wide portfolio of pre-treatment processes and variable selection techniques is available for pattern recognition analysis, following the needs of the acquired MS data structures.
Table 2.
Validation of test set data
| (A)TS-MS stepwise LDA on the targeted dataset | (C) TS-MS untargeted prediction rates | ||||
| Normal | 100% | Normal | 96% | ||
| Tumor | 100% | Tumor | 89% | ||
| Average | 100% | Average | 92.5% | ||
| (B) TS-MS stepwise LDA targeted confusion matrix | (D) TS-MS untargeted confusion matrix | ||||
| Species | Normal | Tumor | Species | Normal | Tumor |
| Normal | 74 | 0 | Normal | 97 | 4 |
| Tumor | 0 | 26 | Tumor | 1 | 8 |
DESI and TS similarities and fit-for-purpose applications
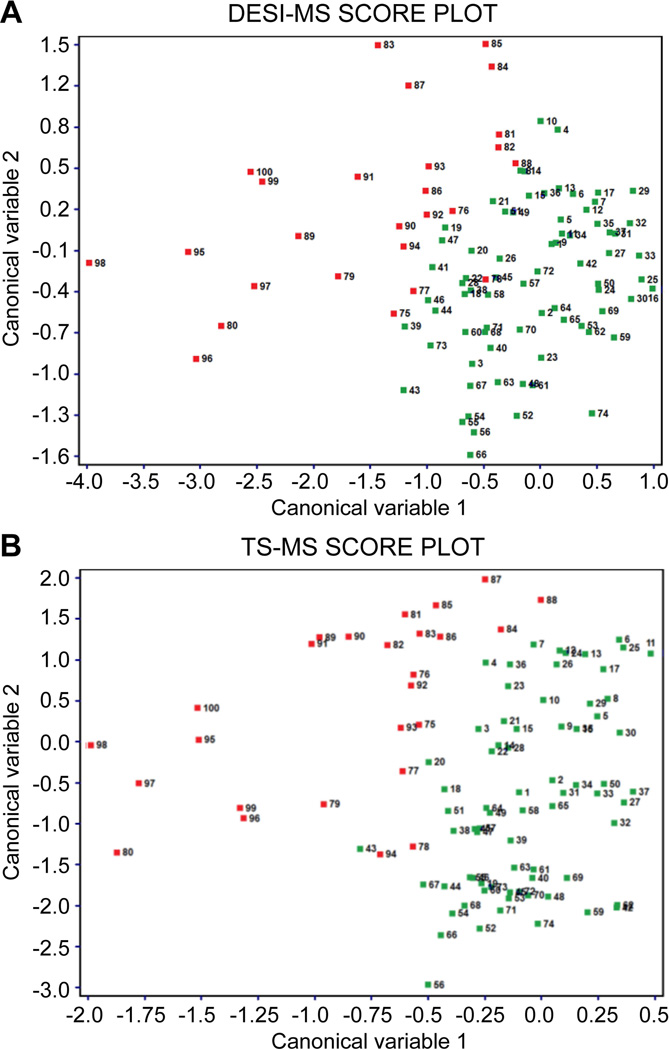
The similarities between DESI and TS mass spectra in providing chemical features associated with a healthy/disease state is of interest. Indeed, although based on similar mechanisms of ion generation (i.e. electrospray-like processes), DESI and TS have specific analytical features that may be used to propose diverse fit-for-purpose strategies of molecular diagnostics. To estimate the correlation between the two sets of MS data, canonical correlation analysis (CCA) was performed on the DESI and TS target datasets (100 samples). Figure 3 shows the samples in the canonical variable space (1 vs. 2) for DESI and TS with a clear separation between tumor (red) and normal tissue samples (dark green). The correlation coefficients for the first three canonical variables are as high as 0.94, 0.76, and 0.68 respectively (Supplemental Figure 2). The high correlation between DESI and TS is also supported by the location of each sample (labelled with the same sample number in both plots) in the CCA space. The loading plots (Supporting Figure 3) show the greatest contribution to the canonical variable computation from principal components 1, 2, and 3 for DESI, whereas principal components 2, 3, and 4 have the highest contribution for TS. Most importantly, the high correlation between DESI and TS data is related to the principal components that carry the biochemical information relevant to separating tumor and normal tissue samples.
Figure 3.
(A) Canonical variable 1 vs. 2 score plot displaying separation of tumor (red objects) and normal tissue samples (dark green objects) for DESI-MS target dataset (B) Canonical variable 1 vs. 2 score plot displaying separation of tumor (red objects) and normal tissue samples (dark green objects) for TS-MS target dataset [Updated figure attached separately]
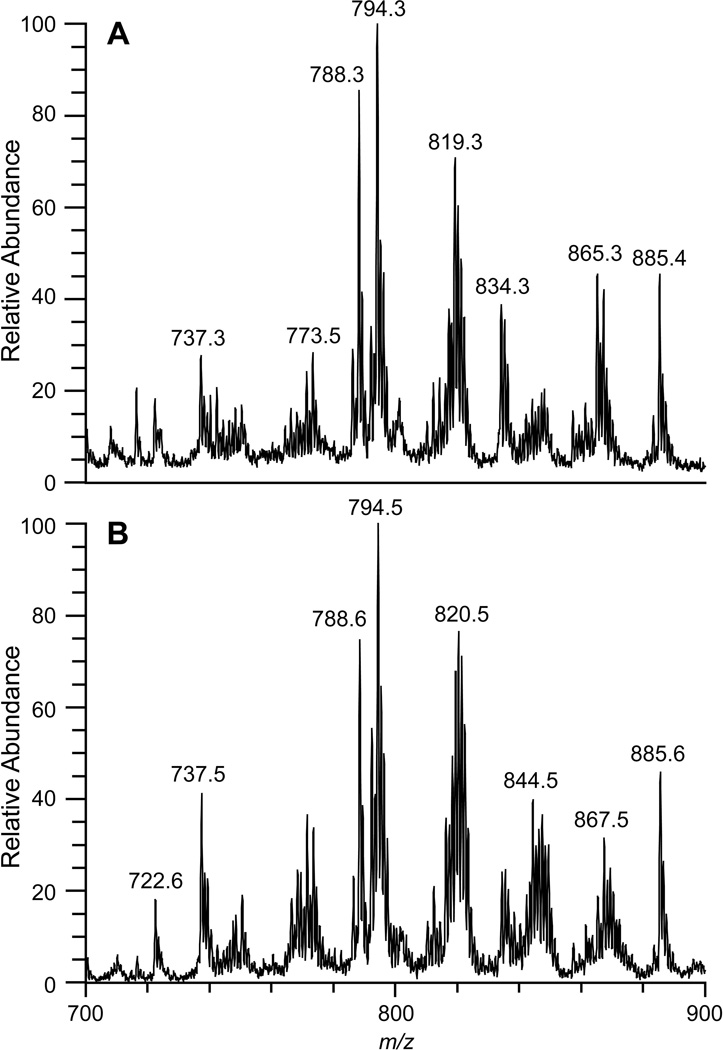
CCA results suggest that the same interpretations regarding the chemical features which characterize tumor and normal tissue can be applied to DESI and TS spectral patterns. This suggests that both methods have the same diagnostic purpose but with different, possibly complementary, implementations as now discussed. One of the most important features of TS is its use of the probe as means of sampling a complex matrix as well as serving as a source of the ions which move into the mass spectrometer upon the addition of solvent and high voltage. This transfer-based mechanism makes TS completely user-guided and applicable to a variety of samples. TS is not limited to tissue sections, but can be used to investigate areas of interest in vivo. Figure 4 shows the degree of similarity of (A) a TS mass spectrum obtained by sampling a bisected radical prostatectomy specimen in vitro and (B) a TS mass spectrum obtained from a frozen tissue section from the training set. The complete TS process of sampling to collecting data can occur within 20 seconds. Similar lipid profiles are obtained for DESI and TS, even though comparisons between ex vivo and in vivo TS analysis, as well as sources of contamination affecting the mass spectra, e.g. due to blood, urine etc., need further investigation.
Figure 4.
(A) TS-MS obtained in vitro from radical prostatectomy case 19, lipid pattern suggests sampled area was cancerous and this agrees with gross pathology findings (B) TS-MS obtained from targeted cancer sample from training set [Updated figure attached separately]
Limited sample size is the main disadvantage of TS-MS. The minimal removal of material from a sensitive source such as tissue is viewed as an advantage for the patient, but it limits the power of the technique by reducing analysis to a few MS scans before the signal is exhausted. This could be overcome by multiple examination of the same area, but each additional sampling requires more time. The limited sample size collected by the TS probe also forces the user to consider more carefully the measurement uncertainty due to sampling, which may bias the analytical results.
TS-MS is envisioned as a surgical tool for disease screening of areas of interest. As a surgical screening tool, TS could help preserve healthy tissue that may have otherwise been resected for histopathological evaluation or help with decisions to remove additional diseased tissue that was observed to be questionable.
Experimental
Study Protocols
Radical prostatectomy specimens #1–19 were obtained from consented patients undergoing treatment at Indiana School of Medicine (Indianapolis, IN) following an IRB approved study. Biopsies were obtained from specimens #1–18 after resection using a disposable biopsy gun (Max-core disposable core biopsy instrument, Bard Biopsy Systems, Tempe, AZ). Biopsies (approximately 4–15 mm × 1 mm × 1 mm) were subsequently frozen, cryosectioned at 15 µm, and thaw mounted to glass microscope slides. Sections were stored at −80° C prior to MS analysis. The same slide analyzed by DESI-MS imaging was H&E stained and examined by an expert pathologist to identify from tissue morphology the presence/absence of prostate cancer. Adjacent sections (separated by ~15 – 60 µm) were analyzed by TS-MS by sampling areas of 1–4 mm2 from regions of known pathology based on the DESI/histopathology assignment of locations as diseased or normal tissue. Note that this procedure was necessary because TS is a destructive sampling method for thin sections. Radical prostatectomy specimen #19 was inked according to IUSM gross pathology protocol then bisected to allow the tissue to be analyzed in vitro. The specimen was analyzed by TS-MS at IUSM using an on-site commercial mass spectrometer. No biopsies were taken from specimen #19 and thus no DESI-MS analysis was performed. Radical prostatectomy specimen #19 results were compared to gross pathological analysis obtained from IUSM.
A total of 100 samples were prepared from patients # 1–12 and used to create a database of tumor and normal tissue mass spectra for both DESI and TS methods. These databases are referred to as targeted datasets, because the data used is directly correlated with histopathological analysis of the tissue. For the DESI targeted datasets, regions outlined as tumor or normal by pathology were selected (~ 4 mm2 on average) and data were averaged to create each data point for subsequent multivariate analysis. In parallel, TS was performed in the regions outlined as tumor or normal tissue from adjacent tissue sections used for DESI and that data was directly used to create the TS targeted dataset. Table S1 shows in detail the number of sections prepared per biopsy and patient. Another 70 biopsies were prepared from the other 6 radical prostatectomy specimens (patients # 13–18) and these were analyzed only by TS-MS to create an evaluation dataset to further test performance. The biopsy sections used to create this independent evaluation dataset were randomly chosen and sampled 1–2 times (depending on the size of the tissue) to create a 110 sample dataset. Also included in the evaluation set were 1–2 randomly selected samples from each patient used in the targeted dataset. Histopathology of these tissue sections was performed after MS analysis on adjacent tissue (spacing ~15–60 µm).
DESI-MS
A laboratory-built DESI ion source, similar to the commercial 2D source from Prosolia, Inc. (Indianapolis, IN, USA) was coupled to a linear ion trap mass spectrometer (LTQ) controlled by XCalibur 2.0 software (ThermoFisher Scientific, Waltham, MA) and used in DESI experiments. The negative ionization mode was used with the automatic gain control (AGC) inactivated. The spray solvent used for DESI-MS was dimethylformamide (DMF)-acetonitrile (ACN) at a 1:1 ratio (v/v), both solvents were purchased from Mallinckrodt Baker Inc. (Phillipsburg, NJ), and delivered at 1.0 µL/min flow rate using the instrument syringe pump. The DESI source parameters were set as follows: capillary temperature 275°C, voltage applied to the stainless steel needle syringe 5 kV, capillary voltage −25 V, tube lens voltage −115 V, capillary incident angle 54°, spray to surface distance ~3 mm, sample to inlet distance ~5 mm, and nitrogen gas at 180 PSI. Prostate tissue sections were analyzed using a moving stage with a lateral scan rate of 303.03 µm/s in horizontal rows separated by a 200 µm vertical step. Instrument scan time was coordinated with scan speed providing ~200×200 µm pixels. Full scan mass spectra were acquired in negative ion mode in the mass range m/z 200–1000. For statistical analysis of the evaluation dataset, a reduced mass range of 700–1000 was used, limiting the biochemical information to complex phospholipids.
Touch spray MS
The TS probe used was a commercially available teasing needle purchased from Fisher Scientific (Pittsburgh, PA, USA)7. Methanol (Mallinckrodt Baker Inc., Phillipsburg, NJ) was used as spray solvent and 1 µL was applied manually via an adjustable pipette (Eppendorf Research-2.5 µL). The LTQ linear ion trap mass spectrometer was also used for the TS experiments with the same operating parameters as those used in the DESI experiments except that the voltage applied to the TS probe was 4 kV and the automatic gain control was active. TS was performed by touching and desorbing material onto a teasing needle from regions of interest of 1–4 mm2. After sampling, the tip of the probe was directed at the inlet of the mass spectrometer and the high voltage and 1 µL of solvent were applied. The extracted analytes were analyzed in a spray time of ~6 seconds. The data acquired within this period were averaged to represent a single data point.
Principal component analysis of target datasets
An in-house program was used to convert the MS data files (.raw) into ASCII files (.txt), which were imported into Matlab (MathWorks, Inc., Natick, USA). Biomap software (http://www.maldi-msi.org) was used to display single ion images (i.e. spatial distribution of a single m/z variable, Supplemental figure 4) with MS intensity represented in false color (normalized to the absolute value). For each DESI image, information was coded as a datacube (X∙Y∙MS), where “X” and “Y” are spatial dimensions and the “MS” domain contains the m/z variables and corresponding intensity (i.e. an entire mass spectrum). Groupings of adjacent pixels, i.e. regions-of-interest (ROIs), representing areas of tumor or normal tissue were selected in the 2D spatial domain (X∙Y), according to pathological evaluations, and the corresponding mass spectra were averaged. The averaged mass spectra from the prostate sections constituted the DESI target dataset. The TS target dataset was built using a list of m/z values and ion abundances from the average mass spectrum (over ~6 s of data acquisition) acquired per sample. The data were then imported into Matlab.
The acquisition step used to increment m/z values was equal to 0.0833, therefore the two data matrices (DESI and TS) consisted of 3601 columns (i.e. m/z values) and 100 rows (i.e. samples: normal tissue sections, n = 74; tumor tissue sections, n = 26). PCA was performed with in-house Matlab routines. PCA is commonly used for exploring complex information contained within mass spectral datasets, allowing consideration of all spectral variables and possible inter-correlations simultaneously25, 31. By means of PCA, the information of the original m/z variables is reorganized and compacted in principal components (PCs). That is, PCA can be used as an unsupervised data compression technique. When dealing with high-dimensional data, the compression of the relevant information is a preliminary step in order to efficiently manage and extract useful features. All MS spectra were normalized by the standard normal variate (SNV) transform, correcting for both baseline shifts and global intensity variations32, 33, and then column centered. The principal components are orthogonal (i.e. uncorrelated) and efficiently describe large fractions of the information. The projections of the data objects onto the PCs are called scores, while the importance of each original variable in defining a certain PC is given by a loading coefficient. Groupings in the score plot indicate similarities among the objects (i.e. samples), based on the information derived from the mass spectra. Both scores and loading values were represented in two-dimensional scatter plots.
Canonical correlation analysis on the target datasets
Canonical correlation analysis (CCA) is a way of measuring the linear relationship between two multidimensional variables (DESI and TS) observed on the same sample collection34, 35. In this study, the two datasets represent the DESI and TS mass spectra recorded from 100 prostate tissue sections. The DESI mass spectral dataset is considered as a reference. CCA rotates the original variables in the two blocks, to obtain some pairs of variables (one for each block), called canonical variables, with maximum correlation between the two blocks. The dimensionality of these new bases is equal to or less than the smallest dimensionality of the two sets of variables. The canonical variables are linear combinations of the original autoscaled (i.e. unitary variance) variables, whose contribution in defining a specific canonical variable can be inferred through the corresponding loading value. The correlation coefficients between the DESI canonical variable and the corresponding TS canonical variable are termed canonical correlation coefficients. Specific details on CCA can be found elsewhere34, 35. In order to overcome the high inter-correlations across m/z values, CCA was performed after PCA, which acts as an unsupervised data compression technique. Two separate PCAs were run on the SNV normalized and column-centered DESI and TS mass spectral data. The first 10 PCs, explaining about 90% of total data variation, were selected and used for CCA. CCA was performed by the free chemometric package V-PARVUS 2010 (University of Genova, Italy, (http://www.csita.unige.it/software/free/other.html).
Linear discriminant analysis on the target and the evaluation sets
Linear discriminant analysis (LDA) was performed as a supervised discriminant classification technique. Discriminant methods look for a delimiter that divides the global domain into a number of regions, each assigned to one of the classes. This delimiter identifies an open region for each class and such regions determine the assignment of the samples to one of the classes30, 36. Model validation (i.e. evaluation of the predictive ability of the model) was performed by means of cross validation (CV)36. For this study, five cross-validation deletion groups were selected, meaning that all the samples (n=100) were divided five times systematically in a training set (objects used for building the classification model) and a test set (remaining objects used to evaluate the predictive ability of the model), with all the samples being in the test set only once. Eventually, the final model was built using all the objects. LDA was applied on the DESI and TS target datasets of SNV normalized mass spectra after compression by PCA, thereby using as variables the principal components (instead of the original mass spectral data). Two classes were modelled: normal tissue (n=74) and tumor (n=26). In more detail, 8 PCs for DESI and 9 PCs for TS were computed each time with the training samples and used for building the classification model. The test sets – samples that did not contribute in the model building process - were used to estimate the global prediction rate for all classes, hence as the CV prediction rate for each class. This is a so-called complete validation strategy36. The CV prediction rate is the percentage of correct predictions on the objects in the CV test sets. The CV confusion matrix shows how many samples belonging to a certain category were correctly/incorrectly assigned by the classification rule to that category. Indeed, in this matrix, each element gives the number of samples of the row category assigned to the column category. When the matrix is diagonal (entries outside the main diagonal are all zero) there is a perfect prediction of all the samples. Note that LDA models used for supervised discriminant classification were built by using 4 to 10 PCs. The models with 8 and 9 PCs, for unsupervised DESI and TS respectively, were chosen as they provided the least number of false results (i.e. highest prediction rates) in cross-validation.
The model built for the TS dataset was used further to predict the normal/disease state of another 110 samples (the evaluation set), whose histopathology was unknown at the time of MS analysis. The subsequent comparison between molecular diagnosis by TS-MS and histological diagnosis further validated the TS-MS model performances. LDA was performed with Matlab using in-house routines.
Conclusions
DESI-MS and TS-MS have been used to differentiate tumor and normal tissue from biopsies taken from 18 radical prostatectomy surgical specimens. The targeted dataset (radical prostatectomy specimen 1–12) was subjected to discriminant classification analysis using LDA, providing an average prediction rate of 97.5% for DESI-MS and 96% for TS-MS. Validation of TS-MS as a non-targeted technique was performed using an external evaluation set (radical prostatectomy specimen 13–18) of unknown prostate specimens, which provided average predictions rates of 92.5%. DESI and TS results are comparable (0.94 correlation coefficient by CCA), recovering the biochemical information, primarily in the lipids, that is responsible for the separation of tumor and normal tissue samples.
Supplementary Material
Acknowledgements
Funding for this research was supported by grants from the National Institute of Health (1R21EB009459-01), the National Institute of Biomedical Imaging and Bioengineering (1R21EB015722-01A1) and the Purdue University Center for Cancer Research. The authors thank Timothy Ratliff for valuable discussions and Dr. Paolo Oliveri (University of Genova, Italy) for proving the in-house Matlab routines. This work was performed within the limits of approved IRBs: Purdue protocol (1203011967) and Indiana University protocol (1205008669).
Footnotes
Footnotes should appear here. These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Notes and references
- 1.National Cancer Institute. Prostate Cancer. [accessed Jul 28, 2014];2014 http://www.cancer.gov/cancertopics/types/prostate.
- 2.Tewari A, Sooriakumaran P, Bloch DA, Seshadri-Kreaden U, Hebert AE, Wiklund P. European Urology. 2012;62:1–15. doi: 10.1016/j.eururo.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 3.Yossepowitch O, Briganti A, Eastham JA, Epstein J, Graefen M, Montironi R, Touijer K. European Urology. 2014;65:303–313. doi: 10.1016/j.eururo.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 4.Che M, Grignon D. In: Prostate Cancer: New Horizons in Research and Treatment. Cher M, Raz A, Honn K, editors. Vol. 81. Springer US; 2002. pp. 3–17. [Google Scholar]
- 5.DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. The Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 6.Takats Z, Wiseman JM, Gologan B, Cooks RG. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 7.Kerian KS, Jarmusch AK, Cooks RG. The Analyst. 2014;139:2714–2720. doi: 10.1039/c4an00548a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Analytical Chemistry. 2011;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiraoka K, Nishidate K, Mori K, Asakawa D, Suzuki S. Rapid Communications in Mass Spectrometry. 2007;21:3139–3144. doi: 10.1002/rcm.3201. [DOI] [PubMed] [Google Scholar]
- 10.Nemes P, Vertes A. Analytical Chemistry. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 11.Eberlin LS, Tibshirani RJ, Zhang J, Longacre TA, Berry GJ, Bingham DB, Norton JA, Zare RN, Poultsides GA. Proceedings of the National Academy of Sciences. 2014;111:2436–2441. doi: 10.1073/pnas.1400274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, Cooks RG, Agar NYR. Cancer Research. 2012;72:645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberlin LS, Norton I, Orringer D, Dunn IF, Liu X, Ide JL, Jarmusch AK, Ligon KL, Jolesz FA, Golby AJ, Santagata S, Agar NYR, Cooks RG. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1215687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zang X, Jones CM, Long TQ, Monge ME, Zhou M, Walker LD, Mezencev R, Gray A, McDonald JF, Fernandez FM. Journal of proteome research. 2014;13:3444–3454. doi: 10.1021/pr500409q. [DOI] [PubMed] [Google Scholar]
- 15.Schwamborn K, Caprioli RM. Nature reviews. Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 16.Seeley EH, Caprioli RM. Proceedings of the National Academy of Sciences. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badu-Tawiah AK, Eberlin LS, Ouyang Z, Cooks RG. Annual Review of Physical Chemistry. 2013;64:481–505. doi: 10.1146/annurev-physchem-040412-110026. [DOI] [PubMed] [Google Scholar]
- 18.Wiseman JM, Puolitaival SM, Takáts Z, Cooks RG, Caprioli RM. Angewandte Chemie. 2005;117:7256–7259. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- 19.Dill A, Eberlin L, Zheng C, Costa A, Ifa D, Cheng L, Masterson T, Koch M, Vitek O, Cooks RG. Anal Bioanal Chem. 2010;398:2969–2978. doi: 10.1007/s00216-010-4259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dill AL, Eberlin LS, Costa AB, Zheng C, Ifa DR, Cheng L, Masterson TA, Koch MO, Vitek O, Cooks RG. Chemistry – A European Journal. 2011;17:2897–2902. doi: 10.1002/chem.201001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masterson T, Dill A, Eberlin L, Mattarozzi M, Cheng L, Beck SW, Bianchi F, Cooks RG. J. Am. Soc. Mass Spectrom. 2011;22:1326–1333. doi: 10.1007/s13361-011-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santagata S, Eberlin LS, Norton I, Calligaris D, Feldman DR, Ide JL, Liu X, Wiley JS, Vestal ML, Ramkissoon SH, Orringer DA, Gill KK, Dunn IF, Dias-Santagata D, Ligon KL, Jolesz FA, Golby AJ, Cooks RG, Agar NYR. Proceedings of the National Academy of Sciences. 2014;111:11121–11126. doi: 10.1073/pnas.1404724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberlin LS, Dill AL, Golby AJ, Ligon KL, Wiseman JM, Cooks RG, Agar NYR. Angewandte Chemie. 2010;122:6089–6092. doi: 10.1002/anie.201001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balog J, Sasi-Szabó L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, Mirnezami R, Dezső B, Damjanovich L, Darzi A, Nicholson JK, Takáts Z. Science Translational Medicine. 2013;5:194ra193. doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
- 25.Pirro V, Eberlin LS, Oliveri P, Cooks RG. The Analyst. 2012;137:2374–2380. doi: 10.1039/c2an35122f. [DOI] [PubMed] [Google Scholar]
- 26.Cully M, You H, Levine AJ, Mak TW. Nature reviews. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 27.Gao N, Zhang Z, Jiang BH, Shi X. Biochemical and biophysical research communications. 2003;310:1124–1132. doi: 10.1016/j.bbrc.2003.09.132. [DOI] [PubMed] [Google Scholar]
- 28.Swinnen JV, Heemers H, van de Sande T, de Schrijver E, Brusselmans K, Heyns W, Verhoeven G. The Journal of steroid biochemistry and molecular biology. 2004;92:273–279. doi: 10.1016/j.jsbmb.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Wong KK, Engelman JA, Cantley LC. Current opinion in genetics & development. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forina M, Oliveri P, Casale M. Chemometr Intell Lab. 2010;102:110–122. [Google Scholar]
- 31.Bro R, Smilde AK. Anal Methods-Uk. 2014;6:2812–2831. [Google Scholar]
- 32.Oliveri P, Casolino MC, Forina M. Advances in food and nutrition research. 2010;61:57–117. doi: 10.1016/B978-0-12-374468-5.00002-7. [DOI] [PubMed] [Google Scholar]
- 33.Fearn T. NIR News. 2009;20:15–16. [Google Scholar]
- 34.Doeswijk TG, Hageman JA, Westerhuis JA, Tikunov Y, Bovy A, van Eeuwijk FA. Chemometr Intell Lab. 2011;107:371–376. [Google Scholar]
- 35.Devaux MF, Robert P, Qannari A, Safar M, Vigneau E. Appl Spectrosc. 1993;47:1024–1029. [Google Scholar]
- 36.Oliveri P, Downey G. Trac-Trend Anal Chem. 2012;35:74–86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.