Abstract
Regular exercise, particularly resistance training (RT), is the only therapy known to consistently improve muscle strength and quality (force per unit of mass) in older persons, but there is considerable variability in responsiveness to training. Identifying sensitive diagnostic biomarkers of responsiveness to RT may inform the design of a more efficient exercise regimen to improve muscle strength in older adults. MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression. We quantified six muscle specific miRNAs (miR-1, -133a, -133b, -206, -208b and -499) in both muscle tissue and blood plasma, and their relationship with knee extensor strength in seven older (age = 70.5 ± 2.5 years) adults before and after 5 months of RT. MiRNAs differentially responded to RT; muscle miR-133b decreased, while all plasma miRNAs tended to increase. Percent changes in knee extensor strength with RT showed strong positive correlations with percent changes in muscle miR-133a, -133b, -206 and with percent changes in plasma and plasma/muscle miR-499 ratio. Baseline level of plasma or plasma/muscle miR-499 ratio further predicts muscle response to RT, while changes in muscle miR-133a, -133b, -206 may correlate with muscle TNNT1gene alternative splicing in response to RT. Our results indicate that RT alters muscle specific miRNAs in muscle and plasma, and that these changes account for some of the variation in strength responses to RT in older adults.
Keywords: microRNA, skeletal muscle, aging, resistance training
Introduction
Mammalian species, and humans in particular, undergo a progressive and gradual loss of skeletal muscle mass and strength with aging, which impairs performance of daily living activities and leads to major medical and socioeconomic problems. Age-related decreases in muscle mass only partially account for muscular weakening with age (Delbono 2003; Delbono 2011; Goodpaster and others 2006), and improvements in muscle strength with resistance training (RT) are not entirely accounted for by muscle hypertrophy with RT (Fiatarone and others 1990; Leenders and others 2013; Tracy and others 1999). This suggests that strength is regulated by cellular and molecular mechanisms that are independent of muscle size; yet these mechanisms have yet to be entirely elucidated. In addition, there is considerable variability of strength responses to RT that can inform the discovery of these mechanisms. Recently, we reported increased specific force and power in single muscle fibers, but not myofiber cross-sectional area, in the vastus lateralis muscle of older adults subjected to a 5-month RT program (Zhang and others 2013). This study indicated that RT altered the Troponin T1 (TNNT1) pre-mRNA alternative splicing (AS) profile in muscle and that muscle strength after RT was related to these gene profiles. The next step is to determine the signals induced by RT to trigger the alternative splicing of TNNT1.
MiRNAs, small (~20–30 nucleotides) noncoding RNAs that inhibit protein translation or enhance mRNA degradation, have been implicated in the response of various organs to a single bout of RT or endurance training (Davidsen and others 2011; Drummond and others 2008; Hamilton and Baulcombe 1999; Reinhart and others 2000; Tonevitsky and others 2013; Zacharewicz and others 2013). Muscle-specific miRNAs (myomiRs) are a group of striated muscle (skeletal and/or cardiac) miRNAs, including miR-1, miR-133a, miR-133b, miR-206, miR-208, miR-208b, miR-486 and miR-499 (Callis and others 2008; McCarthy and Esser 2007; Small and others 2010; van Rooij and others 2008; van Rooij and others 2009). Among these myomiRs, miR-1, miR- 133a/b, and miR-206, make up nearly 25% of miRNA expression in skeletal muscle in both humans and mice (McCarthy 2008; Sempere and others 2004). Epigenetic control of muscle performance in response to overload, muscle innervation, specification of muscle fiber identity, and myoblast proliferation in response to tissue injury, are regulated by specific miRNAs (Dey and others 2011; Moresi and others 2010; Nakasa and others 2010; Small and others 2010; van Rooij and others 2009; Williams and others 2009). Some muscle-enriched miRNAs, such as miR-1, miR-133a, miR-206 and miR-499, can be detected in blood and respond to pathologies in humans and animals (Deng and others 2011; Donaldson and others 2013; Eisenberg and others 2007; McCarthy and others 2007; Mizuno and others 2011). As circulating miRNAs are stable (e.g., resistant to endogenous RNAses) (Mitchell and others 2008), they are considered useful biomarkers (Alevizos and Illei 2010) for disease incidence and prognosis. Interestingly, circulating miRNAs may be taken up by non-muscle cells and thus regulate protein expression, contributing to the pathogenesis of disease (Heneghan and others 2010; Mitchell and others 2008; Wang and others 2010; Wang and others 2009). Thus, miRNAs are promising diagnostic and therapeutic targets (Hoy and Buck 2012; Uhlemann and others 2012; Valadi and others 2007; Weilner and others 2013).
Only a few studies have analyzed whether resistance exercise has an effect on miRNA levels in humans. A study in healthy young men undertaking a five day per week RT program for 12 weeks showed that miRNAs play a role in regulating muscle mass response to RT (Davidsen and others 2011). Additionally, compared to young subjects, older subjects showed smaller decreases in miR-1 expression after an acute bout of resistance exercise (Drummond and others 2008). However, whether chronic RT alters miRNAs in older adults and whether this effect potentially contributes to the magnitude of strength gain with RT is not known. Therefore, the purpose of this study was to measure six muscle specific miRNAs (miR-1, -133a, -133b, -206, -208b and -499) in muscle tissue and blood plasma in older adults before and after five months of RT and to examine the relationship between changes in knee extensor strength with changes in miRNAs or miRNAs at baseline level.
Methods and Materials
Study participants
Participants in this study included older men (n = 3) and women (n = 4) who were recruited from the Piedmont Triad area of North Carolina. All participants were age 65–80 years, non-smoking, not on hormone replacement therapy, sedentary (< 15 min of exercise, 2 times/week) and weight stable (< 5% weight change) for at least six months prior to enrollment. All had normal liver, kidney, pulmonary, and thyroid function; no history of excessive alcohol intake; and no major chronic illness, anemia, or orthopedic impairment as described (Zhang and others 2013). The study was approved by the Wake Forest Institutional Review Board for Human Research and all participants signed informed consent to participate in the study.
Resistance training intervention
The exercise intervention consisted of five months of progressive RT designed to elicit adaptations in skeletal muscle and increase strength and power. To optimize the resistance stimulus for maximal functional gain, the exercise prescription was based on a relative intensity level and progressed with each individual’s strength gain. Participants exercised three days/week under the supervision of two exercise physiologists. Participants walked or cycled slowly for 5–10 minutes to warm-up prior to RT. The resistance exercises were performed on machines (Cybex, Medway, MA and Nautilus, Vancouver, WA), on which the load could be adjusted in small increments. The participants completed four lower extremity exercises, two of which targeted the quadriceps muscles (leg press, knee extension), one that targeted the hamstrings (leg curl), and one that targeted the triceps surae (calf press). The program was based on American College of Sports Medicine guidelines for intensity, number of repetitions, number of sets, and number of days per week (American College of Sports 2009). The one repetition maximum (1RM), that is, the maximal weight that could be lifted with correct form in a single repetition was used to prescribe intensity. Intensity was progressively increased during the first month towards the training goal of three sets of 10 repetitions at 70% 1RM for a given exercise. The 1RM strength testing was repeated every four weeks, and the training loads adjusted to achieve the 70% 1RM goal.
Knee extensor strength
We assessed maximal isokinetic knee extensor strength using an isokinetic dynamometer (Biodex) at a fixed speed (60° per second) with the participant sitting and the hips and knee flexed at 90°. This method is widely considered the gold standard of strength assessment (Martin and others 2006). The 1-week test-retest reliability in our lab is very high (Intra-Class Correlation Coefficient > 0.97). We set start and stop angles at 90° and 30° of knee flexion (range of motion of 60°). We used the peak torque observed during this range of motion as the measure of strength for each knee extension movement.
Body composition measurements
Height and mass were measured to calculate BMI (kg/m2). All participants underwent a total-body scan using dual-energy X-ray absorptiometry (DXA, Model DPX-L, Lunar Corp., Madison, Wisconsin) to determine lean muscle mass and percent body fat before and after RT.
Skeletal muscle biopsy and blood collection
Needle biopsies of the vastus lateralis were collected under local anesthesia with 1% lidocaine. All biopsies were performed in the early morning after an overnight fast. Subjects were asked to refrain from taking aspirin, prescription and over-the-counter non-steroidal anti-inflammatory drugs, or other compounds that may affect bleeding, platelets, or bruising for the week prior to the biopsy, and to refrain from any strenuous activity (including RT) for at least 36 hours prior to the biopsy. Visible blood and connective tissue were removed from the muscle specimen. A muscle portion (30 mg) used for RNA extraction was snap-frozen in liquid nitrogen under RNAse free conditions, and stored at −80°C until analysis. Participants completed their last bout of RT 36–72 hours prior to the post-RT biopsies and blood sampling. Whole blood from participants was collected in sodium EDTA tubes before and after RT and before muscle collection. Centrifugation was then performed at 4°C with an initial spin at 2000 g for 10 minutes to separate the plasma from the buffy coat and red blood cells and was stored at −80°C until analysis. Plasma from two of the subjects was not collected because they did not provide consent to store their blood samples.
RNA isolation and quantitative real-time RT-PCR for miRNA detection
The miRNAs miR-1, miR-133a, miR-133b, miR-206, miR-208b, and miR-499 are enriched in muscles, but typically found at low levels in other tissues (McCarthy 2011). We examined the levels of muscle-specific miRNAs in both muscle and blood plasma using real-time RT–PCR. Total RNA was extracted from human vastus lateralis muscle tissues using the mirVana™ miRNA Isolation Kit (Applied Biosystems, Carlsbad, CA) or from 200 μl plasma using the phenol-free total RNA purification kit (Amresco, Solon, OH) according to the manufacturer’s instructions. RNA samples and the same volume of water (negative control) were reverse-transcribed using TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). The TaqMan miRNA assay was used according to the manufacturer’s instructions to measure the expression of human miR-1, miR-133a, miR-133b, miR-206, miR-208b, miR-499 using pre-developed reagents from ABI (Applied Biosystems) run on an Mx3000 (Stratagene, La Jolla, CA). The U6 small nuclear RNA (RNU6B), an endogenous control, was used to normalize blood plasma and skeletal muscle miRNAs expression (Asaga and others 2011; Ng and others 2009; Song and others 2012; Togliatto and others 2013). Analysis was performed by the comparative threshold cycle (Ct) method under the following cycling conditions: 10 min at 95°C; 65 cycles of 15 sec at 95°C and 1 min at 60°C. Relative abundance of each miRNA was determined from the Ct values using the 2−ΔΔCt method (Livak and Schmittgen 2001) after normalization to RNU6B and all experiments were performed in triplicate. As the cut-off value for low-copy (< 250 copies) circulation miRNAs is approximately 35 PCR cycles (Mestdagh and others 2009), and plasma miR-208b’s Ct value was more than 50 in all subjects, it was considered non-detectable or unreliable, and not reported further.
Statistical analysis
Data analysis was performed with SAS 9.4 (Cary, NC). Percent change ((post-measure – pre-measure)/pre-measure× 100%) was calculated for the outcomes of knee strength, body composition (BMI, %fat and lean mass) and miRNAs to take into account individual differences at baseline. A single group t-test was used to determine intervention effect on each variable. Then we examined the age and gender effect on the percent change of knee strength, body composition and miRNAs after RT. We also used mixed effect models to control for age and gender, where pre- and post-measures were treated as repeated measures. Compound symmetry covariance structure was used to account for within subject correlation. Pearson Product correlations were calculated to determine the strength of the association between pairs of variables. For all tests, statistical significant level was set at 0.05. In acknowledgement of our small sample size, we also report effect sizes (ES) of RT effect calculated with Cohen’s D, where ES = (post-measure – pre-measure)/pooled standard deviation of pre-measure and post-measure.
Results
We noticed that the mixed effect model results did not differ materially from the single group t-test results, so we only report the t-test results. It appeared that age did not affect percent change of knee strength, body composition, or miRNAs, while gender only showed a statistically significant effect on percent change of plasma miR-133b. Due to the small sample size, we did not do any stratified analyses based on gender for plasma miR-133b.
Knee extensor strength, lean muscle mass and body composition changes with RT (Figure 1)
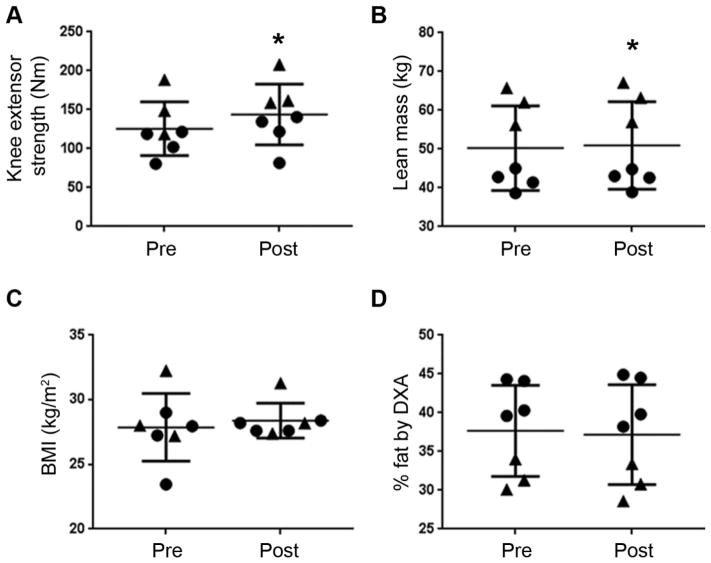
Figure 1. Knee extensor strength changes with RT.
All 7 subjects responded to RT. *, p = 0.0085 for knee strength (A) and p = 0.021 for lean mass (B). BMI and percent body fat were not modified by RT (C, D). Pre: before RT; Post: after RT. (●), women, (▲), men.
All seven subjects showed improvement in knee extensor strength with RT (14.7 ± 9.4%; p = 0.0085; ES = 0.50); however, there was high inter-subject variability (range = 1.04 to 40.1 Nm) (Fig. 1A). Lean muscle mass also increased after RT (1.3 ± 1.1%; p = 0.021; ES = 0.06) (Fig. 1B). BMI and percent body fat were not altered with RT (Fig. 1C, D).
Changes in muscle and plasma miRNAs with RT (Figure 2)
Figure 2. Changes in muscle and plasma miRNAs with RT.
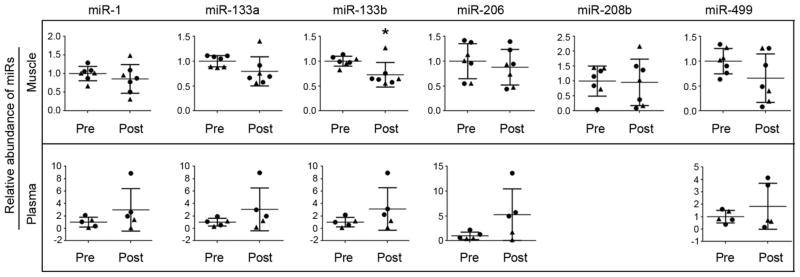
RNU6B normalized miRNAs relative abundance showed a trend to decrease with RT in muscle, and tended to increase with RT in plasma. MiR-133b in muscle tissue was the only one to show a statistically significant decrease after RT. *, p = 0.043. (●), women, (▲), men.
Compared to baseline (before RT), muscle levels of all miRNAs tended to decrease (ES = −0.47, −0.92, −1.46, −0.34, −0.06, −0.88 for miR-1, -133a, -133b, -206, -208b and -499, respectively); however, the decrease was significant only for miR-133b (−26.5 ± 27.5%; p = 0.043; ES = −1.46). The relative abundance of plasma levels of all miRNAs tended to increase with RT (ES = 0.80, 0.83, 0.86, 1.15, 0.61 for miR-1, -133a, -133b, -206 and -499, respectively), but none of these changes were statistically significant (Fig. 2).
Relationships between changes in knee extensor strength and changes in miRNAs (Figure 3 and 4) or miRNAs at baseline level (Figure 5)
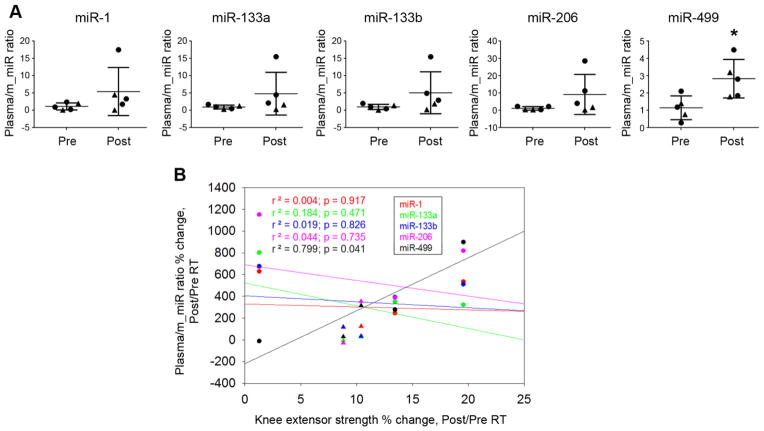
Figure 3. Relationships between changes in knee extensor strength and changes in miRNA.
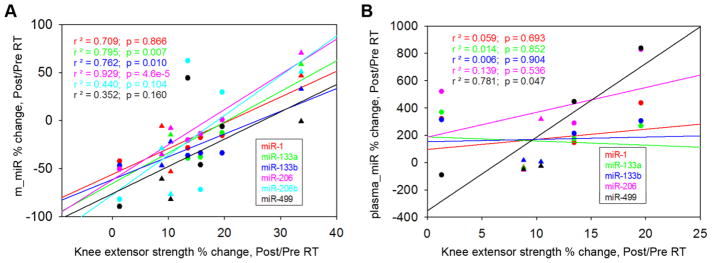
(A) Percent changes in knee extensor strength with RT correlated positively with percent changes in miR-133a, miR-133b, miR-206. (B) Percent change in plasma miR-499 showed a significant positive correlation with the knee extensor strength after RT. (●), women, (▲), men.
Figure 4. Relationships between plasma/muscle miRNA ratio changes and knee extensor strength changes.
(A) Plasma/muscle miR ratios tended to increase with RT and was significant for miR-499 (*, p < 0.05). (B) The percent change in knee extensor strength correlated positively with percent change in plasma/muscle miR-499 ratio. (●), women, (▲), men.
Figure 5. Prediction of muscle response to RT using baseline miRNA measurements.
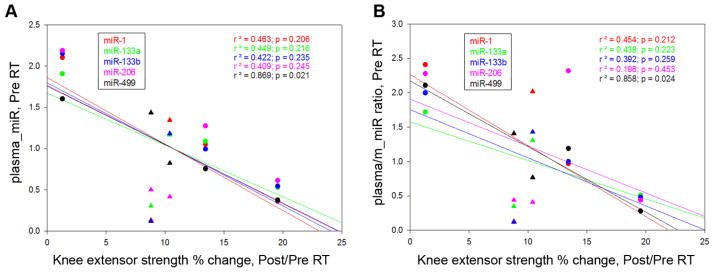
(A) Baseline level plasma miR-499 negatively correlates with percent change in knee extensor strength after RT. (B) Plasma/muscle miR-499 baseline ratio negatively correlates with percent change in knee extensor strength after RT. (●), women, (▲), men.
There were strong positive correlations between percent change in knee strength and percent change in muscle miR-133a, miR-133b, miR-206 (p < 0.01, Fig. 3A). In contrast, percent change in strength only correlated with percent change in one plasma miRNA (miR-499, p < 0.05, Fig. 3B). Consistently, we also observed a significant increase in the plasma/muscle miR-499 ratio with RT (p = 0.021, Fig. 4A); and, percent change in knee strength correlated positively with percent change in the plasma/muscle miR-499 ratio (p = 0.041, Fig. 4B). Notably, baseline level of plasma miR-499 showed significant negative correlation with percent change in strength (p = 0.021. Fig. 5A); yet, none of the baseline level muscle miR showed any positive correlation with percent change in strength (data not shown); For plasma/muscle miR ratio, only that of miR-499 showed a significant negative correlation (p = 0.024) with strength change (Fig. 5B)
Relationship between muscle miRNAs and TNNT1 alternative splicing (Figure 6)
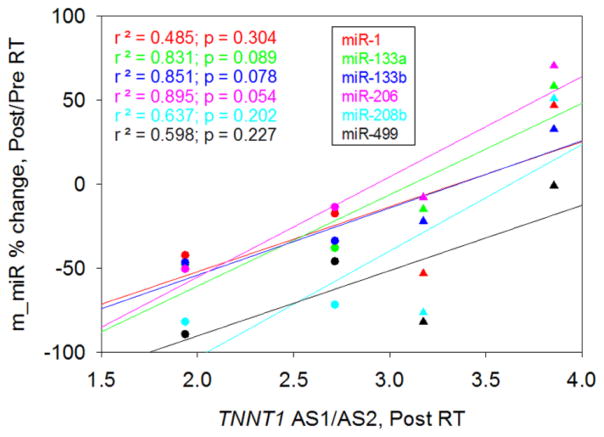
Figure 6. Relationship between muscle miRNAs and TNNT1 AS.
Changes of muscle miR-133a, miR-133b and miR-206 showed a trend of positive correlation with the TNNT1 AS1/AS2 ratio after RT. (●), women, (▲), men.
We recently reported that the TNNT1 AS1/AS2 ratio in skeletal muscle is a potential quantitative biomarker of muscle adaptation to RT in older adults that also reflects enhanced single muscle fiber force in the absence of significant increases in fiber cross-sectional area (Zhang and others 2013). From the seven subjects studied for miRNA and six for TNNT1 pre-mRNA AS, only four were studied for both. Thus, we explored the relationship between percent changes in miRNAs and TNNT1 AS1/AS2 ratio in response to RT. A trend for a positive correlation between percent changes in muscle miR-133a, miR-133b, miR-206 and miR-208b with RT and the TNNT1 AS1/AS2 ratio was observed (Fig. 6).
Discussion
This study is the first to assess muscle-specific miRNAs in older adults undergoing RT and their relationship with changes in knee extensor strength induced by RT. Specifically, we examined the effects of RT on whether muscle-specific muscle and plasma miRNAs were altered with five months of progressive RT in older adults. The major findings are: (1) a trend for muscle-specific miRNAs to decrease in muscle tissue, but increase in plasma with RT; (2) a specific, rather than global, correlation between changes in miRNAs in muscle or blood and changes in knee extensor strength following RT; (3) plasma and muscle miR-499 ratio change was a sensitive marker of increases in knee extensor strength with RT; (4) Baseline level of plasma or plasma/muscle miR-499 ratio predicts knee extensor strength changes in response to RT; (5) a subset of muscle myomiRs correlate with TNNT1 pre-mRNA AS1/AS2 ratio after RT.
Several miRNAs are reported to affect muscle cell function through modulation of proliferation, differentiation, hypertrophy, and nutrient metabolism (Cardinali and others 2009; Chen and others 2006; Dey and others 2011; McCarthy 2008). Acute and sustained endurance and resistance training differentially regulate miRNAs abundance in muscle and/or the circulation in animals and young and older adults (Aoi and others 2013; Baggish and others 2011; Kirby and McCarthy 2013; Russell and others 2013; Sawada and others 2013; Tonevitsky and others 2013; Uhlemann and others 2012; Zacharewicz and others 2013). In the older adults in this study, RT tended to decrease the relative abundance of all six muscle-specific miRNAs in muscle tissue with a statistically significant reduction in miR-133b. The reduction of muscle-specific miRNAs has also been reported after an acute bout of resistance exercise in young and older adults (Drummond and others 2008). In contrast, trends for RT to increase all miRNAs in plasma were observed. These findings indicate that RT may induce muscle to release miRNAs into circulation. A reduction in miR-1 at three- and six-hours post-exercise was observed in both older and younger adults; yet no changes in miR-133a or miR-206 were observed (Drummond and others 2008). Collectively, this and previous studies show that exercise regulates muscle and blood miRNAs levels. Given that muscle miR-133b was the only miRNA to decrease in muscle tissue with RT in our study, we propose that acute resistance exercise (Drummond and others 2008) and chronic RT may affect muscle-specific miRNAs differently. Similarly, changes in plasma miRNAs were also dependent on the type of exercise (Banzet and others 2013).
Most of the miRNAs that we studied are also present in cardiac muscle (Endo and others 2013) and some of them, miR-1, miR-133a and miR-499, have been reported to be increased in circulation in rats and patients with acute myocardial infarction (Wang and others 2010). In addition, miR-208 was upregulated, whereas miR-1 and miR-133a downregulated in myocardial infarction compared to healthy adult and fetal hearts (Bostjancic and others 2010). Contamination from cardiac miRNAs in this study cannot be ruled out, however, it is worth noting that our study participants did not have a history of chronic heart disease, and they did not report any sign of cardiac ischemia during or after the RT sessions, which minimizes potential contamination.
Unlike the miR-1 family which is organized as bicistronic genes (Kusakabe and others 2013), miR-208b and miR-499 are encoded within the slow myosin heavy chain (MHC) genes (van Rooij and others 2009) and thereby are restricted to type I fibers and modulate fiber type (Endo and others 2013; McCarthy and others 2009; van Rooij and others 2009). A transition from fast- to slow-twitch fibers has been reported in aging rodents and humans (Andersen 2003; Lexell 1995), and type-I fibers largely outnumber type-II fibers in our samples from monkey and human vastus lateralis muscles (Choi and others 2013; Zhang and others 2013). Thus, changes in miR-208b and miR-499 are intriguing and particularly relevant for muscle fiber function.
We found strong relationships between changes in knee extensor strength with RT and changes in specific miRNA levels. Individuals with the largest changes in muscle miR-133a, -133b and -206 tended to show the largest changes in knee extensor strength. In addition, variation in knee extensor strength improvement correlated with changes in plasma miR-499 and its plasma/muscle ratio. These data indicate that a set of miRNAs measured in muscle and plasma might be useful biomarkers of muscle function adaptation to RT. Ideally, a biomarker that predicts responsiveness to exercise would require only one biopsy or blood sample, prior to deciding to initiate exercise. When we analyzed the correlation between baseline level miRNAs and percent changes in knee extensor strength, plasma miR-499 showed significant negative correlation. Similarly, plasma/muscle miR-499 ratio also showed a negative correlation with percent change in knee extensor strength. Yet, we did not find any correlation between baseline level muscle miRs and percent change in knee strength. These finding implies that plasma miR-499 or plasma/muscle miR-499 ratio could be a better biomarker for prediction of muscle response to RT in older adults. As the ratio between plasma and muscle miR-499 increased significantly after RT, and it was the only miRNA that showed a statistically significant positive correlation with knee extensor strength, we also propose that miR-499 regulates muscle strength in response to progressive RT in older adults. Muscle miR-133b was significantly reduced with RT, and changes in this miRNA predicted changes in knee extensor strength with RT. Thus, miR-133b may play a major role in regulation of muscle response to RT and also may be a marker for the magnitude of muscle response to RT in older adults.
The positive correlation of miRNA changes with that of the knee extensor strength changes indicates that miRNA changes may be a useful biomarker reflecting the muscle response to RT. Considering the biological function of miRNAs, it is possible that some of the myomiRs are playing key roles in regulating muscle mass and force by modifying muscle specific gene expression and protein translation. One of those potential targets of myomiRs is TNNT1. We recently reported that TNNT1 AS is an indicator of skeletal muscle response to RT in older adults. Specially, TNNT1 AS1/AS2 ratio is a potential quantitative biomarker of skeletal muscle adaptation to RT in older adults, and their profile reflects enhanced single-fiber muscle force in the absence of significant increases in fiber cross-sectional area (Zhang and others 2013). A trend for positive correlations between percent changes in several miRNAs (miR-133a, miR-133b and miR-206) and TNNT1 AS1/AS2 ratio was observed. Additionally, correlation between percent changes in muscle miR-133a, miR-133b and miR-206, and percent changes in knee extensor strength after RT were also observed, which suggests that RT induced changes in muscle strength through regulating a miRNA-TNNT1 AS pathway in older adults. Although a relationship has not been reported, we propose that miRNAs may act on TNNT1 alternative splicing factors.
It should be noted that percent changes in knee extensor strength directly correlated with changes in miR-133a, miR-133b and miR-206, making them potential candidates as biomarkers and therapeutic targets. Muscle specific miRNA downregulation has been reported to regulate muscle atrophy directly or indirectly through inhibiting IGF1/PI3K/Akt axis (Wang 2013). Also, they positively regulate muscle differentiation and reinnervation (Kim and others 2006; Williams and others 2009), which may explain the increased muscle force in the absence of increased muscle cross-sectional area (Zhang and others 2013). Thus, our findings indicate that these myomiRs may play critical roles in regulation of muscle response to RT in older adults through targeting and balancing multiple signaling pathways involved in muscle mass, innervation, and force regulation.
Limitations
Analysis of a larger cohort will help to better establish whether miRNA expression and/or activity influences RT-evoked changes in skeletal muscle strength, whether different exercise modalities differentially regulate miRNA expression and associated cellular pathways, and how miRNA regulation contributes to physiological adaptation to RT. Due to the small number of participants, we were not able to carry out sex-specific analyses. As exercise evokes a whole body response, extending our analysis to non-muscle specific miRNAs might provide a more comprehensive understanding of the neuromuscular system adaptation to RT in older adults. Since miR-1, -133a and -133b are expressed also in the central nervous system (Bak and others 2008), their levels in circulation derive from both muscle and central nervous system. Therefore, the response to RT may result from muscle and neural adaptations. Future studies should include a non-exercise group. Measurements of central drive indices will help to examine the contribution of motor learning (improved motor unit recruitment, better relaxation of antagonist muscles) over repetitive exercise sessions.
Highlights.
Diagnostic biomarkers of muscle responsiveness to RT are lacking
Muscle specific miRNAs differentially responded to RT in muscle and plasma.
MiRNAs may regulate muscle response to RT
Acknowledgments
This work was supported by the National Institutes of Health grants, AG13934 and AG15820 to O.D., AG020583 to B.N, and the Wake Forest Claude D. Pepper Older Americans Independence Center (P30-AG21332).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alevizos I, Illei GG. MicroRNAs as biomarkers in rheumatic diseases. Nature reviews Rheumatology. 2010;6:391–398. doi: 10.1038/nrrheum.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American College of Sports, M. American College of Sports Medicine position stand. Progression models in resistance training for healthy adults. Medicine and science in sports and exercise. 2009;41:687–708. doi: 10.1249/MSS.0b013e3181915670. [DOI] [PubMed] [Google Scholar]
- Andersen JL. Muscle fibre type adaptation in the elderly human muscle. Scand J Med Sci Sports. 2003;13:40–47. doi: 10.1034/j.1600-0838.2003.00299.x. [DOI] [PubMed] [Google Scholar]
- Aoi W, Ichikawa H, Mune K, Tanimura Y, Mizushima K, Naito Y, Yoshikawa T. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Frontiers in physiology. 2013;4:80. doi: 10.3389/fphys.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clinical chemistry. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, Wang TJ, Chan SY. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. The Journal of physiology. 2011;589:3983–3994. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. Rna. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzet S, Chennaoui M, Girard O, Racinais S, Drogou C, Chalabi H, Koulmann N. Changes in circulating microRNAs levels with exercise modality. Journal of applied physiology. 2013;115:1237–1244. doi: 10.1152/japplphysiol.00075.2013. [DOI] [PubMed] [Google Scholar]
- Bostjancic E, Zidar N, Stajer D, Glavac D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology. 2010;115:163–169. doi: 10.1159/000268088. [DOI] [PubMed] [Google Scholar]
- Callis TE, Deng Z, Chen JF, Wang DZ. Muscling through the microRNA world. Experimental biology and medicine. 2008;233:131–138. doi: 10.3181/0709-MR-237. [DOI] [PubMed] [Google Scholar]
- Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, Falcone G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PloS one. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature genetics. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SJ, Shively CA, Register TC, Feng X, Stehle J, High K, Ip E, Kritchevsky SB, Nicklas B, Delbono O. Force-generation capacity of single vastus lateralis muscle fibers and physical function decline with age in African green vervet monkeys. The journals of gerontology Series A, Biological sciences and medical sciences. 2013;68:258–267. doi: 10.1093/gerona/gls143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW, Timmons JA, Phillips SM. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. Journal of applied physiology. 2011;110:309–317. doi: 10.1152/japplphysiol.00901.2010. [DOI] [PubMed] [Google Scholar]
- Delbono O. Neural control of aging skeletal muscle. Aging cell. 2003;2:21–29. doi: 10.1046/j.1474-9728.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- Delbono O. Expression and regulation of excitation-contraction coupling proteins in aging skeletal muscle. Current aging science. 2011;4:248–259. doi: 10.2174/1874609811104030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Chen JF, Wang DZ. Transgenic overexpression of miR-133a in skeletal muscle. BMC musculoskeletal disorders. 2011;12:115. doi: 10.1186/1471-2474-12-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Molecular and cellular biology. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A, Natanek SA, Lewis A, Man WD, Hopkinson NS, Polkey MI, Kemp PR. Increased skeletal muscle-specific microRNA in the blood of patients with COPD. Thorax. 2013;68:1140–1149. doi: 10.1136/thoraxjnl-2012-203129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. American journal of physiology Endocrinology and metabolism. 2008;295:E1333–1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, Flanigan KM, Neely LA, Whitney D, Beggs AH, Kohane IS, Kunkel LM. Distinctive patterns of microRNA expression in primary muscular disorders. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Weng H, Naito Y, Sasaoka T, Takahashi A, Fukushima Y, Iwai N. Classification of various muscular tissues using miRNA profiling. Biomedical research. 2013;34:289–299. doi: 10.2220/biomedres.34.289. [DOI] [PubMed] [Google Scholar]
- Fiatarone MA, Marks EC, Ryan ND, Meredith CN, Lipsitz LA, Evans WJ. High-intensity strength training in nonagenarians. Effects on skeletal muscle. JAMA. 1990;263:3029–3034. [PubMed] [Google Scholar]
- Goodpaster BH, Park SW, Harris TB, Kritchevsky SB, Nevitt M, Schwartz AV, Simonsick EM, Tylavsky FA, Visser M, Newman AB. The loss of skeletal muscle strength, mass, and quality in older adults: the health, aging and body composition study. The journals of gerontology Series A, Biological sciences and medical sciences. 2006;61:1059–1064. doi: 10.1093/gerona/61.10.1059. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Annals of surgery. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- Hoy AM, Buck AH. Extracellular small RNAs: what, where, why? Biochemical Society transactions. 2012;40:886–890. doi: 10.1042/BST20120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. The Journal of cell biology. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby TJ, McCarthy JJ. MicroRNAs in skeletal muscle biology and exercise adaptation. Free radical biology & medicine. 2013;64:95–105. doi: 10.1016/j.freeradbiomed.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe R, Tani S, Nishitsuji K, Shindo M, Okamura K, Miyamoto Y, Nakai K, Suzuki Y, Kusakabe TG, Inoue K. Characterization of the compact bicistronic microRNA precursor, miR-1/miR-133, expressed specifically in Ciona muscle tissues. Gene expression patterns : GEP. 2013;13:43–50. doi: 10.1016/j.gep.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Leenders M, Verdijk LB, van der Hoeven L, van Kranenburg J, Nilwik R, van Loon LJ. Elderly men and women benefit equally from prolonged resistance-type exercise training. J Gerontol A Biol Sci Med Sci. 2013;68:769–779. doi: 10.1093/gerona/gls241. [DOI] [PubMed] [Google Scholar]
- Lexell J. Human aging, muscle mass, and fiber type composition. J Gerontol A Biol Sci Med Sci. 1995;50(Spec No):11–16. doi: 10.1093/gerona/50a.special_issue.11. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martin HJ, Yule V, Syddall HE, Dennison EM, Cooper C, Aihie Sayer A. Is hand-held dynamometry useful for the measurement of quadriceps strength in older people? A comparison with the gold standard Bodex dynamometry. Gerontology. 2006;52:154–159. doi: 10.1159/000091824. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochimica et biophysica acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ. The MyomiR network in skeletal muscle plasticity. Exercise and sport sciences reviews. 2011;39:150–154. doi: 10.1097/JES.0b013e31821c01e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. Journal of applied physiology. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. American journal of physiology Cell physiology. 2007;293:C451–457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA, Peterson CA, Dupont-Versteegden EE. Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiological genomics. 2009;39:219–226. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome biology. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Nakamura A, Aoki Y, Ito N, Kishi S, Yamamoto K, Sekiguchi M, Takeda S, Hashido K. Identification of muscle-specific microRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PloS one. 2011;6:e18388. doi: 10.1371/journal.pone.0018388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. Journal of cellular and molecular medicine. 2010;14:2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Russell AP, Lamon S, Boon H, Wada S, Guller I, Brown EL, Chibalin AV, Zierath JR, Snow RJ, Stepto N, Wadley GD, Akimoto T. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. The Journal of physiology. 2013;591:4637–4653. doi: 10.1113/jphysiol.2013.255695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Kon M, Wada S, Ushida T, Suzuki K, Akimoto T. Profiling of circulating microRNAs after a bout of acute resistance exercise in humans. PloS one. 2013;8:e70823. doi: 10.1371/journal.pone.0070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome biology. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Digestive diseases and sciences. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- Togliatto G, Trombetta A, Dentelli P, Cotogni P, Rosso A, Tschop MH, Granata R, Ghigo E, Brizzi MF. Unacylated ghrelin promotes skeletal muscle regeneration following hindlimb ischemia via SOD-2-mediated miR-221/222 expression. Journal of the American Heart Association. 2013;2:e000376. doi: 10.1161/JAHA.113.000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonevitsky AG, Maltseva DV, Abbasi A, Samatov TR, Sakharov DA, Shkurnikov MU, Lebedev AE, Galatenko VV, Grigoriev AI, Northoff H. Dynamically regulated miRNA-mRNA networks revealed by exercise. BMC physiology. 2013;13:9. doi: 10.1186/1472-6793-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy BL, Ivey FM, Hurlbut D, Martel GF, Lemmer JT, Siegel EL, Metter EJ, Fozard JL, Fleg JL, Hurley BF. Muscle quality. II. Effects Of strength training in 65- to 75-yr-old men and women. Journal of applied physiology. 1999;86:195–201. doi: 10.1152/jappl.1999.86.1.195. [DOI] [PubMed] [Google Scholar]
- Uhlemann M, Mobius-Winkler S, Fikenzer S, Adam J, Redlich M, Mohlenkamp S, Hilberg T, Schuler GC, Adams V. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. European journal of preventive cardiology. 2012 doi: 10.1177/2047487312467902. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends in genetics : TIG. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Developmental cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. European heart journal. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH. MicroRNA in myogenesis and muscle atrophy. Current opinion in clinical nutrition and metabolic care. 2013;16:258–266. doi: 10.1097/MCO.0b013e32835f81b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilner S, Schraml E, Redl H, Grillari-Voglauer R, Grillari J. Secretion of microvesicular miRNAs in cellular and organismal aging. Experimental gerontology. 2013;48:626–633. doi: 10.1016/j.exger.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharewicz E, Lamon S, Russell AP. MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Frontiers in physiology. 2013;4:266. doi: 10.3389/fphys.2013.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Choi SJ, Wang ZM, Birbrair A, Messi ML, Jin JP, Marsh AP, Nicklas B, Delbono O. Human Slow Troponin T (TNNT1) Pre-mRNA Alternative Splicing Is an Indicator of Skeletal Muscle Response to Resistance Exercise in Older Adults. The journals of gerontology Series A, Biological sciences and medical sciences. 2013 doi: 10.1093/gerona/glt204. [DOI] [PMC free article] [PubMed] [Google Scholar]