Abstract
Historically, extraction of bone proteins has relied on the use of demineralization to better retrieve proteins from the extracellular matrix; however, demineralization can be a slow process that restricts subsequent analysis of the samples. Here, we developed a novel protein extraction method that does not use demineralization, but utilizes a methodology from hydroxyapatite chromatography where high concentrations of ammonium phosphate and ammonium bicarbonate are used to extract bone proteins. We report that this method has a higher yield than previously published small-scale extant bone extractions, with and without demineralization. Furthermore, after digestion with trypsin and subsequent HPLC-MS/MS analysis, we were able to detect several extracellular matrix and vascular proteins in addition to collagen I and osteocalcin. Our new method has the potential to isolate proteins in a short period (4 hrs) and provide information about bone proteins that may be lost during demineralization or with the use of denaturing agents.
Keywords: Ammonium phosphate, demineralization free, human bone, collagen I, osteocalcin
Introduction
The study of bone proteins and their modifications has emerged as a promising method to better understand and identify bone diseases (e.g., osteoporosis) [1; 2; 3; 4; 5; 6] as well as provide molecular information for extinct taxa [7; 8; 9; 10; 11; 12; 13; 14; 15; 16; 17; 18; 19; 20; 21; 22; 23]. However, because bone is mineralized, analyzing the protein content in bone is more challenging than other non-mineralized tissue. In particular, protein extraction protocols rely on demineralization of bone followed by protein solubilization (reviewed in [24]). Consequently, the protocols are typically slow, taking days to weeks to perform [24], or may result in unknown breakdown of proteins by hydrolysis. Through these traditional extraction protocols, ~1% or less of the original bone mass is extracted [24], most of which is composed of collagen I.
In contrast to above, several protocols have extracted proteins without the demineralization step [25; 26; 27; 28; 29], but the total yield has been limited to ~3 mg protein/g bone or less [27]. Jiang et al. [27] suggest that demineralization is a critical step for bone protein extraction; however, bone protein extraction with only acid labile surfactant allowed for a extensive bone proteome coverage using mass spectrometry [28]. Salmon et al. [28] further suggest that the method does not fully release mineral specific proteins, but may allow recovery of non-collagenous proteins without demineralization. In fact, Pastorelli et al. [26] identified over 200 gel spots were identified from extraction using only a low concentration phosphate buffer for extraction [26]. Thus a large number of proteins could be extracted from bone without extensive demineralization.
In hydroxyapatite chromatography, proteins are eluted from the hydroxyapatite column with increasing phosphate concentration [30]. Because bone is a composite of hydroxyapatite and protein, we have incorporated the use of higher concentration phosphate buffers, similar to the final concentration used in hydroxyapatite chromatography, to develop a novel bone protein extraction protocol without the use of demineralization.
Materials and Methods
Bone samples
Tibial cortical bone samples were sampled from seven Caucasian cadavers (23F, 25M, 48M, 56M, 79M, 81F, 82M). All samples were previously diagnosed to be free from metabolic bone diseases, HIV, and hepatitis B (National Disease Research Interchange and International Institute for the Advancement of Medicine). No live human subjects were involved in this research study (IRB waiver, Rensselaer Polytechnic Institute).
Protein extraction
Utilizing phosphate elution principles from hydroxyapatite chromatography [30; 31], we made either 400 mM ammonium phosphate dibasic (Sigma-Aldrich) or 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate (Sigma-Aldrich). To determine differences between the two extraction solutions, we performed a number of initial tests on bone obtained from a 48-year-old male. Bone samples (100 mg each; fragmented to ~1mm3) were extracted in 600 μL of solutions of 400 mM ammonium phosphate dibasic or 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate after homogenization using stainless steel beads in a Bullet Blender (Next Advance). Because this is a tube based homogenization method, particle size was not measured. Aliquots were taken at 4, 8, and 24 hrs to evaluate the amount of time necessary to extract protein for each solution.
After initial set of tests, we repeated the extraction on ~50 mg of bone with 400 mM ammonium phosphate, 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate, 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4M guanidine HCl (GuHCl) for a fixed period of 24 hours only. Temperature was varied at 4°C, room temperature, or 75°C to determine the effects of temperature on extraction. Lastly, an additional ~50 mg of bone was extracted at 75°C with 200 mM ammonium bicarbonate for 24 hrs for comparison to the ammonium phosphate extractions. The 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4M GuHCl extraction was only tested at 75°C.
After establishing the method with the highest yields, ~50 mg of bone from other cadaveric donors were extracted using the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate extraction for 24 hrs at 75°C. Protein concentration was determined using Coomassie (Bradford) Assay Kit (Thermo-Scientific) with BSA as a protein standard, and all samples were desalted using micro dialysis (3500 MWCO regenerated cellulose; Fisher Scientific) against nanopure water [32] for 4 days.
To evaluate if proteolysis occurs during the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate or the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4M GuHCl extraction process, additional 50 mg aliquots of the 48M samples were homogenized with the inclusion of 10 μg/mL Halt Protease Inhibitor (Thermo-Scientific) and incubated for 24 hr at 75°C.
Mass spectrometry
The 400 mM ammonium phosphate dibasic extraction and all 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate samples were reduced with 10 mM dithiothreitol for 1 hour at room temperature followed by alkylation using 30 mM iodoacetamide for 1 hour in the dark. Proteins were digested overnight with Trypsin Gold (Promega) at 37°C (1:100 trypsin:protein). Peptide samples were prepared for mass spectrometry using a C18 stage tip [33]. After binding to the C18 disk, samples were washed with 50 μL of 0.1% formic acid and eluted using 20 μL of 80% acetonitrile, 0.1% formic acid. Samples were partially dried in air to remove excess acetonitrile and resuspended to a final volume of 15 μL in 0.1% formic acid. Prepared peptides were separated using an Agilent 1200 Series HPLC with a Thermo Scientific BioBasic C18 (2.1 mm ID, 100 mm column length, 5 μm particle size) for 75 minutes using either of the following gradients: 1) 2% B 0–5 min, 30% B 5–15 min, 60% B 15–60 min, 95% B 60.01–64 min, 2% B 64.01–75 min or 2) 2% B 0–5 min, 30% B 5–35 min, 60% B 35–60 min, 95% B 60.01–64 min, 2% B 64.01–75 min where A is 0.1% formic acid and B is 100 acetonitrile, 0.1% formic acid. Eluted peptides were characterized on a LTQ-Orbitrap XL (Thermo Scientific). The top 2 peaks were fragmented using either collision induced dissociation (CID) or higher energy collisional dissociation (HCD) in the Orbitrap or the top 5 peaks were fragmented with CID and analyzed in the ion trap. All samples were analyzed by mass spectrometry in triplicate.
Peak lists (mgf) were created in MassMatrix Mass Spectrometric File Conversion Tools v. 3.2. Peak lists were searched against Swissprot and a decoy database using Mascot 2.3 (Matrix Science). The following parameters were set for each search: taxonomy was set to Homo sapiens; enzyme = trypsin; up to 3 missed cleavages; variable modifications: carbamidomethyl (C), deamidation (NQ), carboxy (E), oxidation (MKP); static modifications: none; peptide tolerance = 10 ppm; fragment tolerance = 0.5 Da; and peptide charge = 2+, 3+, 4+. Peptide results were filtered using Percolator at p<0.05. Peptides with nonsensical post-translational modifications (e.g., carboxyglutamic acid (Gla) on non-Gla containing proteins) were filtered by hand.
Statistics
To evaluate the differences in protein yield between extraction types, one-way ANOVA was performed in SigmaStat for Windows 2.03 (SPSS Inc.). Significance was set at p<0.05.
Results
Protein extraction
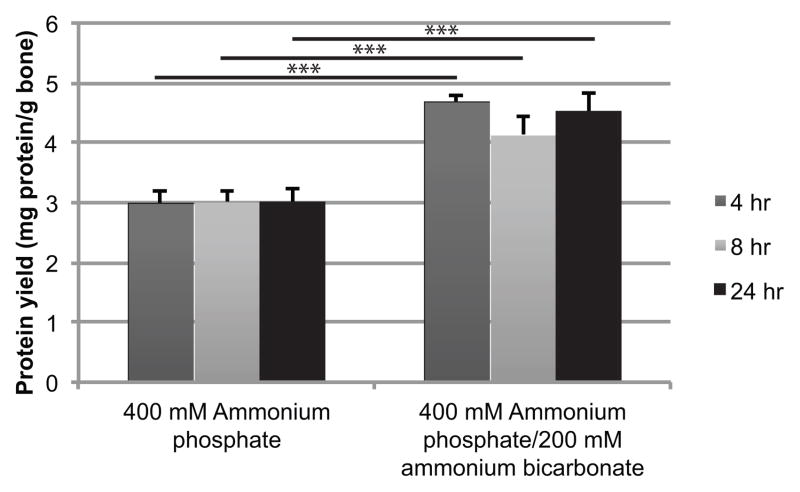
The 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate had a significantly greater yield than 400 mM ammonium phosphate dibasic alone for all times (p<0.001; Fig. 1). No variation in yield was observed between times.
Figure 1.
Time series (4, 8, 24 hr) for 400 mM ammonium phosphate and 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate extracted at room temperature. ***p<0.001
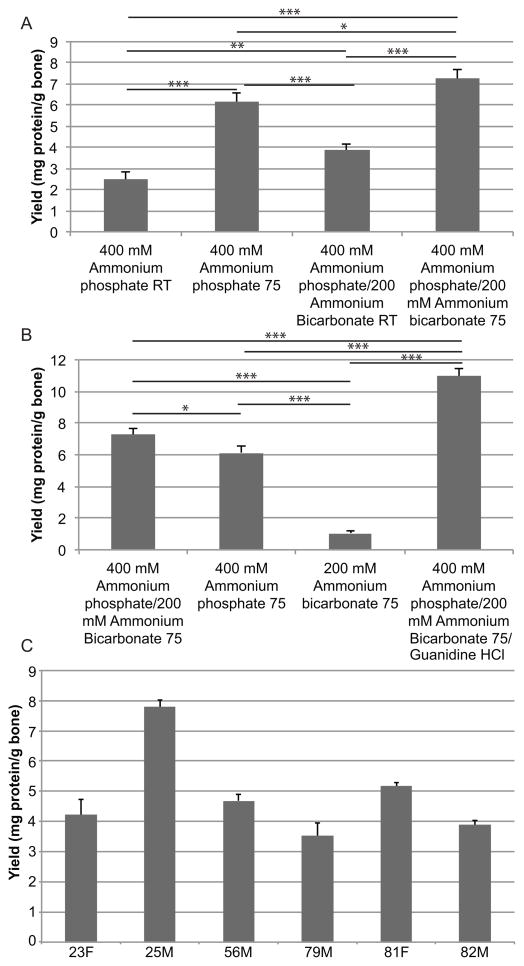
Temperature change resulted in a significant increase (p<0.001) in protein concentration for both the 400 mM ammonium phosphate dibasic and 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate solutions (Fig. 2A). Very little yield (1.05 ± 0.16 mg of protein/g bone) was detected after extraction with 200 mM ammonium bicarbonate at 75°C (Fig. 2B); much less than either ammonium phosphate extraction (p<0.001). The highest yield was obtained with the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4M GuHCl extraction (p<0.001).
Figure 2.
A) Protein yield from a 48-year-old Caucasian male donor bone for 400 mM ammonium phosphate dibasic at room temperature (RT) and 75°C and 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate at RT or 75°C. B) Protein yield for at 75°C for 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate, 400 mM ammonium phosphate dibasic, 200 mM ammonium bicarbonate, and 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4M guanidine HCl. C) Protein yield for all other human samples extracted using 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate at 75°C. *p<0.05 **p<0.01 ***p<0.001
Extractions from bones obtained from the other cadaveric donors resulted in yields between 3.53 ± 0.42 and 7.79 ± 0.23 mg protein/g bone (Fig. 2C).
Mass Spectrometry
For both extractions from 48-yr donor, peptides from collagen I were the most abundantly detected (Table 1, Table S1). Osteocalcin and ceruloplasmin were only detected in the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate extractions using the top 2 CID method (Table 1), and actin, serum albumin, and apolipoprotein A-1 were only detected in the 400 mM ammonium phosphate dibasic extraction also only using the top 2 CID method. Hemoglobin, vimentin, and fibrinogen gamma chain peptides were detected for both 400 mM ammonium phosphate dibasic and 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate (Table 1). Osteocalcin was detected in the 400 mM ammonium phosphate dibasic extraction when using the top 5 CID fragmentation method.
Table 1.
Proteins detected for the 48-year-old male donor bone for all temperatures and times on the initial tests of 400 mM ammonium phosphate dibasic and ammonium phosphate dibasic/200 mM ammonium bicarbonate in alphabetical order.
| 400 mM ammonium phosphate dibasic | 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate |
| Actin, aortic smooth muscle | Ceruloplasmin |
| Apolipoprotein A-I | Collagen alpha-1(I) chain |
| Collagen alpha-1(I) chain | Collagen alpha-2(I) chain |
| Collagen alpha-2(I) chain | Fibrinogen gamma chain |
| Fibrinogen gamma chain | Hemoglobin subunit alpha |
| Hemoglobin subunit beta | Hemoglobin subunit beta |
| Histone H2A | Osteocalcin |
| Serum albumin | Vimentin |
| Vimentin |
While as few as 4 hrs of extraction is possible for the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate method, we utilized 24 hrs for the individual ages to maximize the amount and types of protein extracted for mass spectrometry. In all samples for the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate extraction fragmented using the top 5 method, collagen I alpha-2 and alpha-1 were consistently the most abundant and second most abundant protein chains detected, respectively (Table S2). Osteocalcin was detected by Mascot for all samples (Table S1). Several other proteins were also detected (e.g., vitronectin, lumican, biglycan; Table S2). For all samples, 7.3 ± 2.4 proteins, 939.1 ± 185.8 total peptides, and 128.4 ± 19.1 unique peptides were detected using this extraction and mass spectrometry method. After using protease inhibitors, 9 proteins were identified for the 48M whereas only 5 were identified in the non-inhibited sample.
The 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4M GuHCl extraction resulted in the greatest number of protein identifications (as many at 20 unique accession numbers; Table S2). Collagen I alpha 1 and 2 and osteocalcin were the highest scoring proteins for this extraction, consistent with the other 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate extraction. Other matrix proteins (e.g., lumican, biglycan, collagen III, vitronectin, osteomodulin) were also detected (Table S2).
Discussion
Because extraction remains at a low level of total yield for extant bone [24; 27], bone protein extraction remains a major challenge in fully understanding the proteome of bone beyond the isolation and characterization of individual proteins [34; 35; 36; 37] Using methodology derived from hydroxyapatite chromatography for elution, we have developed a small scale technique to extract protein from extant bone that has yields higher than other small scale, previously reported non-demineralization and demineralization extractions (Fig. 3). This result is especially evident when heating was included during incubation. However, unlike other non-demineralization extractions [26; 27; 29], our methodology does not require the use of denaturing agents (e.g., guanidine HCl [27] or Rapigest [28]), potentially leading to a better understanding of bone proteins in a more native conformation or without the loss of denatured crosslinks. However, the addition of 4M guanidine HCl to the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate buffer resulted in an increase in the number of protein identifications by increasing the solubility of matrix proteins that may not be soluble in the without denaturation. The inclusion or exclusion of guanidine HCl to the extraction buffer can provide the flexibility to examine proteins in a more native or completely denatured states broadening the application of this extraction methodology.
In accordance with other bone proteome and extraction studies [24; 25; 26; 27; 29], peptides from collagen I were the most commonly detected (total: alpha 1: 376 ± 50.7; alpha 2: 469.6 ± 63.4 peptides; unique: alpha 1: 95.7 ± 10.8, alpha 2: 94.9 ± 9.0 peptides) by mass spectrometry because collagen I is by far the most abundant protein in bone [38]. The detection of collagen I peptides still remains a problem in understanding the bone proteome because it can block the detection of lower abundance proteins from the extracellular matrix (ECM), bone cells, and vasculature. However, osteocalcin (the second most abundant protein; [38]), several proteins derived from vasculature (e.g., hemoglobin, serum albumin, fibrinogen), and vimentin from cells within the bone were still detected, suggesting that the use of ammonium phosphate dibasic and ammonium bicarbonate can extract a variety of proteins beyond collagen I and allow for their detection. This observation is further bolstered by samples of various ages that show additional proteins from the ECM (e.g., biglycan, collagen III, lumican, vitronectin, tenascin, osteomodulin, chondroadherin). Osteocalcin [36] and osteomodulin were the only mineral specific proteins we detected (Table 1, S1, S2). However, this result is promising because it implies that our extraction methodology can interact with the hydroxyapatite surface sufficiently to dissociate mineral proteins. Because osteocalcin was only detected consistently in the extraction containing ammonium bicarbonate, our results suggest that bicarbonate can disrupt the carboxyl interaction between osteocalcin and the mineral surface. Future pre-HPLC fractionation (e.g., strong ion exchange, phosphopeptide enrichment) might be necessary to fully characterize these protein samples beyond a basic shotgun approach. This is especially true for peptides from the acidic mineral associated proteins (e.g., osteocalcin, osteopontin) that may coelute with more basic peptides (e.g., from collagen I) resulting in limited ionization [39]. Additionally, while we only observe a few additional identifications with the addition of protease inhibitors (Table S2), these may be critical to more widely characterize the bone proteome using this methodology without loss of post-translational modifications or production of non-tryptic peptides resulting in more complicated database searching.
Our study has limitations. While we detect fewer total proteins than some previous studies [26; 27], this may be a result of our chromatographic separation and peptide fragmentation than the extraction method itself. SDS-PAGE gels (data not shown) indicate a large amount of protein across the molecular weight range for the heated extractions using both the 400 mM ammonium phosphate dibasic and 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate. Thus, other proteins are also present in these extractions beyond those that are detected by mass spectrometry. We were also unable to detect osteopontin, but this may be the product of phosphorylation resulting in peptide suppression during coelution with other non-phosphorylated peptides [40]. Additional sample processing (e.g., immobilized metal affinity chromatography [40]) may be necessary to identify this important ECM protein. Our total protein yield for both the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate and 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate/4M GuHCl are some of the highest reported for any bone type (e.g., long bones, skull bones), especially at 75°C for the 400 mM ammonium phosphate dibasic/200 mM ammonium bicarbonate, without demineralization and is a similar level to previously published methods including demineralization on canine parietal bone [27].
Supplementary Material
Acknowledgments
We would like to thank D. Zagorevski for RPI Proteomic Core access, F. Kazi for laboratory assistance, and C. Drouet, C. Combes, D. Laurencin and C. Rey for helpful discussions. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01 AR49635. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional funding was provided by an internal grant from RPI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tang SY, Zeenath U, Vashishth D. Effects of non-enzymatic glycation on cancellous bone fragility. Bone. 2007;40:1144–1151. doi: 10.1016/j.bone.2006.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vashishth D. Advanced glycation end-products and bone fractures. IBMS BoneKEy. 2009;6:268–278. doi: 10.1138/20090390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karim L, Vashishth D. Heterogeneous Glycation of Cancellous Bone and Its Association with Bone Quality and Fragility. PLoS One. 2012;7:e35047. doi: 10.1371/journal.pone.0035047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poundarik AA, Diab T, Sroga GE, Ural A, Boskey AL, Gundberg CM, Vashishth D. Dilatational band formation in bone. Proceedings of the National Academy of Sciences. 2012;109:19178–19183. doi: 10.1073/pnas.1201513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sroga GE, Vashishth D. Effects of bone matrix proteins on fracture and fragility in osteoporosis. Current osteoporosis reports. 2012;10:141–50. doi: 10.1007/s11914-012-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito M, Marumo K. Collagen cross-links as a determinant of bone quality: a possible explanation for bone fragility in aging, osteoporosis, and diabetes mellitus. Osteoporosis International. 2010;21:195–214. doi: 10.1007/s00198-009-1066-z. [DOI] [PubMed] [Google Scholar]
- 7.Cappellini E, Jensen LJ, Szklarczyk D, Ginolhac Al, da Fonseca RAR, Stafford TW, Holen SR, Collins MJ, Orlando L, Willerslev E, Gilbert MTP, Olsen JV. Proteomic Analysis of a Pleistocene Mammoth Femur Reveals More than One Hundred Ancient Bone Proteins. Journal of Proteome Research. 2012;11:917–926. doi: 10.1021/pr200721u. [DOI] [PubMed] [Google Scholar]
- 8.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PLF, Fumagalli M, Vilstrup JT, Raghavan M, Korneliussen T, Malaspinas AS, Vogt J, Szklarczyk D, Kelstrup CD, Vinther J, Dolocan A, Stenderup J, Velazquez AMV, Cahill J, Rasmussen M, Wang X, Min J, Zazula GD, Seguin-Orlando A, Mortensen C, Magnussen K, Thompson JF, Weinstock J, Gregersen K, Roed KH, Eisenmann V, Rubin CJ, Miller DC, Antczak DF, Bertelsen MF, Brunak S, Al-Rasheid KAS, Ryder O, Andersson L, Mundy J, Krogh A, Gilbert MTP, Kjaer K, Sicheritz-Ponten T, Jensen LJ, Olsen JV, Hofreiter M, Nielsen R, Shapiro B, Wang J, Willerslev E. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 9.Schweitzer MH, Suo Z, Avci R, Asara JM, Allen MA, Arce FT, Horner JR. Analyses of Soft Tissue from Tyrannosaurus rex Suggest the Presence of Protein. Science. 2007;316:277–280. doi: 10.1126/science.1138709. [DOI] [PubMed] [Google Scholar]
- 10.Schweitzer MH, Zheng W, Organ CL, Avci R, Suo Z, Freimark LM, Lebleu VS, Duncan MB, Vander Heiden MG, Neveu JM, Lane WS, Cottrell JS, Horner JR, Cantley LC, Kalluri R, Asara JM. Biomolecular Characterization and Protein Sequences of the Campanian Hadrosaur B. canadensis. Science. 2009;324:626–631. doi: 10.1126/science.1165069. [DOI] [PubMed] [Google Scholar]
- 11.Schweitzer MH, Zheng W, Cleland TP, Bern M. Molecular analyses of dinosaur osteocytes support the presence of endogenous molecules. Bone. 2013;52:414–423. doi: 10.1016/j.bone.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Schweitzer M, Hill CL, Asara JM, Lane WS, Pincus SH. Identification of Immunoreactive Material in Mammoth Fossils. Journal of Molecular Evolution. 2002;55:696–705. doi: 10.1007/s00239-002-2365-6. [DOI] [PubMed] [Google Scholar]
- 13.Buckley M, Collins M, Thomas-Oates J, Wilson JC. Species identification by analysis of bone collagen using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2009;23:3843–3854. doi: 10.1002/rcm.4316. [DOI] [PubMed] [Google Scholar]
- 14.Buckley M, Whitcher Kansa S, Howard S, Campbell S, Thomas-Oates J, Collins M. Distinguishing between archaeological sheep and goat bones using a single collagen peptide. Journal of Archaeological Science. 2010;37:13–20. [Google Scholar]
- 15.Buckley M, Kansa S. Collagen fingerprinting of archaeological bone and teeth remains from Domuztepe, South Eastern Turkey. Archaeological and Anthropological Sciences. 2011;3:271–280. [Google Scholar]
- 16.Buckley M, Larkin N, Collins M. Mammoth and Mastodon collagen sequences; survival and utility. Geochimica et Cosmochimica Acta. 2011;75:2007–2016. [Google Scholar]
- 17.Richter KK, Wilson J, Jones AKG, Buckley M, van Doorn N, Collins MJ. Fish 'n chips: ZooMS peptide mass fingerprinting in a 96 well plate format to identify fish bone fragments. Journal of Archaeological Science. 2011;38:1502–1510. [Google Scholar]
- 18.Buckley M. A Molecular Phylogeny of Plesiorycteropus Reassigns the Extinct Mammalian Order 'Bibymalagasia'. PLoS One. 2013;8:e59614. doi: 10.1371/journal.pone.0059614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadsworth C, Buckley M. Proteome degradation in fossils: investigating the longevity of protein survival in ancient bone. Rapid Communications in Mass Spectrometry. 2014;28:605–615. doi: 10.1002/rcm.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrom PH, Gandhi H, Strahler JR, Walker AK, Andrews PC, Leykam J, Stafford TW, Kelly RL, Walker DN, Buckley M, Humpula J. Unraveling the sequence and structure of the protein osteocalcin from a 42 ka fossil horse. Geochimica et Cosmochimica Acta. 2006;70:2034–2044. [Google Scholar]
- 21.Buckley M, Anderung C, Penkman K, Raney BJ, Götherström A, Thomas-Oates J, Collins MJ. Comparing the survival of osteocalcin and mtDNA in archaeological bone from four European sites. Journal of Archaeological Science. 2008;35:1756–1764. [Google Scholar]
- 22.Ostrom PH, Schall M, Gandhi H, Shen TL, Hauschka PV, Strahler JR, Gage DA. New strategies for characterizing ancient proteins using matrix-assisted laser desorption ionization mass spectrometry. Geochimica et Cosmochimica Acta. 2000;64:1043–1050. [Google Scholar]
- 23.Nielsen-Marsh CM, Ostrom PH, Gandhi H, Shapiro B, Cooper A, Hauschka PV, Collins MJ. Sequence preservation of osteocalcin protein and mitochondrial DNA in bison bones older than 55 ka. Geology. 2002;30:1099–1102. [Google Scholar]
- 24.Cleland TP, Voegele K, Schweitzer MH. Empirical Evaluation of Bone Extraction Protocols. PLoS One. 2012;7:e31443. doi: 10.1371/journal.pone.0031443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Y, Liu J, Wang S, Wang H, Shi F, Xiong L, He W, Peng X. Functional proteome of bones in rats with osteoporosis following ovariectomy. Life Sciences. 2005;76:2893–2901. doi: 10.1016/j.lfs.2004.10.059. [DOI] [PubMed] [Google Scholar]
- 26.Pastorelli R, Carpi D, Airoldi L, Chiabrando C, Bagnati R, Fanelli R, Moverare S, Ohlsson C. Proteome analysis for the identification of in vivo estrogen-regulated proteins in bone. Proteomics. 2005;5:4936–4945. doi: 10.1002/pmic.200401325. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Ye M, Liu G, Feng S, Cui L, Zou H. Method development of efficient protein extraction in bone tissue for proteome analysis. J Proteome Res. 2007;6:2287–94. doi: 10.1021/pr070056t. [DOI] [PubMed] [Google Scholar]
- 28.Salmon CR, Tomazela DM, Ruiz KG, Sr, Foster BL, Paes Leme AF, Sallum EA, Somerman MJ, Nociti FH., Jr Proteomic analysis of human dental cementum and alveolar bone. Journal of Proteomics. 2013;91:544–555. doi: 10.1016/j.jprot.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerstenfeld LC, Feng M, Gotoh Y, Glimcher MJ. Selective extractability of noncallagenous proteins from chicken bone. Calcified Tissue International. 1994;55:230–235. doi: 10.1007/BF00425880. [DOI] [PubMed] [Google Scholar]
- 30.Freitag R, Hilbrig F. Isolation and purification of recombinant proteins, antibodies and plasmid DNA with hydroxyapatite chromatography. Biotechnology journal. 2012;7:90–102. doi: 10.1002/biot.201100015. [DOI] [PubMed] [Google Scholar]
- 31.Hou Y, Morrison CJ, Cramer SM. Classification of protein binding in hydroxyapatite chromatography: synergistic interactions on the molecular scale. Analytical Chemistry. 2011;83:3709–16. doi: 10.1021/ac103336h. [DOI] [PubMed] [Google Scholar]
- 32.Sroga GE, Karim L, Colón W, Vashishth D. Biochemical Characterization of Major Bone-Matrix Proteins Using Nanoscale-Size Bone Samples and Proteomics Methodology. Molecular & Cellular Proteomics. 2011;10:M110.006718. doi: 10.1074/mcp.M110.006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protocols. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 34.Franzén A, Heinegård D. Extraction and purification of proteoglycans from mature bovine bones. Biochemistry Journal. 1984;224:47–58. doi: 10.1042/bj2240047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franzén A, Heinegård D. Isolation and characterization of two sialoproteins present only in bone calcified matrix. Biochemistry Journal. 1985;232:715–724. doi: 10.1042/bj2320715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gundberg CM, Hauschka PV, Lian JB, Gallop PM. Osteocalcin: Isolation, characterization, and detection. In: Finn W, Kivie M, editors. Methods in Enzymology. Academic Press; 1984. pp. 516–544. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg H, Sodek J. Purification of mineralized tissue-associated osteopontin. Journal of tissue culture methods. 1994;16:211–215. [Google Scholar]
- 38.Hauschka PV, Wians FH. Osteocalcin-hydroxyapatite interaction in the extracellular organic matrix of bone. The Anatomical Record. 1989;224:180–188. doi: 10.1002/ar.1092240208. [DOI] [PubMed] [Google Scholar]
- 39.Tang H, Arnold RJ, Alves P, Xun Z, Clemmer DE, Novotny MV, Reilly JP, Radivojac P. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–e488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- 40.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr Opin Chem Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.