Abstract
Objective
The recently described endocrine functions of osteoblasts raise questions about their transcriptional regulation. Thus far, this aspect of osteoblast biology has been addressed only by examining the role of transcription factors binding to specific cis-acting elements in the promoter of the Osteocalcin gene.
Methods
In contrast, the role of chromatin remodeling enzymes, such as histone deacetylases (HDACs), in this process has not as yet been thoroughly understood.
Results
Here we show that through its expression in osteoblasts, one class II HDAC molecule, HDAC4, favors Osteocalcin expression, and as a result, the physiological functions regulated by osteocalcin such as spatial learning, memory, male fertility and insulin secretion. Molecular and genetic evidence indicates that through its expression in osteoblasts HDAC4 fulfills these long-range functions in part by stabilizing the transcription factor ATF4. Remarkably, through its expression in osteoblasts, HDAC4 also enhances appetite, a physiological function that is not regulated by osteocalcin.
Conclusions
These results provide a more in depth molecular understanding of the regulation of the endocrine functions of the osteoblast, and suggest the existence of additional hormones synthesized by osteoblasts that also regulate appetite.
Keywords: HDAC4, Osteoblasts, Cognition, Endocrine functions, Appetite
1. Introduction
A recent significant advance in bone biology has been the demonstration that osteoblasts are endocrine cells secreting at least two hormones, FGF23 and osteocalcin [1–3]. These two hormones influence a wide range of physiological processes ranging from the phosphate metabolism of FGF23 [1,2], to insulin secretion and glucose homeostasis, testosterone synthesis and male fertility, brain development and cognitive functions for osteocalcin [3–7]. One of the questions raised by this growing body of work is to determine how the expression of these two hormones is controlled. Because of our long-term interest in the biology of this hormone [1–3,7], we have addressed this question by studying the regulation Osteocalcin expression.
Previous work from other laboratories and ours has identified several transcription factors binding to the promoters of the Osteocalcin genes and regulating their expression in the mouse. These include Runx2, which binds to the OSE2 cis-acting element present in the promoter of the Osteocalcin genes [1,2,8,9], ATF4 that binds to the OSE1 element in the same promoters [3–7,9,10], and FoxO1 [11] and AP-1 [12]. For all these transcription factors, there is genetic evidence that they affect the endocrine functions of osteocalcin [11,13–15].
A second level of complexity in the regulation of gene expression is represented by chromatin structure. Class II histone deacetylases (HDACs) are a class of chromatin remodeling enzymes that have a poorly active deacetylase domain, but a long N-terminal domain to which transcription factors can bind. As a result, class II HDACs have often been proposed to link extracellular cues to the genome of a given cell [16–21]. Consistent with this view of the functions of class II HDACs, we recently provided evidence that the accumulation of one deacetylase in osteoblasts, HDAC4, is controlled by sympathetic signaling, which favors HDAC4 translocation into the nucleus of osteoblasts, its association with ATF4, and consequently, the regulation of Rankl expression by ATF4 [22].
Given the known regulation of Osteocalcin expression by ATF4, these data raise the prospect that HDAC4 might also enhance the expression of Osteocalcin. Conceivably, HDAC4 may also regulate the expression of other as-yet-unidentified osteoblast-derived hormones, the existence of which was suggested by the discrepancies between the metabolic abnormalities seen in Osteocalcin−/− mice and in mice lacking osteoblasts [23]. The present study was initiated to address these questions.
2. Results
2.1. HDAC4 regulates Osteocalcin expression
As a first approach to determine if HDAC4 could affect some of the long-range functions exerted by the osteoblast, we measured Osteocalcin expression in mice lacking Hdac4 only in osteoblasts (Hdac4osb−/−). As shown in Figure 1A, Osteocalcin expression in bone was decreased nearly 4-fold in Hdac4osb−/− compared with control mice and more moderately, and not significantly, in Hdac4osb+/- mice (Figure 1A,B). This regulation of Osteocalcin expression by class II HDACs appeared to be specific to HDAC4, as mice lacking another class II HDAC, HDAC5, did not experience any decrease in Osteocalcin expression (Figure 1A). To determine if HDAC4 regulates Osteocalcin expression in part through its association with ATF4 [22], we generated compound heterozygous mice lacking one copy of Hdac4 and one copy of Atf4 in osteoblasts only. As shown previously, this gene deletion strategy removed Hdac4 and Atf4 from osteoblasts but not from any other tissues tested (Figure 1C) [13,22]. Osteocalcin expression was significantly decreased in Atf4osb+/−; Hdac4osb+/− mice although not to the same extent as in Hdac4osb−/− mice (Figure 1B). This latter observation suggests that HDAC4 may interact with additional transcription factors besides ATF4 to regulate Osteocalcin expression. Nevertheless, in agreement with this decrease in Osteocalcin expression, the circulating level of all forms of osteocalcin, including its undercarboxylated i.e., active, form were decreased in a statistically significant manner in Hdac4osb−/− and Atf4osb+/−;Hdac4osb+/− but not in Hdac4osb+/− or Atf4osb+/− mice when compared with control mice (Figure 1D). These data indicate that HDAC4 is a regulator of Osteocalcin expression in part because it enhances the activity of ATF4 in osteoblasts. This regulation of Osteocalcin expression by HDAC4-ATF4 complex appears to be independent of the regulation of HDAC4 activity by the sympathetic nervous system, since Atf4 expression is normal in Adrb2osb−/− mice (data not shown) and isoproterenol does not regulate Osteocalcin expression [24].
Figure 1.

(A) Analysis of Osteocalcin expression in 2-month-old Hdac4f/f (n = 11), Hdac4osb−/−(n = 11) bones. Results are presented as fold changes compared to levels seen in bones of Hdac4f/f or WT mice. (B) Analysis of Osteocalcin expression in 2-month-old control (n = 8), Atf4osb+/− (n = 2), Hdac4osb+/− (n = 3), Atf4osb+/−;Hdac4osb+/− (n = 4) bones. Results are presented as fold changes compared to levels seen in bones of control mice. (C) Analysis of Atf4 and Hdac4 expression in 2-month-old control (n = 8), Atf4osb+/− (n = 2), Hdac4osb+/− (n = 3), Atf4osb+/−;Hdac4osb+/− (n = 4) bones. Results are presented as fold changes compared to levels seen in bones of Hdac4f/f or control mice. (D) Circulating levels of total and undercarboxylated osteocalcin in Hdac4f/f (n = 11), Hdac4osb−/− (n = 11), control (n = 8), Atf4osb+/− (n = 2), Hdac4osb+/− (n = 3), Atf4osb+/−;Hdac4osb+/− (n = 4) mice. Results are given as means ± SEM. *p < 0.05 by Student's t-test.
2.2. HDAC4 regulates spatial learning through its expression in osteoblasts
In view of the data presented above, we analyzed whether physiological processes known to be regulated by osteocalcin were affected in Hdac4osb−/− mice. Since the decrease in circulating osteocalcin levels remains moderate in the Hdac4osb−/− mice, we began this analysis by evaluating spatial learning and memory, because of all the functions ascribed to osteocalcin, this is the most clearly affected in Osteocalcin+/− mice [5]. For that purpose, control and Hdac4osb−/− mice were subjected to a Morris Water Maze test that analyzes the ability of mice to use spatial cues to locate a submerged platform. Spatial learning was assessed over 3 trials per day for 10 days. As shown in Figure 2, Hdac4osb−/− mice that demonstrate a more than 50% decrease in circulating osteocalcin levels, experienced an impairment to learn that was comparable in severity to what was observed in Osteocalcin+/− mice [5]. We remain aware, however, that HDAC4 through its expression in osteoblasts, could regulate the expression of other hormones affecting spatial learning and memory.
Figure 2.

(A) Morris water maze test performed over 10 days. The graph shows the time (seconds) needed for 3 month-old Hdac4f/f (n = 8) and Hdac4osb−/− (n = 9) mice to localize the platform in the swimming area. Results are given as means ± SEM. *p < 0.05 by Student's t-test.
2.3. HDAC4 regulates male fertility through its expression in osteoblasts
Because this is another physiological function that is severely hampered by the absence of osteocalcin [4] we next studied male fertility in Hdac4osb−/− mice. Testis size and weight were significantly decreased in Hdac4osb−/− male mice at 2 months of age compared to control littermates (Figure 3A–B). The epididymis and seminal vesicle weight, as well as sperm counts, were also decreased in Hdac4osb−/− male mice, as were the circulating levels of testosterone (Figure 3C–F).
Figure 3.
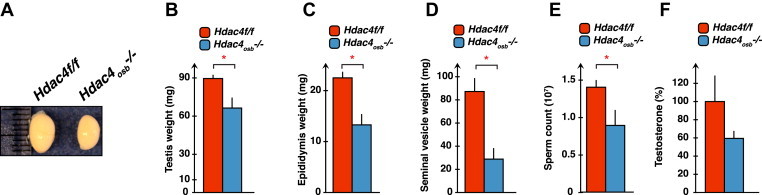
(A–F) Testis size and weight, epididymes and seminal vesicle weights, sperm count and testosterone levels in 2-months-old Hdac4osb−/− (n = 5–8) and Hdac4f/f (n = 5–9) mice. Results are given as means ± SEM. *p < 0.05 by Student's t-test.
2.4. HDAC4, through its expression in osteoblasts, regulates energy metabolism in an osteocalcin-dependent and independent manner
In the last set of experiments, we asked if Hdac4, through its expression in osteoblasts, was regulating glucose metabolism. In support of this idea, the circulating levels of insulin were decreased in Hdac4osb−/− and Atf4osb+/−; Hdac4osb+/− but not in Hdac5−/− compared with control mice (Figure 4A,B). In addition, insulin content in the pancreas, as well as β-cell area, were significantly decreased in Hdac4osb−/− compared to Hdac4f/f control mice (Figure 4C–D). As a result, insulin secretion, as measured by a glucose stimulated insulin secretion test, was decreased in Hdac4osb−/− mice compared with control littermates (Figure 4E). All these results are consistent with the notion that HDAC4 regulates glucose metabolism in an osteocalcin-dependent manner. However, unlike what is observed in Osteocalcin−/− mice [3], insulin sensitivity as determined by an insulin tolerance test was not affected by the deletion of Hdac4 from osteoblasts (Figure 4F). Furthermore, circulating levels of GLP1, another regulator of insulin sensitivity [25,26], were normal in Hdac4osb−/− mice (Figure 4G). In addition to these disturbances of glucose metabolism, appetite, a function that is not regulated by osteocalcin [3], was significantly decreased in Hdac4osb−/− mice (Figure 4H). Consequently, body weight was also reduced in Hdac4osb−/− mice (Figure 4I). This decrease in appetite was not caused by an increase in leptin levels in Hdac4osb−/− mice (Figure 4J).
Figure 4.
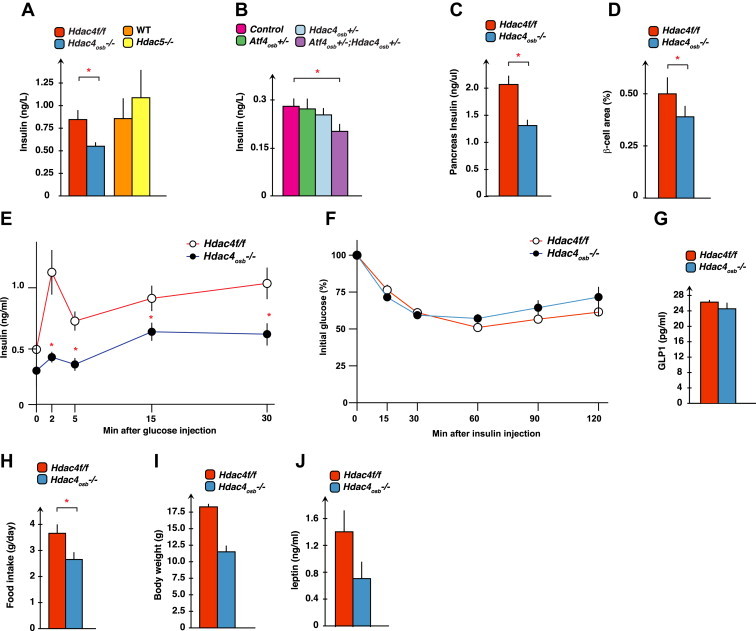
(A) Serum insulin levels in 2-month-old Hdac4f/f (n = 11), Hdac4osb−/− (n = 11), wild-type (n = 8) and Hdac5−/− (n = 8) mice. (B) Serum insulin levels in control (n = 8), Atf4osb+/− (n = 3), Hdac4osb+/− (n = 3), Atf4osb+/−;Hdac4osb+/− (n = 4) mice. (C) Insulin content in the pancreata of 2-month-old Hdac4f/f (n = 7) and Hdac4osb−/− (n = 8) mice. (D) Histomorphometric analysis of β-cell area (%) in pancreata of 2-month-old Hdac4f/f (n = 7) and Hdac4osb−/− (n = 8) mice. (E) Glucose-stimulated insulin secretion (GSIS) test in 2-month-old Hdac4f/f (n = 7) and Hdac4osb−/− (n = 6) mice. (F) Insulin tolerance test (ITT) in 2 months-old Hdac4f/f (n = 12) and Hdac4osb−/− (n = 7) mice. (G) Serum GLP1 levels in 2-month-old Hdac4f/f (n = 5), Hdac4osb−/− (n = 5). (H) Food intake per day. Results are given as means ± SEM. *p < 0.05 by Student's t-test. (I) Body weight of 2-month-old Hdac4f/f (n = 5), Hdac4osb−/− (n = 6) mice. (J) Serum leptin levels in 3-month-old Hdac4f/f (n = 5), Hdac4osb−/− (n = 4) mice. Results are given as means ± SEM. *p < 0.05 by Student's t-test.
Taken together, these results indicate that through its expression in osteoblasts, HDAC4 regulates Osteocalcin expression, and many of the endocrine functions this hormone regulates. Through its expression in osteoblasts, Hdac4 also regulates another metabolic function, appetite, that is not affected by osteocalcin [3]. This latter result indicates that osteoblasts may secrete additional hormone(s) regulating other aspects of energy metabolism such as appetite.
3. Discussion
In this study, we show that the class II HDAC molecule HDAC4 regulates, through its expression in osteoblasts, physiological processes as diverse as spatial learning and memory, male fertility, insulin secretion and appetite. Many, but not all of these functions are regulated by the bone-derived hormone osteocalcin and indeed, Osteocalcin expression and circulating levels are lower in Hdac4osb−/− mice than in control mice. These results provide a better molecular understanding of the regulation of the endocrine functions of osteoblasts.
Although they belong to the large family of histone deacetylases, class II HDACs do not usually fulfill a deacetylase function [16–21]. Instead, through their long N-terminal domain, they can interact with transcription factors, such as members of the MEF2 family, Runx2 or ATF4 in osteoblasts [22,27]. This is why they have been proposed to act as bridges between extracellular cues and the genome of osteoblasts. For instance, HDAC4 integrates sympathetic signaling in osteoblasts, and consequently regulates osteoclast differentiation [22]. The ability of HDAC4 to interact with and increase the activity of ATF4 was of interest to us, since we have shown previously that ATF4 regulates Osteocalcin expression and therefore the endocrine functions of osteoblasts [13].
As one would anticipate given its interaction with ATF4 in osteoblasts, HDAC4 is necessary for proper Osteocalcin expression in these cells to maintain normal circulating levels of osteocalcin, and as a result for a variety of functions ascribed to this hormone. Remarkably, the regulation of the endocrine functions of osteoblasts by HDAC4 is specific to this molecule and is not shared by another closely related class II HDAC, HDAC5. Although HDAC4 fulfills some of its functions in an ATF4-dependent manner, our results do not rule out the possibility that HDAC4 could regulate Osteocalcin expression and function by interacting with other transcription factors affecting the expression of Osteocalcin. Osteocalcin circulating levels are lower in Hdac4osb−/− and Atf4osb+/−;Hdac4osb+/− mice but not all the functions ascribed to osteocalcin are affected in these two mutant mouse strains. For instance, anxiety or insulin sensitivity was normal in these mutant mice. One explanation for these observations may be that to be affected, these functions require even lower levels of circulating osteocalcin than the impaired functions observed in Hdac4osb−/− mice.
In addition, at least one physiological function, the control of appetite, affected in mice lacking Hdac4 in osteoblasts only is not regulated by osteocalcin in vivo [3]. Therefore, these results suggest that HDAC4 regulates the expression of another osteoblast-secreted hormone regulating appetite. These observations are consistent with Yoshikawa et al., who also observed osteocalcin-independent regulation of energy metabolism in their model of osteoblast ablation [23]. Thus, the results presented in this study imply that the osteoblast is a richer endocrine cell than what has already been described. As such, they are an incentive to embark on a systematic search for other osteoblast-derived hormones that could regulate appetite.
4. Material and methods
4.1. Mouse generation
Hdac4 floxed mice were generated by inserting loxP sites flanking exon 6 that results in an out-of-frame mutation in the Hdac4 allele (provided by Dr. E. Olson and Dr. R. Bassel-Duby, UT Southwestern, Dallas, Texas) [28]. Hdac4osb−/− mice were generated by intercrossing Hdac4f/f mice with Runx2-Cre transgenic mice [29]. Hdac5−/− mice were generated as previously reported [30]. Briefly, the 5′ region of Hdac5 exon 3 was fused in-frame with the lacZ cDNA and neomycin resistance cassette under the control of the PGK promoter to the 5′ region of exon 3, therefore placing a β-galactosidase reporter gene under the control of endogenous Hdac5 promoter (provided by Dr. E. Olson and Dr. R. Bassel-Duby, UT Southwestern, Dallas, Texas). Atf4 conditional allele was generated by placing a floxed neo cassette upstream of exon2 and a loxP site downstream of exon3 [13]. Atf4osb+/−;Hdac4osb+/− mice were generated by crossing Atf4f/f or Atf4f/+ mice with Hdac4osb+/− mice. Floxed mice and Cre expressing mice littermates (indicated as control in figures) were used as control mice for Atf4osb+/−;Hdac4osb+/−. Mouse genotypes were determined by PCR. All procedures involving animals were approved by the IACUC and conform to the relevant regulatory standards.
4.2. Metabolic studies
For insulin tolerance test (ITT) mice were fasted for 4 h and then insulin (0.44 U/kg) was given with intraperitoneal injection (IP). Blood glucose levels were measured at indicated time points using the Accu-Chek active system (Roche). For glucose-stimulated insulin secretion (GSIS) test, mice were fasted for 16 h then injected IP with 3 g/kg of glucose. Blood was collected from the tails of these mice at indicated time points and serum insulin levels were assessed using the Ultra-sensitive Insulin ELISA kit (Crystal Chem). Food intake was measured using metabolic cages as the daily change of food weight.
4.3. Sperm counts
Caudal epididymides were minced in 1 ml PBS and the number of cells released was counted after 1 h. The total sperm count was assessed in the final suspension by using a hemocytometer [31].
4.4. Hormone measurements
Circulating testosterone levels were measured by radioactive immunoassay (Alpco). Random insulin levels in mice were measured in tail blood collected and assayed with the mouse/human insulin ELISA (Mercodia). Total and undercarboxylated osteocalcin levels were quantified using a previously described ELISA [32]. Circulating leptin and GLP1 levels in mice were measured by specific ELISA (Milipore EZML-82K and Raybiotech EIA-GLP1).
4.5. Histology
Pancreatic tissues were fixed in 10% neutral formalin, embedded in paraffin, and sectioned at 5 μm; sections were subjected to immunohistochemistry using an anti-insulin antibody and counterstained with hematoxylin and eosin (H&E). To evaluate cell sizes or numbers, five to ten sections (each 50 μm apart) were analyzed using a 40× objective on a Leica microscope outfitted with a CCD camera (SONY). β-cell area represents the surface positive for insulin immunostaining divided by the total pancreatic surface.
4.6. Gene expression
RNA isolation, cDNA preparation and quantitative PCR analysis were carried out following standard protocols. Briefly, total RNA was extracted, DNase I-treated and reverse transcribed with random primers using the Superscript III cDNA synthesis kit (Invitrogen, Carlsbad, CA). The cDNA samples were then used as templates for qPCR analysis that was performed using the Taq SYBR Green supermix with ROX (Biorad, Hercules, CA) and specific primers on a Biorad CFX-Connect apparatus. The following primers were used: Osteocalcin 5′-AAGCAGGAGGGCAATAAGGT-3′ and 5′-TTTGTAGGCGGTCTTCAAGC-3′ Expression levels of all qPCR reactions were normalized using Hprt expression levels as internal control for each sample.
4.7. Morris Water Maze test
Animals were transported to the testing room in their home cages, and left undisturbed for at least 30 min prior to the first trial. The maze comprised of a large swimming pool (150 cm diameter) filled with water (23 °C) made opaque with non-toxic white paint. The pool was located in a brightly lit room filled with visual cues, including geometric figures on the walls of the maze demarking the four fixed starting positions of the trials, at (0°, 90°, 180°, and 270°). A 15 cm round platform was hidden 1 cm beneath the surface of the water at a fixed position. Each daily trial block consisted of four swimming trials, with each mouse starting from the same randomly chosen starting position. The starting position varied between days. On days 1–2, mice that failed to find the platform within 2 min were guided to the platform. They had to remain on the platform for 15 s before they were returned to their cages. Mice were not guided to the platform after day 2, and the time it took them to reach the platform over repeated trials (3 trails/day for the next 10 days) was recorded as a measure of spatial learning.
4.8. Statistical analyses
Results are given as means ± standard error of the mean (SEM). Statistical analyses were performed using unpaired, two-tailed Student's t-test. In all figures, error bars represent SEM and * represents p < 0.05.
Acknowledgments
We thank Dr. P. Ducy for valuable suggestions and critical reading of the manuscript, L. Khrimian for language editing. Dr. R Bassel-Duby and Dr. E Olson for generously providing the Hdac4fl/fl, Hdac5−/− mice, and Dr. J Tuckerman for generously providing the Runx2-Cre mice. This work was supported by grant AR045548 from the National Institutes of Health (G.K.).
Conflict of interest
The authors declare no competing financial interest.
References
- 1.Fukumoto S., Yamashita T. FGF23 is a hormone-regulating phosphate metabolism—Unique biological characteristics of FGF23. Bone. 2007;40:1190–1195. doi: 10.1016/j.bone.2006.12.062. [DOI] [PubMed] [Google Scholar]
- 2.Shimada T., Mizutani S., Muto T., Yoneya T., Hino R., Takeda S. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6500–6505. doi: 10.1073/pnas.101545198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee N.K., Sowa H., Hinoi E., Ferron M., Ahn J.D., Confavreux C. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oury F., Sumara G., Sumara O., Ferron M., Chang H., Smith C.E. Endocrine regulation of male fertility by the skeleton. Cell. 2011;144:796–809. doi: 10.1016/j.cell.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oury F., Khrimian L., Denny C.A., Gardin A., Chamouni A., Goeden N. Maternal and offspring pools of osteocalcin influence brain development and functions. Cell. 2013;155:228–241. doi: 10.1016/j.cell.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferron M., Wei J., Yoshizawa T., Del Fattore A., DePinho R.A., Teti A. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 2010;142:296–308. doi: 10.1016/j.cell.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karsenty G., Oury F. Biology without walls: the novel endocrinology of bone. Annual Review of Physiology. 2012;74:87–105. doi: 10.1146/annurev-physiol-020911-153233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 9.Ducy P., Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Molecular and Cellular Biology. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H.C., Schinke T. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 11.Rached M.-T., Kode A., Silva B.C., Jung D.Y., Gray S., Ong H. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. Journal of Clinical Investigation. 2010;120:357–368. doi: 10.1172/JCI39901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozec A., Bakiri L., Jimenez M., Schinke T., Amling M., Wagner E.F. Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. Journal of Cell Biology. 2010;190:1093–1106. doi: 10.1083/jcb.201002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshizawa T., Hinoi E., Jung D.Y., Kajimura D., Ferron M., Seo J. The transcription factor ATF4 regulates glucose metabolism in mice through its expression in osteoblasts. Journal of Clinical Investigation. 2009;119:2807–2817. doi: 10.1172/JCI39366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kode A., Mosialou I., Silva B.C., Joshi S., Ferron M., Rached M.-T. FoxO1 protein cooperates with ATF4 protein in osteoblasts to control glucose homeostasis. Journal of Biological Chemistry. 2012;287:8757–8768. doi: 10.1074/jbc.M111.282897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bozec A., Bakiri L., Jimenez M., Rosen E.D., Catalá-Lehnen P., Schinke T. Osteoblast-specific expression of Fra-2/AP-1 controls adiponectin and osteocalcin expression and affects metabolism. Journal of Cell Science. 2013;126:5432–5440. doi: 10.1242/jcs.134510. [DOI] [PubMed] [Google Scholar]
- 16.Verdin E., Dequiedt F., Kasler H.G. Class II histone deacetylases: versatile regulators. Trends in Genetics. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 17.Haberland M., Montgomery R.L., Olson E.N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger S.L. Histone modifications in transcriptional regulation. Current Opinion in Genetics & Development. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 19.Yang X.-J., Grégoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Molecular and Cellular Biology. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 21.Schreiber S.L., Bernstein B.E. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 22.Obri A., Makinistoglu M.P., Zhang H., Karsenty G. HDAC4 integrates PTH and sympathetic signaling in osteoblasts. Journal of Cell Biology. 2014 doi: 10.1083/jcb.201403138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshikawa Y., Kode A., Xu L., Mosialou I., Silva B.C., Ferron M. Genetic evidence points to an osteocalcin-independent influence of osteoblasts on energy metabolism. Journal of Bone and Mineral Research. 2011;26:2012–2025. doi: 10.1002/jbmr.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinoi E., Gao N., Jung D.Y., Yadav V., Yoshizawa T., Myers M.G.J. The sympathetic tone mediates leptin's inhibition of insulin secretion by modulating osteocalcin bioactivity. Journal of Cell Biology. 2008;183:1235–1242. doi: 10.1083/jcb.200809113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drucker D.J., Philippe J., Mojsov S., Chick W.L., Habener J.F. Glucagon-like peptide I stimulates insulin gene expression and increases cyclic AMP levels in a rat islet cell line. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:3434–3438. doi: 10.1073/pnas.84.10.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreymann B., Williams G., Ghatei M.A., Bloom S.R. Glucagon-like peptide-1 7-36: a physiological incretin in man. Lancet. 1987;2:1300–1304. doi: 10.1016/s0140-6736(87)91194-9. [DOI] [PubMed] [Google Scholar]
- 27.Vega R.B., Matsuda K., Oh J., Barbosa A.C., Yang X., Meadows E. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Potthoff M.J., Wu H., Arnold M.A., Shelton J.M., Backs J., McAnally J. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. Journal of Clinical Investigation. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rauch A., Seitz S., Baschant U., Schilling A.F., Illing A., Stride B. Glucocorticoids suppress bone formation by attenuating osteoblast differentiation via the monomeric glucocorticoid receptor. Cell Metabolism. 2010;11:517–531. doi: 10.1016/j.cmet.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Molecular and Cellular Biology. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dakhova O., O'Day D., Kinet N., Yucer N., Wiese M., Shetty G. Dickkopf-like1 regulates postpubertal spermatocyte apoptosis and testosterone production. Endocrinology. 2009;150:404–412. doi: 10.1210/en.2008-0673. [DOI] [PubMed] [Google Scholar]
- 32.Ferron M., Wei J., Yoshizawa T., Ducy P., Karsenty G. An ELISA-based method to quantify osteocalcin carboxylation in mice. Biochemical and Biophysical Research Communications. 2010;397:691–696. doi: 10.1016/j.bbrc.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]


