Abstract
Vancomycin resistant enterococci (VRE) constitute a challenging problem in health care institutions worldwide. Novel methods to rapidly identify resistances are highly required to ensure an early start of tailored therapy and to prevent further spread of the bacteria. Here, a spectroscopy-based rapid test is presented that reveals resistances of enterococci towards vancomycin within 3.5 hours. Without any specific knowledge on the strain, VRE can be recognized with high accuracy in two different enterococci species. By means of dielectrophoresis, bacteria are directly captured from dilute suspensions, making sample preparation very easy. Raman spectroscopic analysis of the trapped bacteria over a time span of two hours in absence and presence of antibiotics reveals characteristic differences in the molecular response of sensitive as well as resistant Enterococcus faecalis and Enterococcus faecium. Furthermore, the spectroscopic fingerprints provide an indication on the mechanisms of induced resistance in VRE.
The rising resistances of pathogens towards antibiotics, both in hospital- and community-acquired infections, represent a significant problem in human health-care as the variety of powerful drugs is on the decline. The increasing resistances are mainly caused by the extensive use of antibiotics during the past decades1. However, also today treatment starts with a calculated therapy applying broad-spectrum antibiotics as detailed microbiological analysis of the pathogen and its resistogram takes at least 24 hours due to time consuming cultivation steps2,3. Sepsis is one example, where the survival rate of the patients after 12 hours without appropriate therapy is below 20 percent4. Therefore, novel rapid and sensitive methods to detect resistances are urgently needed in order to save patient's life by timely initiation of tailored therapy, to prevent further spread of infection by efficient eradication of the pathogen and to slow down the rise of further resistances by the use of narrow but effective antibiotics. Recent approaches include matrix-assisted laser ionization time-of-flight mass spectrometry (MALDI-TOF MS)5,6, molecular biology based techniques, such as different polymerase-chain-reaction (PCR) methods, DNA microarrays7 and high-throughput sequencing technologies (also named next-generation sequencing; NGS), and biochemical activity assays8. All of these techniques promise to shorten time to diagnosis, but still hold significant drawbacks, such that they require specific sample preparations (MALDI-TOF MS), laborious optimizations of protocols (PCR), still suffer from sufficient reproducibility (real-time PCR) or limited analytical capacity (MALDI-TOF MS)9. In this contribution, a novel Raman spectroscopy-based approach is presented that can detect antibiotic resistances within 3.5 hours. The proof of concept is demonstrated on vancomycin resistant enterococci (VRE) that have become a challenging nosocomial problem.
Enterococci are Gram positive, facultatively anaerobic oval cocci which form chains of various lengths. They are natural colonizers of the gastrointestinal tract. However, they can also cause a range of infections including pelvic infections, endocarditis, neonatal infections and urinary tract infections (UTIs), covering a spectrum from uncomplicated infections to life threatening health care associated sepsis. In regions, where VREs are not yet the predominant cause of enterococcal infections, the glycopeptide vancomycin is a crucial antibiotic for the treatment of serious infections caused by enterococci unresponsive to many other available antimicrobial agents10. Vancomycin inhibits the cell-wall synthesis by binding to the C-terminal D-alanyl-D-alanine (D-ala-D-ala) residues of peptidoglycan pentapeptide precursors and thus preventing peptidoglycan growth and assembly. Although the outbreak of glycopeptide resistant enterococci in 1986 was very surprising, today it is well-known that vancomycin resistances are common in nature and VREs are nowadays encountered in hospitals worldwide11,12,13. The two most frequently encountered resistant genotypes are called vanA and vanB where the terminal D-ala of the pentapeptide is replaced with D-lactate (D-lac) producing high-level resistances with minimal inhibition concentrations (MIC) up to 1024 μg/ml14,15.
Raman spectroscopy, a powerful vibrational spectroscopic technique, holds a high potential to significantly improve microbiological diagnosis16,17. In the past, the highly specific molecular information contained in the Raman spectra was successfully utilized to identify bacteria in patient's samples18,19,20,21,22,23,24, even on a single bacterial cell level. Furthermore, antibiotic-bacteria interactions could be detected and described by means of Raman spectroscopy25,26,27,28,29. However, the detection of bacterial resistances early in time without any time-consuming overnight cultivation steps remains a challenging task. In order to enable analysis directly from suspensions like patients' body fluids an integrated dielectrophoresis (DEP)-Raman approach has been developed in our previous study18. Using the forces on dielectric particles in spatially non-uniform electric fields, the bacteria are captured in micron-sized regions of the device where they are Raman spectroscopically characterized. As intrinsic molecular information is probed by the inelastic scattering of light, no external label is required and only small sample volumes are necessary, making Raman spectroscopy a very attractive technique for medical diagnosis.
In this study, the DEP-Raman setup was used to develop a general method to identify vancomycin resistance in enterococci with high accuracy within only 3.5 hours without the need of any information on strain identity. Therefore, the method provides a high potential to be general applicable for enterococci strains and shows many advantages compared to most gene- or antibody-based arrays that require special knowledge of the geno- or phenotypes of the resistant bacteria. In the presented method, the sample to be characterized is divided into two parts: one part is treated with vancomycin, the second part is used as control. Both parts are incubated, and within 120 minutes the sample is analyzed on the integrated DEP-Raman device (Figure 1a and 1b). The Raman spectroscopic signature of the treated sample and the control change characteristically depending whether the bacteria are sensitive or resistant towards vancomycin. A three level chemometric model based on Partial Least Squares Regression (PLS) in combination with Linear Discriminant Analysis (LDA) is used to extract those molecular changes and distinguishes resistant and sensitive enterococci (Figure 1c). The PLS-regularized LDA model is optimized to detect specific changes due to the effect of vancomycin on the bacteria and is, therefore, independent of the enterococcus strain. This was demonstrated by using two different E. faecalis and two different E. faecium strains, one being a patient isolate. Independence from the actual enterococci species and strain is of utmost importance when transferring the method to clinical application as each patient will carry its own strain.
Figure 1. Scheme of the spectroscopic method to identify VRE.
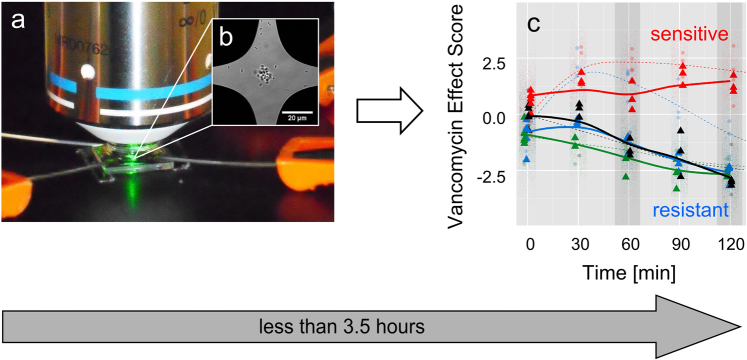
(a) Bacteria suspensions are treated with vancomycin and after 0, 30, 60, 90, and 120 min the samples are analyzed on a combined DEP-Raman setup. (b) On the chip, the bacteria are dielectrophoretically captured in a micrometer-sized region and characterized by means of Raman spectroscopy. (c) The Raman spectra are projected into a statistical model and reveal whether the pathogens are sensitive (positive vancomycin effect score) or resistant (negative vancomycin effect score) towards vancomycin. Starting with the antibiotic treatment the whole procedure takes about 2 h. Including the pre-cultivation, vancomycin resistant enteroccoci can be identified within 3.5 hours.
Results
Raman spectra reveal the interaction of vancomycin with sensitive and resistant bacteria
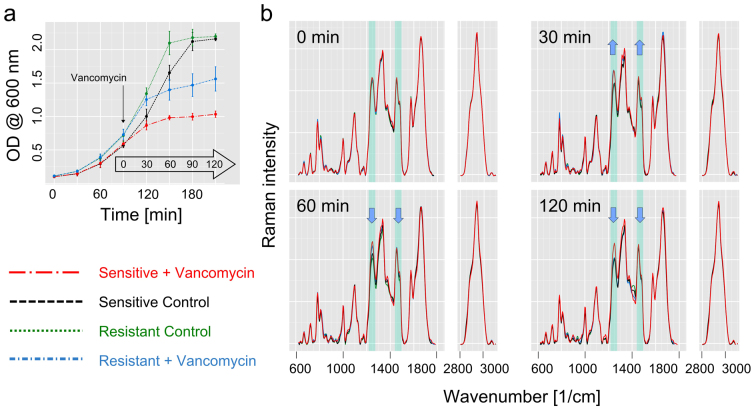
As vancomycin acts on the cell wall synthesis, bacteria suspensions were pre-incubated under defined conditions to assure they were in the exponential growth phase. Figure 2a shows the mean growth curves with standard deviations for six independently repeated measurements with one sensitive and one resistant (VanB, MIC 32 μg/ml) Enterococcus faecalis strain. The bacteria entered the exponential growth phase where their specific growth rate was constant very reproducibly. In the presented experiments, the Raman based assay was started after 90 minutes when the bacteria were in the midst of their exponential growth phase: One aliquot per strain was treated with 10 μg/ml vancomycin, a second aliquot served as a control without vancomycin treatment. Immediately after addition of the antibiotics (0 min) and after that, every 30 minutes up to 2 hours, the bacteria in the suspension were characterized by means of Raman spectroscopy on the integrated dielectrophoresis chip (Figures 1a, b). The respective mean Raman spectra for the time points 0, 30, 60 and 120 min after antibiotic treatment are presented in Figure 2b. The spectra recorded at 90 min are not shown since they do not contribute new information. All Raman spectra show typical features of Raman spectra of bacteria: nucleic acid contributions can be found at 785 cm−1 (PO2−, cytosine, thymine), 1096 cm−1 (PO2− stretching), 1250 cm−1 (adenine), 1339 cm−1 (adenine), 1485 cm−1 (guanine, adenine) and 1578 cm−1 (guanine, adenine); vibrational contributions from proteins show up at 1004 cm−1 (phenylalanine), around 1250 cm−1 with the broad amide III band and around 1660 cm−1 with the broad and prominent amide I band30,31. At the time point 0 min, the spectra of control and treated bacteria look very similar. This shows that the pure presence of vancomycin in the solution does not alter the Raman spectra and an interaction of the drug with the bacteria is necessary in order to observe an influence within the bacterial Raman spectra. Already 30 min after the addition of vancomycin, first changes within the bacteria Raman spectra can be observed. In Figure 2b, exemplarily the two vibrational bands around 1250 cm−1 and around 1485 cm−1 that change during the observation time are highlighted. Thirty minutes after vancomycin treatment both Raman bands show increased relative intensities in the treated cultures, both for the sensitive and the resistant strains, as compared to the untreated controls. At 60 min and even more at 120 min, the Raman spectra of the resistant strain change and become more similar to the spectra of the untreated control samples, well separated from the Raman spectra of the treated sensitive strain. This clearly shows the interaction of the drug with the bacteria. While the bacteria of the sensitive E. faecalis strain were permanently affected by the antibiotic, the E. faecalis bacteria with VanB resistance could adapt to the presence of vancomycin in activating their resistance mechanism. This is in good agreement with previous biochemical studies32,33,34,35,36: In resistant bacteria vancomycin also binds to the membrane sensor VanS triggering autophosphorylation and subsequent transfer of the phosphate group to the response regulator VanR. Accumulated phosphorylated VanR activates the promoters of the van gene cluster which ultimately leads to a replacement of the terminal D-alanine in the peptidoglycan precursors with a D-lactate. With this substitution, the resistant bacteria can avoid the toxic effects of the antibiotic and – after the short time it takes to convert the metabolism - continue to grow again as can also be seen in the Raman spectra. The effect of vancomycin interaction and induction of resistance is also indirectly reflected in the growth curve obtained by monitoring the optical density of the suspension (Figure 2a), however without providing the important molecular information contained in the Raman spectra (Figure 2b).
Figure 2. Spectroscopic analysis of E. faecalis at different time points with and without antibiotic treatment.
(a) Bacterial growth curves (average of 9 independent biological replicates) of the sensitive strain with vancomycin treatment (red big chain line) and without (black dashed line) and of the resistant strain with vancomycin treatment (blue small chain line) and without (green dotted line). Vancomycin is added in the exponential growth phase (10 μg/ml). At the indicated time points 0, 30, 60 and 120 min after addition of vancomycin samples were analyzed on the integrated DEP-Raman device. (b) Raman mean spectra at four different time points. Immediately after vancomycin addition (t = 0 min) all Raman spectra look very similar and no significant spectral differences can be identified, proving that the pure presence of vancomycin has no influence on the spectra. After 30 min, spectral changes are observed. In the highlighted spectral regions (at 1236 cm−1 and between 1450 and 1475 cm−1) spectral differences between the vancomycin treated and the untreated control samples are visible. The blue arrows indicate the trend of relative changes for the treated resistant strain. While after 30 min of vancomycin treatment, the low-level resistant E. faecalis strain (MIC = 32 μg/ml) shows similar changes as the sensitive strain, after 60 min, its spectral features become more similar to the control samples, and finally, after 120 min, separation between resistant and sensitive strain is achieved.
A statistical model can differentiate resistant and sensitive enterococci with high accuracy
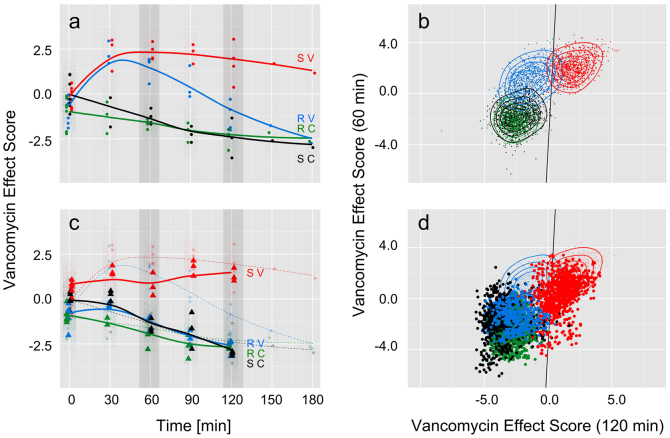
In order to use the specific information contained in the Raman spectra to predict vancomycin resistance of various enterococci strains, multivariate statistical analysis methods were applied to build a combined Partial Least Squares Regression and Linear Discriminant Analysis (PLS-LDA) model based on the Raman data. This model was specifically trained to the spectral changes caused by the inhibiting action of the drug by using the Raman data of the sensitive E. faecalis strain. Excluding the resistant strain ensured that the model learned the vancomycin effect as opposed to learning strain differences. We found a vancomycin effect score that distinguishes with high accuracy between treated and untreated enterococci (Figure 3a). Prominent contributions in the corresponding vancomycin effect score coefficients (loadings are depicted in supplementary Fig. S1) are in agreement with the highlighted changes of the Raman bands in Figure 2b and indicate that the chemical information is responsible for clustering and not baseline deviations or noise. When the data of two different E. faecalis strains at time point 0 min, i.e., immediately after vancomycin addition, are projected into the model, treated and untreated samples are not discriminated. The vancomycin effect scores form overlapping point clouds. This confirms again, that the pure presence of the drug does not cause the differentiation and vancomycin needs to interact with the enterococci to cause the observed spectral changes. Furthermore, the overlapping scores at time point 0 minutes for the resistant and the sensitive E. faecalis strain proof that the vancomycin effect score is largely independent of the specific strain. This is of utmost importance when unknown samples are analyzed where the exact strain is unknown. At later time points (30 min and later), the reaction towards antibiotic treatment is clearly visible: while the untreated controls of the resistant and the sensitive strain always have a negative vancomycin effect scores, the vancomycin treated bacterial samples show positive vancomcin effect scores. Thirty minutes after vancomycin treatment the sensitive and the resistant strain cannot be distinguished anymore. However, already after 60 min of vancomycin treatment the resistance mechanism in the VanB strain is successfully induced causing a continuous movement of the vancomycin effect scores from the positive side to the negative side. For the used resistant VanB strain with a MIC close to the used drug concentration (MIC 32 μg/ml vs. 10 μg/ml vancomycin) a full separation between sensitive and resistant strains is achieved three hours after vancomycin treatment. At that time point the scores of the treated resistant strain have reached the scores of the untreated controls.
Figure 3. Three-level chemometric model to differentiate resistant and sensitive enterococci.
(a) Vancomycin effect scores of E. faecalis strains plotted over time: PLS-LDA model trained with Raman spectra of treated (red, S V) and untreated (black, S C) sensitive strain after at least 30 min of vancomycin incubation. All data of t = 0 min and data of treated (blue, R V) and untreated (green, R C) resistant strain is projected. At t = 0 min all scores are close together and overlapping, indicating the same starting conditions. At t = 30 min sensitive and the low-level resistant strain show equal vancomycin effect values, treated and untreated strains can be separated. At 60 min the vancomycin effect score of the resistant strain starts to separate from the sensitive strain and reaches between 90 and 120 min negative vancomycin effect values, whereas the sensitive strain remains at positive vancomycin effect score values. The control samples all show up with negative vancomycin effect values and resistant and sensitive strain are not separated, indicating, that the model does not separate between strains but with respect to vancomycin response. (b) Time dependency of the vancomycin effect scores is included in the model by pairing vancomycin effect scores of E. faecalis at 60 min and 120 min and plotting against each other. A LDA is performed with these new scores and the separation line between treated sensitive and resistant strain is drawn in black. Both groups are very well separated from each other. (c) Data of E. faecium (treated and untreated, sensitive and resistant strains, same color code as above) is projected into the vector space spanned by the PLS-LDA shown in (a). In contrast to the low level resistant E. faecalis strain in (a) the high level resistant E. faecium strain immediately separates from the sensitive one and behaves like the control samples. (d) The 60 min and 120 min scores in figure (c) are paired to include time dependency in the model.
Considering the time-dependent changes of the vancomycin effect score shortens the diagnosis time
While a full separation between the sensitive and resistant strains is achieved only three hours after vancomycin treatment, the development of the vancomycin effect scores with time holds the potential to predict vancomycin resistance earlier. Therefore, the time-dependence of the vancomycin effect scores was included in the data analysis by pairing the vancomycin effect scores of time point 60 min with those scores of time point 120 min. Figure 3b shows that already after 2 hours of vancomycin treatment the point clouds of resistant and sensitive E. faecalis are very well separated, while the untreated controls of different E. faecalis strains are indistinguishable. The three level PLS-LDA-LDA model can distinguish the sensitive from the resistant strain with high accuracy. Leave-one-batch-out validation, where the model is tested with data of an independent biological replicate, yields > 86% sensitivity and 93% specificity with respect to the prediction of vancomycin resistance in E. faecalis or vice versa 93% sensitivity and 87% specificity with respect to the prediction of vancomycin sensitive E. faecalis on a single spectra level (Table 1). Thus, the antibiotic resistance can be recognized with high accuracy within two hours after admission of vancomycin, or 3.5 hours if the preliminary cultivation of the bacterial sample is taken into account. Both, acquisition of the spectra and prediction of the final result by the three level model is only a matter of seconds.
Table 1. Confusion table for predicting vancomycin resistance in enterococci. The table summarizes the classification of independent enterococci spectra after the performance of all three levels of the statistic model. The numbers of correctly identified data are found on the main diagonal, and the numbers of false classified data on the off diagonal. For E. faecalis a sensitivity of 87% and specificity of 93% are achieved, for E. faecium sensitivity and specificity are 99% and 77%, respectively.
| E. faecalis | E. faecium | |||
|---|---|---|---|---|
| Predicted → | ||||
| True ↓ | Resistant | Sensitive | Resistant | Sensitive |
| Resistant | 782 | 118 | 899 | 1 |
| Sensitive | 59 | 841 | 206 | 694 |
The statistical model correctly predicts vancomycin resistance also in other enterococci species as shown for E. faecium
The generalization power of the classification was further evaluated by testing it with two new strains from a different enterococci species, namely E. faecium. As the spectroscopy-based model is specifically trained on the differences in the Raman bands caused by the action of vancomycin, the characteristic spectral contributions of the individual enterococci strains are subtracted out. Therefore, the model is not limited to the analysis of vancomycin resistance in E. faecalis, but can also be used to reveal vancomycin resistance in E. faecium. Figures 3c and 3d show the projection of the Raman data of two E. faecium strains, one sensitive and one resistant strain with high level vancomycin resistance (MIC > 1024 μg/ml) into the model trained with E. faecalis (the corresponding Raman spectra can be found in Supplementary Fig. S2). The untreated control samples of both E. faecium strains are almost indistinguishable from the untreated controls of E. faecalis for each time point of the analysis, suggesting that the model is independent of the enterococci strain and even species. The treated sensitive E. faecium strain reveals the same behavior as observed for the sensitive E. faecalis strain and can be found with positive vancomycin effect scores. This indicates that sensitivity to vancomycin can be recognized by means of Raman spectroscopy even for unknown species. The treated E. faecium strain with high level vanA resistance shows vancomycin effect scores that are at no time point distinguishable from the untreated controls. This indicates that high level vancomycin resistance could be recognized even earlier than the previously discussed low level vancomycin resistance. The presented spectroscopy-based method can correctly determine the vancomycin resistance of enterococci within 3.5 hours even for resistant strains, where the MIC is only 1.5–3× the administered vancomycin concentration. The three level PLS-LDA-LDA model can correctly recognize resistances with 99% sensitivity from single Raman spectra recorded of E. faecium treated with vancomycin (Figure 3d and Table 1). The obtained specificity of about 77% reveals that every fourth Raman spectrum of the sensitive strain is not recognized as such. This means, with the current model the patient will not receive vancomycin if the patient carries a resistant enterococcus strain, but might - in a few cases - receive an alternative treatment which might not have been necessary, but will also cure the patient. Nevertheless, the model is expected to improve if more enterococci strains and species as well as more biological replicates are included in the training data set. However, it should be noted, that despite the fact, that the E. faecium were grown in slightly different media, the vancomycin effect model was not confused.
Discussion
In this contribution a straightforward, fast and accurate procedure based on Raman spectroscopy is presented to determine the antibiotic resistance of enterococci against vancomycin. This procedure has the potential to be modified to detect other antibiotic resistances as well. No time-consuming sample preparation is necessary as the bacteria can be analyzed directly in suspension. This is important for real world samples, such as patient's urine samples.
Collecting one hundred high-quality Raman spectra of the bacteria captured on the integrated Raman-DEP chip takes only about 1.5 minutes. Once the statistical classification model is set up, computation of the subsequent statistical analysis yields the resistance within a few seconds. For vancomycin resistant enterococci it was shown, that already two hours after addition of vancomycin the Raman spectra can reveal vancomycin resistance with high accuracy. The presented method can be combined with previously reported spectroscopy-based pathogen identification methods18, significantly speeding up medical diagnosis and thus reducing the time to select tailored antibiotic therapy from several days to about four hours.
Another advantage of the proposed method is its high potential to be applicable for different enterococci strains as it selectively monitors the bacteria's interaction with the drug and does not require information on strain identity or special knowledge of the geno- or phenotypes as it is the case for many gene- or antibody-based arrays. It was shown for four different enterococci strains that the three level statistical analyses removes strain specific information from the Raman spectra, so that the analysis of different strains that are unknown to the model is possible. Further enterococci strains, also with different resistance mechanisms, will be included in the model to pave the way for a spectroscopic analysis of patients' samples which can carry their own bacterial strains with their own resistance pattern. When combined with an identification data base18,19, such a Raman spectroscopy-based rapid test could enable full analysis of the patient's urine sample in one preparation step within few hours on one device: First, the urine sample will be applied on the chip and the pathogen identified based on its characteristic Raman spectroscopic fingerprint; second, based on this information, the antibiotic is selected and applied (via microfluidics) and subsequently, the antibiotic susceptibility is revealed as described in this manuscript. Using Raman spectroscopy as a method to probe the antibiotic susceptibility offers several advantages compared to other methods, such as e.g., just monitoring the growth characteristics via optical density (OD) measurements. While OD measurements only reveal the quantity of biomass in the suspension, Raman spectra yield a complete molecular fingerprint which can also be applied if mixed cell populations are present (as it happens in a few cases where infections with more than one causing pathogen are observed). Raman spectroscopy can identify which of the two species is reacting to the antibiotics and which is not. Furthermore, Raman spectroscopy holds the potential to reveal the resistance mechanism. This is of particular importance when different resistance mechanisms would result in different treatment. While in this manuscript the proof of principle is given only for one fixed vancomycin concentration, different drug concentrations can be applied following the described experiments. It is subject of ongoing research and beyond the scope of this manuscript to develop measuring algorithms how to extract the minimal inhibition concentration from the time dependent Raman data.
In order to calculate the positive predictive values (PPV) and the negative predictive values (NPV) for the presented spectroscopic method, the most recent available data for the prevalence of vancomycin resistant E. faecium in the USA (2009) are obtained from the “Center for Disease Dynamics, Economics & Policy (CDDEP)” in Washington DC and exemplarily taken into account. The prevalence is an external parameter which cannot be controlled, but has to be kept in mind when a diagnostic test is applied37. With a prevalence of 75% of the E. faecium patient isolates being VREs, a PPV of 93% and a NPV of 96% are obtained. This means that 93 out of 100 patient's samples for which the test reveals “resistant” are truly resistant. On the other hand, if the test yields vancomycin sensitivity, the result will be correct in about 96 of each 100 tests.
In future experiments, we plan to increase data set with a larger variety of strains with different MICs and to include data from a large number of real patient's samples to cover the variety of real world samples. Given the low number of biological replicates available for the present proof-of-concept study, this should result in a substantial increase in ruggedness as well as sensitivity and specificity of the test which will then be suitable also in situations with low prevalence of VRE.
Methods
Bacterial sample
Enterococcus faecalis ATCC® 29212™ (MICvancomycin = 1–4 μg/ml, vancomycin sensitive) and E. faecalis ATCC® 51299™ (MICvancomycin = 32 μg/ml, VRE) were cultivated overnight in CASO medium (Roth GmbH) at 37°C. Enterococcus faecium DSMZ 20477 (MICvancomycin = 1 μg/ml, sensitive) and the E. faecium patient isolate HSVanA from the Jena University Hospital (MIC > 1024 μg/ml, VRE) were grown in LB medium (Roth GmbH).
Sample preparation
Bacteria suspensions were diluted to an optical density of 0.1 corresponding to 108 bacteria per milliliter. Each suspension was divided into two aliquots and cultivated under standard conditions (37°C, CASO medium for E. faecalis and LB medium for E. faecium) to reach the exponential growth phase. Bacterial growth was monitored by measuring the optical density at 600 nm (Agilent Cary 60 UV/Vis spectrometer). After 90 min (E. faecalis) or 60 min (E. faecium), one aliquot was treated with vancomycin (vancomycin hydrochloride hydrate, Sigma-Aldrich) to give a final concentration of 10 μg/ml, while the second aliquot per sample served as a control without antibiotic treatment. Immediately after addition of the drug (time point 0 min) and after 30, 60, 90 and 120 minutes, 1 ml of each aliquot was taken for Raman spectroscopic analysis. The bacteria were washed twice with 0.5× PBS buffer solution (Roth GmbH) by centrifuging them with a relative centrifugal force of 11,500 g for 1.5 min and afterwards resuspended in 0.5× PBS. The optical density was adjusted to 0.3 and admitted to the combined DEP-Raman-setup for further analysis. In total nine independent biological replicates were prepared for the E. faecalis strains and six for E. faecium strains. For each biological replicate three different time points were recorded and analyzed each day, i.e., time points 0, 30 and 90 min or time points 0, 60 and 120 min. Time points 150 min and 180 min were only carried out to check the continuation of the observed behavior in the model (Figure 3a).
Dielectrophoresis equipment
Dielectrophoresis chips with four gold electrodes defining a quadrupole array with a diametric distance of 40 μm were produced in-house as described previously18. They were used to capture bacteria directly from suspension and hold them on a defined position for Raman spectroscopic analysis (Figure 1b). A HAMEG HMF 2550 arbitrary function generator was used to generate an alternating current (AC) voltage of 4 Vpp and 1 MHz. Tungsten wire (99.95% metals basis, VWR International) was used to contact the electrodes of the DEP-chip.
Micro-Raman setup
Raman spectroscopic analysis was performed with a CRM 300 WITec micro-Raman setup (WITec, Ulm) equipped with a 600 lines/mm grating. A frequency-doubled cw Nd-YAG laser beam with an excitation wavelength of 532 nm and a power of 15 mW before passing the objective was focused on the sample via a 60× Nikon water immersion objective with NA 1.0. The 180° backscattered light was detected by a back illuminated CCD camera (DV401 BV, Andor Technology Ltd, Belfast) with 1024 × 127 pixels cooled to −60°C.
Raman spectroscopic analysis on the combined DEP-Raman setup
Ten μl of the washed bacterial suspension in PBS was injected into a 200 μl PBS droplet on the combined DEP-Raman setup (Figure 1a) so that a stream of bacteria passed the center of the chip and a highly efficient capture of the bacteria was achieved. Within less than one minute the bacteria accumulated in the center of the DEP chip, visible as a bacterial cloud with a diameter of about 10–20 μm (Figure 1b). The laser was focused onto the bacteria and a series of 100 Raman spectra with an integration time of 1 s per spectrum was recorded. This was repeated three times per measuring time point in a sequence measuring alternating the treated sample and the untreated control, so that from each independent biological replicate 300 spectra were available per time point.
Data analysis
Statistical analysis was performed in programming language “R” (R Core Team; 2014; http://www.r-project.org/) using “ggplot2”38 for graphical display. Package hyperSpec (Beleites C., Sergo V.; 2014; http://hyperspec.r-forge.r-project.org/) was used to import and pre-process the spectra as follows. Wavenumber correction was carried out using the position of the Rayleigh line in each spectrum. Dimensionality of the data set was reduced by subjecting spectra to a smoothing “loess” interpolation (package “MASS”39) onto an evenly spaced wavenumber axis with data point spacing 5 cm−1 and target degrees of freedom corresponding to 10 cm−1 spectral resolution. Baseline was corrected using smoothing splines (equivalent degrees of freedom: 10) with a rubberband approach for automatic finding of support points separately in the fingerprint region between 600 and 1850 cm−1 and the CH-stretching region above 2800 cm−1. Then, the spectra were truncated to the regions 600–1800 and 2800–3100 cm−1. Next, the spectra were area normalized to an average intensity of 1 in the CH-stretching region. The resulting data set consisted of 302 data points per spectrum. 14200 spectra were collected from nine biological replicates of the two E. faecalis strains. 9600 spectra were collected from six biological replicates of the two E. faecium strains and preprocessed exactly like the E. faecalis spectra.
A Partial Least Squares Regression (PLS) was carried out as the first level of the statistical model to provide regularization for the subsequent Linear Discriminant Analysis (LDA)40. Internally, PLS describes the data by a small number (in our case 2) of latent variables that vary according to the property of interest, here the effect of the vancomycin treatment. Each spectrum was projected onto these latent variables, yielding the PLS scores (Supplementary Fig. S3). Using these scores, the LDA performed the classification by first projecting into a discriminant space and then predicting posterior class membership probabilities assuming Gaussian distribution. Calculations were implemented as an R package, “cbmodels” (Beleites C; 2014), which is based on packages “pls” (Mevik B., Wehrens R., Liland K.; 2013; http://CRAN.R-project.org/package=pls) for the PLS and “MASS” for LDA taking further care on proper centering so that PLS-LDA retains bilinear properties and provides combined PLS-LDA coefficients (analogue to the PLS regression and LDA coefficients).
To ensure that the PLS-LDA model cannot learn to recognize the strains and thus also the sensitivity associated with the strain, the training data consists only of the sensitive strain. In addition, only spectra acquired after at least 30 min of exposure to vancomycin were used for training of the PLS-LDA projection. In contrast, the third level of the PLS-LDA-LDA model used the predicted “vancomycin effect scores” of the PLS-LDA for spectra of treated samples only, but now of both the sensitive and resistant strain. It should be noted that due to the experimental schedule, only three of the biological replicates of the E. faecalis and three of the biological replicates of the E. faecium measurements have data for both 60 and 120 min of vancomycin treatment. Only this subset of the experiments could be used for the third level of the modeling.
Two validation experiments were carried out. In a first step, a leave-one-batch-out validation was carried out within the E. faecalis data. Each batch was independently prepared at a different day, thus corresponding to biological replicates whereas the spectra within each batch are only technical replicates (repeated measurements). In order to ensure statistical independence of the test batch, all three levels of the model were re-calculated for the current training batches and applied to the test batch. All pre-processing steps were independent on the spectra level, thus re-calculation was not necessary. In the second validation experiment, the model trained on all E. faecalis data was applied to predict E. faecium spectra. These spectra are independent at species level, and samples were prepared and measured at different days from the E. faecalis measurements.
Author Contributions
U.N. and U.S. designed the research, U.S., C.A. and U.G. performed the research, C.B. developed statistical tools and algorithms, U.S. and C.B. analyzed the data, W.P. provided bacteria, W.F. and J.P. provided analytical tools, U.H. fabricated the dielectrophoresis chips, U.S. and U.N. wrote the paper. All authors read, improved and reviewed the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
The financial support by the BMBF via the Integrated Research and Treatment Center “Center for Sepsis Control and Care” (CSCC, FKZ 01EO1002) is highly acknowledged. We thank Jürgen Kunert for support in chip design.
References
- Andersson D. I. & Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8, 260–271 (2010). [DOI] [PubMed] [Google Scholar]
- Wiegand I., Hilpert K. & Hancock R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard–Eleventh Edition. CLSI document M02-A11 32 (2012). [Google Scholar]
- Kumar A. et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34, 1589–1596 (2006). [DOI] [PubMed] [Google Scholar]
- Hrabak J., Chudackova E. & Walkova R. Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometry for Detection of Antibiotic Resistance Mechanisms: from Research to Routine Diagnosis. Clin Microbiol Rev 26, 103–114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry in Clinical Microbiology. Clin Infect Dis 57, 564–572 (2013). [DOI] [PubMed] [Google Scholar]
- Pulido M. R., Garcia-Quintanilla M., Martin-Pena R., Miguel Cisneros J. & McConnell M. J. Progress on the development of rapid methods for antimicrobial susceptibility testing. Journal of Antimicrobial Chemotherapy 68, 2710–2717 (2013). [DOI] [PubMed] [Google Scholar]
- Poirel L., Bonnin R. A. & Nordmann P. Rapid identification of antibiotic-resistant bacteria: how could new diagnostic tests halt potential endemics? Expert Review of Molecular Diagnostics 13, 409–411 (2013). [DOI] [PubMed] [Google Scholar]
- Lupo A., Papp-Wallace K. M., Sendi P., Bonomo R. A. & Endimiani A. Non-phenotypic tests to detect and characterize antibiotic resistance mechanisms in Enterobacteriaceae. Diagnostic Microbiology and Infectious Disease 77, 179–194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootoolal J., Neu J. & Wright G. D. Glycopeptide antibiotic resistance. Annu Rev Pharmacol 42, 381–408 (2002). [DOI] [PubMed] [Google Scholar]
- Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis 42, S25–S34 (2006). [DOI] [PubMed] [Google Scholar]
- Arias C. A. & Murray B. E. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10, 266–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Costa V. M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011). [DOI] [PubMed] [Google Scholar]
- Hill C. M. et al. Specificity of Induction of the vanA and vanB Operons in Vancomycin-Resistant Enterococci by Telavancin. Antimicrob Agents Ch 54, 2814–2818 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold H. S. Vancomycin-resistant enterococci: Mechanisms and clinical observations. Clin Infect Dis 33, 210–219 (2001). [DOI] [PubMed] [Google Scholar]
- Krafft C., Steiner G., Beleites C. & Salzer R. Disease recognition by infrared and Raman spectroscopy. Journal of Biophotonics 2, 13–28 (2009). [DOI] [PubMed] [Google Scholar]
- Wachsmann-Hogiu S., Weeks T. & Huser T. Chemical analysis in vivo and in vitro by Raman spectroscopy-from single cells to humans. Current Opinion in Biotechnology 20, 63–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder U. C. et al. Combined Dielectrophoresis-Raman Setup for the Classification of Pathogens Recovered from the Urinary Tract. Anal Chem 85, 10717–10724 (2013). [DOI] [PubMed] [Google Scholar]
- Kloss S. et al. Culture Independent Raman Spectroscopic Identification of Urinary Tract Infection Pathogens: A Proof of Principle Study. Anal Chem 85, 9610–9616 (2013). [DOI] [PubMed] [Google Scholar]
- te Witt R. et al. Good Performance of the SpectraCellRA System for Typing of Methicillin-Resistant Staphylococcus aureus Isolates. J Clin Microbiol 51, 1434–1438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harz M. et al. Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy. Journal of Biophotonics 2, 70–80 (2009). [DOI] [PubMed] [Google Scholar]
- Maquelin K. et al. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J Clin Microbiol 41, 324–329 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusciano G., Capriglione P., Pesce G., Abete P., Carnovale V. & Sasso A. Raman spectroscopy as a new tool for early detection of bacteria in patients with cystic fibrosis. Laser Physics Letters 10 (2013). [Google Scholar]
- Clarridge J. E. III, Harrington A. T., Roberts M. C., Soge O. O. & Maquelin K. Impact of Strain Typing Methods on Assessment of Relationship between Paired Nares and Wound Isolates of Methicillin-Resistant Staphylococcus aureus. J Clin Microbiol 51, 224–231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer U. et al. Towards a detailed understanding of bacterial metabolism - Spectroscopic characterization of Staphylococcus epidermidis. Chemphyschem 8, 124–137 (2007). [DOI] [PubMed] [Google Scholar]
- Lopez-Diez E. C., Winder C. L., Ashton L., Currie F. & Goodacre R. Monitoring the mode of action of antibiotics using Raman spectroscopy: Investigating subinhibitory effects of amikacin on Pseudomonas aeruginosa. Anal Chem 77, 2901–2906 (2005). [DOI] [PubMed] [Google Scholar]
- Athamneh A. I. M., Alajlouni R. A., Wallace R. S., Seleem M. N. & Senger R. S. Phenotypic Profiling of Antibiotic Response Signatures in Escherichia coli Using Raman Spectroscopy. Antimicrob Agents Ch 58, 1302–1314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz T. J., Taylor D. S., Polage C. R., Krol D. M., Lane S. M. & Chan J. W. Effect of Cefazolin Treatment on the Nonresonant Raman Signatures of the Metabolic State of Individual Escherichia coil Cells. Anal Chem 82, 2703–2710 (2010). [DOI] [PubMed] [Google Scholar]
- Liu T. Y. et al. Functionalized arrays of Raman-enhancing nanoparticles for capture and culture-free analysis of bacteria in human blood. Nat Commun 2 (2011). [DOI] [PubMed] [Google Scholar]
- Notingher I. Raman Spectroscopy cell-based Biosensors. Sensors-Basel 7, 1343–1358 (2007). [Google Scholar]
- Notingher I., Verrier S., Romanska H., Bishop A. E., Polak J. M. & Hench L. L. In situ characterisation of living cells by Raman spectroscopy. Spectrosc-Int J 16, 43–51 (2002). [Google Scholar]
- Arthur M. ANTIBIOTICS Vancomycin sensing. Nat Chem Biol 6, 313–315 (2010). [DOI] [PubMed] [Google Scholar]
- Cetinkaya Y., Falk P. & Mayhall C. G. Vancomycin-resistant enterococci. Clin Microbiol Rev 13, 686–707 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G., Thaker M. N., Koteva K. & Wright G. Glycopeptide antibiotic biosynthesis. J Antibiot 67, 31–41 (2014). [DOI] [PubMed] [Google Scholar]
- Kahne D., Leimkuhler C., Wei L. & Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chemical Reviews 105, 425–448 (2005). [DOI] [PubMed] [Google Scholar]
- Walsh F. M. & Amyes S. G. B. Microbiology and drug resistance mechanisms of fully resistant pathogens. Current Opinion in Microbiology 7, 439–444 (2004). [DOI] [PubMed] [Google Scholar]
- Buchen L. Missing the mark. Nature 471, 428–432 (2011). [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis Springer, New York (2009). [Google Scholar]
- Venables W. N. & Ripley B. D. Modern Applied Statistics with S, 4th edn. Springer, New York (2002). [Google Scholar]
- Barker M. & Rayens W. Partial least squares for discrimination. J Chemometr 17, 166–173 (2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information