Abstract
Candida albicans is a pleiomorphic fungus that forms mixed species biofilms with Streptococcus gordonii, an early colonizer of oral cavity surfaces. Activation of quorum sensing (QS; intercellular signalling) promotes monospecies biofilm development by these micro-organisms, but the role of QS in mixed species communities is not understood. The comCDE genes in S. gordonii encode a sensor–regulator system (ComDE), which is activated by the comC gene product (CSP, competence stimulating peptide) and modulates expression of QS-regulated genes. Dual species biofilms of S. gordonii ΔcomCDE or ΔcomC mutants with C. albicans showed increased biomass compared to biofilms of S. gordonii DL1 wild-type with C. albicans. The ΔcomCDE mutant dual species biofilms in particular contained more extracellular DNA (eDNA), and could be dispersed with DNase I or protease treatment. Exogenous CSP complemented the S. gordonii ΔcomC transformation deficiency, as well as the ΔcomC-C. albicans biofilm phenotype. Purified CSP did not affect C. albicans hyphal filament formation but inhibited monospecies biofilm formation by C. albicans. The results suggest that the S. gordonii comCDE QS-system modulates the production of eDNA and the incorporation of C. albicans into dual species biofilms.
Introduction
Streptococci are major constituents of the oral microbiome (Dewhirst et al., 2010) and mitis group streptococci, such as Streptococcus gordonii, Streptococcus oralis and Streptococcus mitis, are prominent amongst early-stage colonizers of the oral cavity (Nyvad & Kilian, 1990). These species of streptococci interact with a range of other oral micro-organisms, thus establishing a foundation for the building of biofilm communities (Wright et al., 2013). S. gordonii forms biofilms with Porphyromonas gingivalis (Lamont et al., 2002), Actinomyces oris (Palmer et al., 2003), Fusobacterium nucleatum (Foster & Kolenbrander, 2004) and Aggregatibacter actinomycetemcomitans (Liu et al., 2011) in dual or mixed species systems, and also produces dual species biofilms with the fungus Candida albicans (Bamford et al., 2009; Dutton et al., 2014).
C. albicans is a pleiomorphic fungus that can colonize mucosal surfaces and prosthetic materials (e.g. dentures and catheters) throughout the human body. The incidence of candidiasis continues to expand, and Candida is the fourth leading cause of nosocomial bloodstream infections in the USA (Wisplinghoff et al., 2004). In the UK, there was an increase in candidiasis infections of 37 % in the period 2003–2007, and C. albicans is reported to cause 48 % of all Candida-related bloodstream infections (Public Health England, 2013). The interactions of oral streptococci with C. albicans are of particular interest because there is evidence that the bacteria can enhance biofilm formation by C. albicans (Xu et al., 2014a) and fungal pathogenicity (Xu et al., 2014b). S. gordonii cells adhere to hyphal filaments of C. albicans via cell surface adhesin SspB interacting with hyphal cell wall protein Als3 (Nobbs et al., 2010; Silverman et al., 2010). The SspB–Als3 interaction is involved in biofilm development in which bacteria and fungi cooperate at the cellular and molecular level to generate a robust mixed microbial community (Whitmore & Lamont, 2011). Community composition may be regulated by extracellular signalling molecules such as autoinducer-2 (AI-2) (McNab et al., 2003; Bamford et al., 2009), farnesol (Bamford et al., 2009; Lu et al., 2014), N-acetyl-d-glucosamine (Naseem et al., 2011) and homoserine lactones (Hogan et al., 2004). In addition, the quorum-sensing (QS) molecule, competence stimulating peptide (CSP), produced by Streptococcus mutans inhibits hypha formation by C. albicans (Jarosz et al., 2009). Paradoxically, C. albicans induces the CSP associated QS-system of S. mutans in dual species biofilms (Sztajer et al., 2014).
The induction of competence for DNA uptake and transformation in S. gordonii involves a complex regulatory network similar to that in Streptococcus pneumoniae (Vickerman et al., 2007) and shares some similarities to that in S. mutans (Son et al., 2012). Competence regulation in these organisms employs a ComABCDE system that generates the CSP (comC gene product). In S. gordonii, pre-CSP is a 50 aa polypeptide that is cleaved at a double glycine to generate the mature 19 aa residue CSP (Håvarstein et al., 1996). This peptide is then exported out of the cell via an ABC-binding cassette transporter encoded by comA and comB. Once a critical concentration of environmental CSP is reached, it is detected by the two-component system ComDE, comprising a membrane-bound histidine kinase (ComD), which phosphorylates the intracellular response regulator ComE (Håvarstein et al., 1996). ComE activates a signalling cascade that includes upregulation of the master regulator SigX (designated ComR in S. gordonii; Heng et al., 2006), an alternate sigma factor that controls expression of late competence genes encoding DNA uptake and recombination machinery (Lee & Morrison, 1999; Piotrowski et al., 2009).
In this paper we investigated the role of competence induction and CSP formation by S. gordonii in the formation of dual species biofilms with C. albicans. We show that competence for transformation is a factor playing an important role in the development of early-stage biofilms of S. gordonii and C. albicans. In addition, evidence is presented showing that CSP produced by S. gordonii inhibits biofilm formation by C. albicans; therefore, CSP could normally act as a check on C. albicans load in dual species biofilms.
Methods
Strains and growth conditions.
Strains and plasmids utilized are listed in Table S1 (available in the online Supplementary Material). S. gordonii wild-type strain DL1 (Challis) and isogenic mutants were cultured in BHY medium [Brain Heart Infusion Broth (LabM) containing 5 g yeast extract l−1] under stationary conditions in a candle jar at 37 °C. C. albicans strains were maintained on Sabouraud glucose agar (LabM) and suspension cultures were grown in YPD medium [10 g yeast extract l−1, 20 g neopeptone l−1, 2 % (w/v) glucose] at 37 °C with vigorous aeration. A defined medium (YPT-Glc) supported the growth of S. gordonii and C. albicans and comprised 20 mM NaH2PO4 (pH 7.0), 1× yeast nitrogen base, 1 g Bactotryptone l−1 and 0.4 % (w/v) glucose (Dutton et al., 2014). Antibiotics were used at the following concentrations: 100 µg ampicillin ml−1; 100 µg spectinomycin (Sp) ml−1; 5 µg erythromycin ml−1. Competence development by S. gordonii was induced in BHY medium containing 1 % fetal calf serum and 0.1 % (w/v) glucose according to Haisman & Jenkinson (1991). Exogenous synthetic CSP (DVRSNKIRLWWENIFFNKK) was included where appropriate in the concentration range 0.1–10 µg CSP ml−1.
Generation of the S. gordonii ΔcomCDE mutant.
Deletion of the entire competence locus in S. gordonii was achieved by allelic exchange with spectinomycin resistance cassette aad9. A schematic representation of the mutagenesis strategy is shown in Fig. S1. In brief, a chromosomal DNA fragment (540 bp) immediately upstream of comC and a fragment (603 bp) downstream from comE were amplified by PCR with the primer pairs comCDE.F1/comCDE.R1 and comCDE.F2/comCDE.R2 (Table S2), respectively, using Expand Long Template PCR System (Roche). The flanking sequences were ligated by PCR, generating an amplimer (comCDEflank) with a central unique BamHI site that was cloned into pGEM-T (Promega) in E. coli JM109. The aad9 cassette with its own promoter and transcription terminator (1082 bp) was amplified from pFW5 (Podbielski et al., 1996) with terminal BamHI sites and cloned into the unique BamHI site within the vector pGEM-T+comCDEflank. The resulting construct (pGEM-T+comCDEflank–aad9; 2272 bp) was confirmed by restriction enzyme digestion (SacI and PstI) and DNA sequencing. The comCDEflank–aad9 fragment was then amplified by PCR and transformed into S. gordonii with selection for Sp resistance. Transformants were screened by PCR with the primer pair comCDE.F1/comCDE.R2 and a representative transformant, confirmed by DNA sequencing of the chromosomal PCR product, was designated UB2346.
Generation of the ΔcomC mutant.
In a similar way as described above, flanking DNA fragments of comC were prepared by PCR with the primer pairs comCDE.F1/comC2.R1 (690 bp) and comC2.F2/comC2.R2 (626 bp) (Fig. S2 and Table S2). The aad9 cassette (devoid of promoter and transcriptional terminator sequence) was PCR-amplified from the pFW5 vector using the primers aad9.2F and aad9.2R (752 bp) (Table S2). The comC flanking fragments and aad9 cassette were joined together by overlapping PCR using the primers comCDE.F1 and comC2.R2 (Fig. S2). The resulting amplimer (comCflank–aad9; 2114 bp) was cloned into the pGEM-T vector in E. coli and confirmed by sequencing. The fragment was then reamplified, transformed into S. gordonii and Sp-resistant transformants were screened by PCR using the primer pair comCDE.F1/comC2.R2 and identified based on size of the PCR product (2069 bp compared to 1469 bp in wild-type strain). A representative transformant was selected, the chromosomal region sequenced to confirm authenticity, and the strain was designated UB2660.
Preparation of saliva.
Unstimulated whole saliva was collected from a minimum of six healthy adults who provided written informed consent, as approved by the National Research Ethics Committee Central Oxford C (#08/H0606/87+5). Saliva samples were pooled on ice, treated with 2.5 mM DTT for 10 min and then centrifuged to clarify (10 000 g, 10 min). The supernatant was removed, diluted to 10 % with distilled water and filter-sterilized through a 0.45 µm nitrocellulose membrane.
Preparation of microbial cells.
S. gordonii strains were grown in BHY medium for 16 h at 37 °C. Cells were harvested by centrifugation (5000 g, 10 min), suspended in YPT-Glc and adjusted to OD600 0.05 (2.0×107 cells ml−1). C. albicans was grown in YPD for 16 h at 37 °C, cells were harvested by centrifugation (5000 g, 5 min), and suspended in YPT-Glc at OD600 0.1 (1×106 cells ml−1).
Biofilm formation.
Sterile glass coverslips (13 or 19 mm diameter) in wells of 24- or 12-well polystyrene plates (Greiner) were incubated with 10 % saliva for 16 h at 4 °C. Saliva was removed and the coverslips were washed twice with PBS [0.01 M K2HPO4-KH2PO4 (pH 7.0), 2.7 mM KCl, 0.137 M NaCl]. For monospecies biofilms, cells (0.5 or 1 ml S. gordonii or C. albicans) were added to wells containing saliva-coated coverslips and incubated for 1 h at 37 °C with gentle agitation (50 r.p.m.) in a humid environment. Non-adhered cells were removed, fresh YPT-Glc (0.5 or 1 ml) was added to each well and biofilms were grown for a further 6 h. Coverslips were then recovered, washed twice with PBS and air-dried. For dual species biofilms, C. albicans cells (0.5 or 1 ml) were added to wells of 24- or 12-well plates containing saliva-coated coverslips, and the plates were incubated for 1 h at 37 °C. Non-adherent C. albicans were then removed by aspiration, S. gordonii cells (0.5 or 1 ml accordingly) were added and the plates were incubated again for 1 h at 37 °C. Non-adhered cells were then removed from the wells, fresh YPT-Glc (0.5 or 1 ml) was added to each well and plates were incubated for a further 6 h. Coverslips were then removed, washed with PBS and air-dried.
Biofilm assays.
Biofilms were stained with 0.1 % safranin, washed with distilled water until excess stain was removed and air-dried. Coverslips were then inverted and mounted onto microscope slides, and biofilms visualized with a Leica DMLB light microscope with attached colour view camera, using CellD imaging software (Olympus Soft Imaging Solutions). Biomass quantification required release of safranin stain with 10 % acetic acid for 15 min, transfer of 0.1 ml portions to a 96-well plate and measurement at A490 (Lembke et al., 2006) with an iMark microplate reader (Bio-Rad). All studies were performed in triplicate and mean biomass levels calculated from three independent experiments.
To estimate the numbers of S. gordonii cells present in dual species biofilms the biomass was removed from the coverslip with a cell scraper into YPT-Glc medium, the coverslip was vortex mixed in PBS and the suspensions were combined. The suspension was serially diluted 10-fold in YPT-Glc and plated onto BHYN agar containing 30 µg nystatin ml−1 (to prevent growth of C. albicans). Plates were incubated for 24 h at 37 °C and colonies were counted to estimate S. gordonii c.f.u. per biofilm.
For enzyme treatment of biofilms, coverslips with deposited biofilms were incubated with 100 U lyticase ml−1, 4.2 U proteinase K ml−1, 100 U DNase I ml−1 or 0.002 U neuraminidase ml−1 for 1 h at 37 °C. Activity of neuraminidase was confirmed with 2-O-(o-nitrophenyl)-α-d-N-acetylneuraminic acid substrate (Brittan et al., 2012). No enzyme and heat-inactivated (30 min at 80 °C) enzyme controls were included. Biofilms were then estimated for biomass with safranin stain or examined by CLSM as described below.
To measure cell-free DNA, biofilms were scraped from coverslips as described above into 0.5 ml TE buffer (10 mM Tris/HCl, pH 7.5 and 1 mM EDTA), centrifuged (10 000 g, 5 min) and supernatant DNA concentration was calculated from A260 measurements using a UV–Vis spectrophotometer (Shimadzu). Nucleic acid purity was indicated by the A260/A280 value, where a ratio between 1.7–2.0 was accepted as pure.
S. gordonii CSP (DVRSNKIRLWWENIFFNKK) was synthesized commercially (GenicBio) along with a scrambled CSP (sCSP; DKRFKWWILKVFNSNEINR) with identical amino acid residue composition but random sequence. To assess the effects of these peptides on C. albicans biofilms or on dual species biofilms, YPT-Glc medium was supplemented with up to 10 µg peptide ml−1. Biomass values for non-attached cells were calculated by collecting the planktonic phases into microcentrifuge tubes, harvesting by centrifugation (5000 g, 7 min) and mixing the pellet with 0.002 % safranin. After 5 min the cells were centrifuged, washed several times in PBS and then suspended in 10 % acetic acid to solubilize the stain. The suspensions were clarified by centrifugation and then A490 measurements were utilized to determine relative biomass.
CLSM.
Biofilms on coverslips were fixed with 0.5 ml paraformaldehyde (4 %) in PBS for 1 h at room temperature. Coverslips were rinsed with PBS and stained with FITC for 30 min in the dark (Dutton et al., 2014). In some experiments, C. albicans SBC156, derivative of SC5314 expressing red fluorescent protein (RFP) (Milne et al., 2011) was utilized. Biofilms were visualized using a Leica TCS-SP5 confocal imaging system attached to a Leica DMIRBE inverted microscope. Images were observed using ×40 or ×63 oil immersion objective lens with 488 nm excitation wavelength to excite FITC or at 561 nm to excite RFP. Z-slices were obtained every 0.5 µm. The data were analysed using Volocity image analysis software (Improvision).
Wide field microscopy.
To visualize extracellular DNA (eDNA), biofilms were fixed with paraformaldehyde as described above, washed twice with PBS and blocked for 45 min with 2 % BSA in PBS. Biofilms were incubated for 45 min with mouse anti-dsDNA antibody (1 : 1000 dilution; Abcam), washed with 0.5 % BSA in PBS and then incubated for 45 min in the dark with goat anti-mouse Alexa594 (1 : 1000 dilution; Life Technologies). Biofilms were washed and then the coverslips were placed face down onto Vectashield mounting medium on a glass slide. Phase-contrast and Alexa 594 fluorescence images were captured using a Leica DMI6000 microscope with a ×40 (NA 1.25) oil immersion lens, DFC365FX CCD camera and Leica acquisition software. For all microscopy studies images were recorded from at least five fields taken at random.
Statistics.
Data were analysed using GraphPad Prism, version 5.0. All data are presented as the mean±sd of at least three independent experiments. For normally distributed data, comparisons were tested with Student’s t-test. The two-tailed Mann–Whitney U-test was used for comparisons between groups. P values <0.05 were considered statistically significant.
Results
Effects of competence (comCDE) operon deletion on early biofilm formation
The effects of comC or comCDE deletions on biofilm formation have been investigated in S. mutans (Li et al., 2002), but not in S. gordonii. Biofilms of ΔcomC or ΔcomCDE mutants produced after 6 h incubation were compared with those produced by wild-type S. gordonii DL1 (Fig. 1). The streptococcal strains each formed a contiguous layer of cells over the surface of each of the coverslips, but the com operon mutant biofilms had more compact morphologies. Dual species biofilms of S. gordonii and C. albicans generally had denser morphologies and increased biomass compared to the single species biofilms (Fig. 1) (Bamford et al., 2009). The ΔcomC and ΔcomCDE mutant strains had significantly increased dual species biofilm biomass values compared to wild-type (Fig. 1). In the ΔcomC–C. albicans biofilms, the hyphal filaments appeared to be clustered (Fig. 1).
Fig. 1.
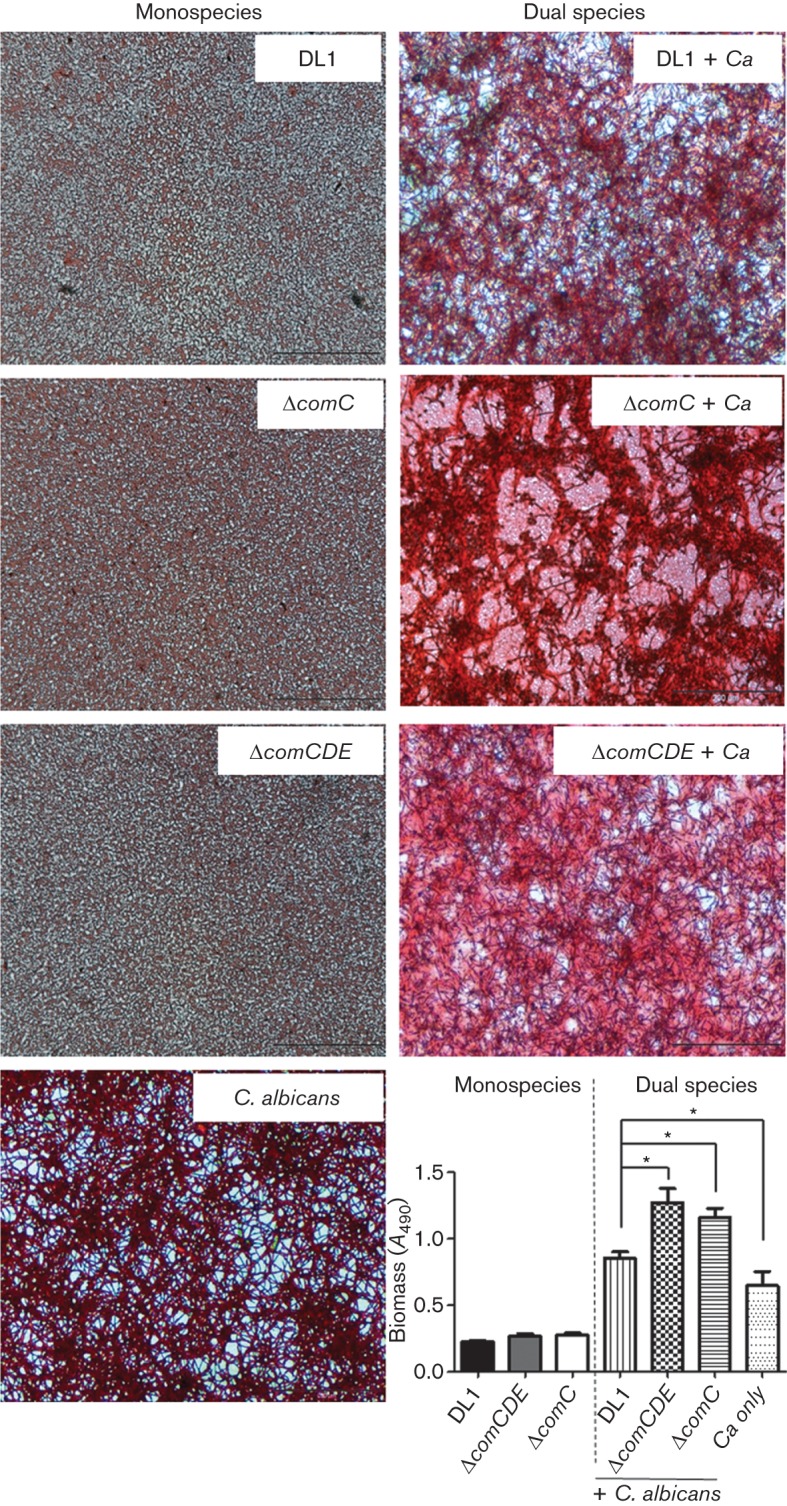
S. gordonii monospecies or dual species biofilms with C. albicans. Biofilms were grown on saliva-coated coverslips for 6 h, safranin-stained and viewed by transmitted light microscopy. Biomass measurements (A490) were taken following release of stain with acetic acid. DL1, S. gordonii wild-type; Ca, C. albicans. *P<0.05 (n = 3). Bar, 50 µm.
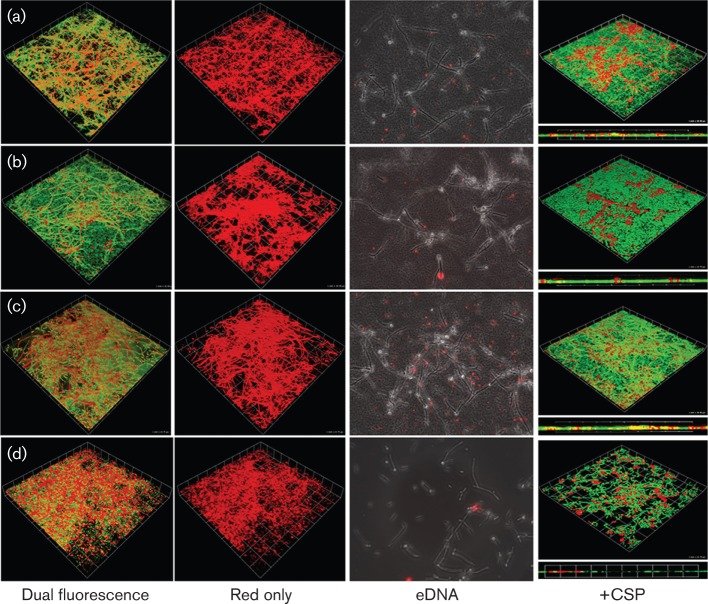
In S. gordonii monospecies biofilms, the ΔcomC and ΔcomCDE mutants both formed more closely knitted mats than S. gordonii wild-type (Fig. 2). Biofilm depth (thickness) was measured as 5.5–6.0 µm for S. gordonii monospecies biofilms. In dual species biofilms for CLSM we utilized a C. albicans SC5314 derivative expressing RFP (Table S1). In these biofilms the salivary pellicle surface was colonized by S. gordonii and a network of C. albicans hyphal filaments (Fig. 3a). By comparison with monospecies C. albicans biofilms (Fig. 3d), more true hyphae or longer hyphal filaments were observed in the dual species biofilms. The S. gordonii ΔcomC and ΔcomCDE mutants formed denser dual species biofilms compared to S. gordonii wild-type biofilms (Fig. 3b, c) and the ΔcomCDE–C. albicans biofilm in particular produced matrix (Fig. 3c). By subtracting the green (FITC) channel using the imaging software it was possible to see that the C. albicans hyphae that formed within the com mutant–C. albicans biofilms were less evenly spread across the surfaces and more coalesced (Fig. 3b, c). The average thickness values for the biofilms (Fig. 3) were ordered ΔcomC– or ΔcomCDE–C. albicans>wild-type–C. albicans>C. albicans. There were no statistically significant differences between c.f.u. values (per coverslip) for S. gordonii wild-type and ΔcomCDE mutant in monospecies (6.4±0.8×106 versus 5.1±0.7×106 c.f.u.) or dual species (1.5±0.3×108 versus 2.2±0.2×108 c.f.u.) biofilms. This implied that the differences in biomass between dual species biofilms with these strains may primarily reflect changes in relative proportions of C. albicans and matrix. Unfortunately, meaningful c.f.u. values for C. albicans could not be obtained because of the filament morphology and aggregation. Overall, these results suggested that deletion of the com genes relieved some form of repression mechanism normally operating on dual species biofilm formation.
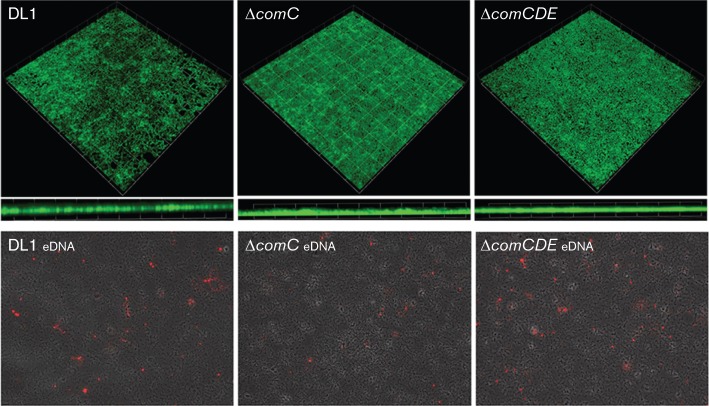
Fig. 2.
Architecture and eDNA deposition patterns of 6 h monospecies biofilms of S. gordonii DL1 wild-type, ΔcomC mutant and ΔcomCDE mutant. Uppermost panels show CLSM 3D (xyz stack) and xz (thickness) images of biofilms stained with FITC. Maximum thickness (depth) measurements for a representative biofilm were: S. gordonii DL1, 6.6 µm; S. gordonii ΔcomC, 5.5 µm; S. gordonii ΔcomCDE, 5.6 µm. Grid square, 25 µm. Lower panels show wide field images of biofilms producing patches of eDNA immunofluorescently labelled with Alexa594 (red).
Fig. 3.
Architecture and eDNA deposition patterns of 6 h dual species biofilms of S. gordonii with C. albicans expressing RFP. (a) S. gordonii DL1 wild-type–C. albicans, (b) ΔcomC–C. albicans, (c) ΔcomCDE–C. albicans, (d) C. albicans. Biofilms were stained with FITC. Dual fluorescence: images taken using both laser channels to visualize C. albicans RFP+ (red) and FITC-labelled S. gordonii (green). Red only: images taken with the red channel only, showing the C. albicans biofilm component. Panels are 3D (xyz stack) images. Maximum thickness (depth) measurements for a representative biofilm were: (a) 13.6 µm, (b) 17.1 µm, (c) 18.1 µm, (d) 7.3 µm. Grid square, 25 µm. eDNA: wide field images of biofilms producing patches of eDNA immunofluorescently labelled with Alexa594 (red). +CSP: effect of S. gordonii CSP on C. albicans monospecies or dual species biofilms. Each panel comprises a 3D (xyz) and xz (thickness) CLSM image of biofilms grown on saliva-coated coverslips for 6 h at 37 °C in the presence of 10 µg CSP ml−1. Grid square, 38.9 µm (a, b, c) and 25 µm (d).
Extracellular polymeric substance (EPS) production
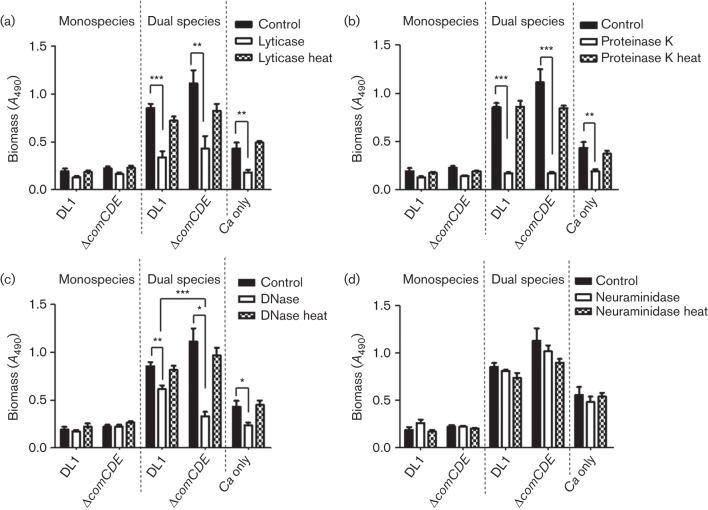
The matrix materials present within streptococcal biofilms include eDNA (Liu & Burne, 2011; Taff et al., 2013; Xu & Kreth, 2013), proteins (Liao et al., 2014) and polysaccharides (when grown in the presence of sucrose) (Falsetta et al., 2014). C. albicans biofilms contain ~50 % protein, as well as mannan, glucan and eDNA (Zarnowski et al., 2014). Incubation of monospecies biofilms of S. gordonii with DNase I, proteinase K or lyticase (β-1,3-glucan hydrolase) resulted in small but not statistically significant effects on biomass (Fig. 4). By contrast, significant reductions in biomass were seen for dual species biofilms with either wild-type or ΔcomCDE mutant following incubation with the three enzymes (Fig. 4). Lyticase treatment reduced biomass by >50 % (Fig. 4a), while proteinase K digestion reduced biomass by >80 % (Fig. 4b). This is consistent with proteins representing a large proportion of biofilm matrix (Flemming & Wingender, 2010). Controls of heat-inactivated enzymes had no significant effects upon biofilm biomass nor did neuraminidase treatment (Fig. 4).
Fig. 4.
Enzymic treatments of monospecies or dual species biofilms. Biofilms were grown for 6 h on saliva-coated coverslips, washed and incubated with enzyme for 1 h at 37 °C. Total biomass was quantified by safranin staining and A490 measurements following release of stain with acetic acid. (a) 100 U lyticase ml−1, (b) 4.2 U proteinase K ml−1, (c) 100 U DNase I ml−1 and (d) 0.002 U neuraminidase ml−1. Filled column, control/no treatment; open column, enzymic treatment; hatched column, treatment with heat-killed enzyme. DL1, S. gordonii wild-type; Ca, C. albicans. P values: *<0.05, **<0.01, ***<0.001 (n = 4).
Incubation of dual species biofilms of S. gordonii wild-type–C. albicans or ΔcomCDE–C. albicans with DNase I led to 25 or 75 % reductions, respectively, in biomass (Fig. 4) and this was supported by CLSM of the biofilms (Fig. 5). Both S. gordonii and C. albicans components were removed from DNase I-treated biofilms (Fig. 5), although more streptococci (green) remained in the S. gordonii wild-type–C. albicans biofilms (Fig. 5c). The C. albicans only biofilms were also disrupted by DNase I treatment (Fig. 5e).
Fig. 5.

Representative CLSM micrographs of monospecies or dual species biofilms following incubation with DNase I. Biofilms of S. gordonii DL1 wild-type or ΔcomCDE mutant, with or without C. albicans expressing RFP, were grown for 6 h on saliva-coated coverslips, washed and incubated with DNase I for 1 h at 37 °C and stained with FITC. (a) S. gordonii DL1 wild-type (maximum thickness measurements for a representative biofilm, 6.18 µm), (b) S. gordonii ΔcomCDE (5.23 µm), (c) S. gordonii DL1–C. albicans RFP+ (8.59 µm), (d) S. gordonii ΔcomCDE–C. albicans RFP+ (6.12 µm) and (e) C. albicans RFP+ (5.58 µm). (c–e) (i) Left side are images of RFP and FITC laser channels combined; (ii) corresponding right side images show the RFP channel only (C. albicans). Grid square, 25 µm.
eDNA within biofilms
To estimate eDNA concentrations within biofilms, matrix fractions associated with each biofilm were collected by washing them with TE buffer and measuring A260. No statistically significant differences in eDNA concentrations were observed for monospecies biofilms of S. gordonii DL1 wild-type and ΔcomCDE mutant (Fig. 6). However, the ΔcomCDE–C. albicans dual species biofilms contained more eDNA than the S. gordonii DL1–C. albicans biofilms (Fig. 6).
Fig. 6.

Concentrations of soluble eDNA extracted from monospecies biofilms or S. gordonii dual species biofilms with C. albicans. DNA was extracted as described in Methods and measured at A260. DL1, S. gordonii wild-type; Ca, C. albicans. *P<0.05 (n = 3).
To visualize eDNA in biofilms we utilized anti-dsDNA antibodies and immunofluorescence. The ΔcomCDE mutant dual species biofilms contained multiple deposits of eDNA compared with the DL1– or ΔcomC–dual species biofilms (Fig. 3). Overall, the eDNA deposits were enhanced in the dual species biofilms compared to monospecies biofilms (Fig. 2) and there was little difference between monospecies biofilms of the S. gordonii strains. C. albicans secreted little or no eDNA in monospecies or dual species biofilms under the experimental conditions (Fig. 3).
Effect of CSP on dual species biofilms
The increased biomass observed for S. gordonii ΔcomC–C. albicans compared to wild-type–C. albicans biofilms (Fig. 1) was reduced to equivalent DL1 wild-type–C. albicans biomass in the presence of 10 µg CSP ml−1, but not with sCSP (Fig. 7). The biomass of ΔcomCDE–C. albicans dual species biofilms was also reduced by addition of CSP (Fig. 7). Inhibition of biofilm formation by C. albicans alone in the presence of CSP may be seen in CLSM images (Fig. 3) and this inhibition was supported by biomass measurements (Fig. 7). Exogenous CSP complemented the ΔcomC mutant deficiency in developing competence for DNA-mediated transformation (Fig. S3c), but did not complement the ΔcomCDE mutant (Fig. S3c). Thus, the effect of CSP on dual species biofilm formation was due to, at least in part, a direct effect of CSP on the biofilms, since the ΔcomCDE mutant was unable to respond to CSP signalling.
Fig. 7.
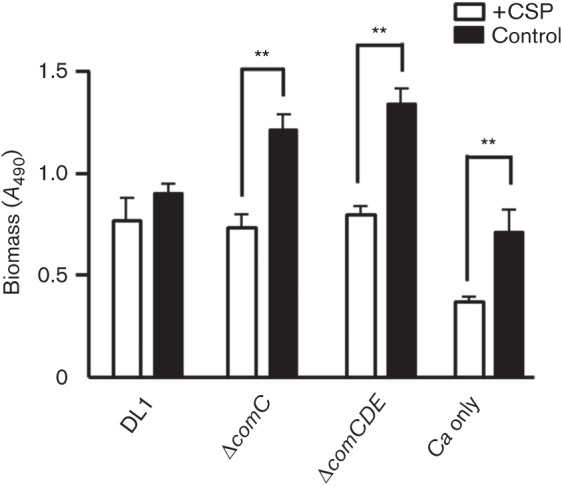
Effect of S. gordonii CSP on biomass of S. gordonii and C. albicans monospecies or dual species biofilms. Biofilms were grown on saliva-coated coverslips for 6 h, with 10 µg CSP ml−1 (+CSP) or 10 µg sCSP ml−1 (Control), safranin-stained and biomass measurements (A490) were taken following release of stain with acetic acid. DL1, S. gordonii wild-type; Ca, C. albicans. **P<0.01 (n = 3).
Effects of CSP on C. albicans
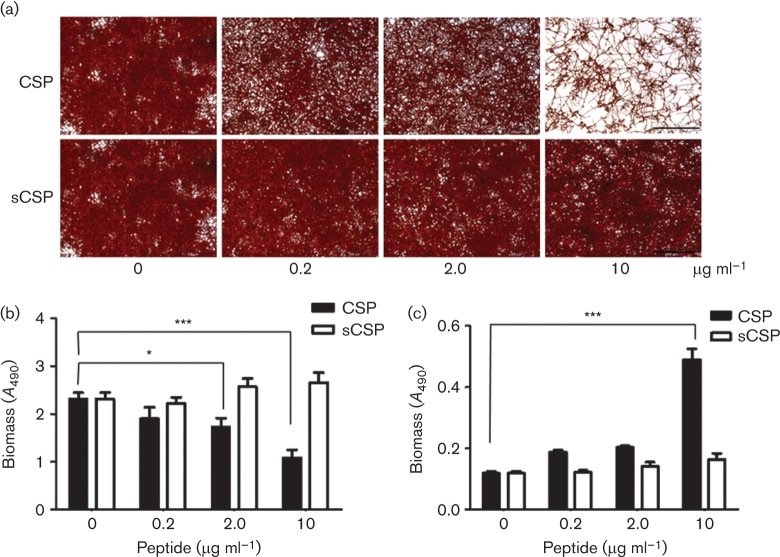
The addition of synthetic S. gordonii CSP to C. albicans biofilms showed a dose-dependent inhibition of C. albicans monospecies biofilm formation (Fig. 8). In the presence of 2 µg CSP ml−1 there was 35 % reduction in biomass, while at 10 µg CSP ml−1 biomass was reduced ~60 % (Fig. 8b). In separate experiments, CSP did not inhibit planktonic growth of C. albicans or hypha formation. After 4 h incubation of C. albicans in YPT-Glc medium, with or without 10 µg CSP ml−1, 80 % of cells formed hyphae and mean hyphal length was 23±4.0 µm. No significant effects on biomass production or biofilm architecture were seen with equivalent concentrations of sCSP (Fig. 8a, b). We then measured biomass of material present in the planktonic phases of these biofilms by safranin stain assay. There was a threefold increase in planktonic phase biomass of biofilm cultures grown with 10 µg CSP ml−1 compared to those grown with sCSP at the same concentration (Fig. 8c). Therefore, reduced biomass of C. albicans biofilms formed in the presence of CSP could be accounted for by increased biomass in the planktonic phase. This indicated that CSP inhibited biofilm development by affecting attachment or promoting dispersal of C. albicans cells. Overall these results suggested that the production of CSP by S. gordonii modulated incorporation of C. albicans into dual species biofilms.
Fig. 8.
Effects of S. gordonii CSP or sCSP on C. albicans biofilm formation. (a) Transmitted light micrographs of C. albicans biofilms grown for 6 h on saliva-coated coverslips in the presence of CSP or sCSP at concentrations indicated. Bar, 200 µm. (b) C. albicans biofilm biomass quantified by safranin stain assay following growth in the presence of CSP or sCSP. (c) Corresponding C. albicans planktonic phase biomass following growth with CSP or sCSP, determined by safranin stain assay. *P<0.05, ***<0.001 (n = 4).
Discussion
Mitis group streptococci are early colonizers of oral cavity surfaces and form the basis of many of the polymicrobial communities found in the mouth and nasopharynx (Aas et al., 2005; Kanasi et al., 2010; Shak et al., 2013; Xu et al., 2014a). Such communities may also contain C. albicans, which is carried by ~40 % of the healthy adult population (Clayton & Noble, 1966), and it is suggested that synergistic interactions between C. albicans and streptococci can promote carriage as well as disease (Dutton et al., 2014; Falsetta et al., 2014; Xu et al., 2014b). The establishment and development of such communities involves small molecule signalling between microbial components, which maximizes metabolic efficiency and controls competition of species. The major signalling molecules produced by Gram-positive cocci are peptides, including CSPs and bacteriocins. There is evidence that peptides produced by S. mutans (Jarosz et al., 2009) and Enterococcus faecalis (Cruz et al., 2013) are inhibitory to filamentation by C. albicans. The ComCDE systems in S. gordonii, S. pneumoniae and S. mutans function as QS autoregulatory control systems, modulating gene expression patterns in different ways for the three streptococcal species (Vickerman et al., 2007; Mashburn-Warren et al., 2010; Son et al., 2012; Merritt & Qi, 2012). Since S. gordonii and C. albicans form fully integrated dual species biofilm communities (Dutton et al., 2014) we investigated the possible role of the ComCDE system in their regulation.
We generated two deletion mutants in the comCDE system. The ΔcomC mutant carried an aad9 cassette encoding spectinomycin resistance, engineered such that read-through into comDE occurred. This was confirmed by showing that the non-transformable phenotype of the mutant was complemented by addition of CSP (Fig. S3c). The other mutant carried a deletion across comCDE and was non-responsive to CSP (Fig. S3c). When grown in dual species biofilms for 6 h, both com mutant–C. albicans dual species biofilms were increased in biomass over and above the S. gordonii wild-type–C. albicans biofilms. The com mutants alone formed biofilms that were slightly denser than S. gordonii wild-type biofilms but there were no significant differences with respect to depth, biomass or growth rates (Fig. S3a, b) in planktonic phase. This is unlike the effects of comCDE mutations in S. mutans that resulted in the formation of abnormal biofilms (Li et al., 2002).
The dual species biofilms of ΔcomCDE–C. albicans had visible differences in matrix architecture and more extensive arrays of C. albicans hyphal structures. This led us to believe that elevated EPS and numbers of C. albicans were responsible for increased biomass. The explanation was in part supported by showing that the biofilms could be ~70 % disrupted by incubation with DNase I. We also obtained evidence that the ΔcomCDE–C. albicans biofilms contained a greater amount of eDNA and that they produced significantly more eDNA deposits. Incubation of the biofilms with lyticase led to ~50 % reduced biomass, suggesting the presence of β-glucans, which are derived from C. albicans cell wall turnover (Xie et al., 2012). The EPS of C. albicans biofilms comprises >50 % protein, branched-chain mannans, mannan–glucan complexes and a small proportion (~5 %) of eDNA (Zarnowski et al., 2014). The EPS of C. albicans biofilms is known to contribute to antifungal drug resistance (Taff et al., 2013).
Major components of EPS in oral streptococcal biofilms are α-linked glucans, fructans, proteins and eDNA. Glucan and fructan polymers are produced by the activities of glycosyltransferases, which hydrolyse glucosidic linkages in disaccharides (e.g. sucrose) or trisaccharides and transfer the appropriate monosaccharide to form α-linked polymers. Since we utilized a monosaccharide as carbon and energy source in our studies, no glucans would be produced (Ricker et al., 2014). It has been suggested that release of eDNA by streptococcal cells during early stages of biofilm formation could involve a specialized active release mechanism (Xu & Kreth, 2013) via lysis-independent membrane vesicles (Liao et al., 2014). Our experiments do not enable us to distinguish between release or lysis as mechanisms for eDNA deposition. We have also not been able to determine the relative proportions of eDNA in dual species biofilms derived from S. gordonii versus C. albicans, but our evidence suggests eDNA emanates mainly from the streptococci. In development of competence, extensive cell lysis occurs to release DNA (Wei & Håvarstein, 2012; Okshevsky & Meyer, 2013; Xu & Kreth, 2013), but the ΔcomCDE mutant did not develop competence, even when CSP was supplied. We also have no evidence for significant differences in eDNA production by S. gordonii wild-type and com mutant strains. The production of increased eDNA by the ΔcomCDE mutant therefore is a response to the presence of C. albicans.
The CSP from S. gordonii inhibited C. albicans biofilm formation, but did not inhibit hyphal filament formation in planktonic phase. Thus, under normal conditions in which both micro-organisms are growing in proximity, we suggest that CSP directly modulates C. albicans biofilm through interfering with cell–cell adherence or activating dispersal. The mechanism by which CSP interacts with C. albicans is also unknown, but shows some specificity, since sCSP had no significant effect on the biofilms. Unlike S. mutans CSP (Jarosz et al., 2009), which has bacteriocin activity, the S. gordonii CSP does not inhibit C. albicans hypha formation. Neither peptide shows any direct antifungal activity and the CSPs have no sequence homology (S. gordonii DVRSNKIRLWWENIFFNKK and S. mutans SGSLSTFFRLFNRSFTQALGK). In future studies we aim to determine the optimal component of S. gordonii CSP that will inhibit biofilm formation. Such a peptide or mimetic might be effective in controlling C. albicans biofilm formation or carriage.
In summary, we provide evidence that S. gordonii CSP can potentially control C. albicans in dual species biofilms. Localized concentrations of CSP within biofilms in which the bacteria and fungi are in close contact (Dutton et al., 2014) could be biologically effective in this respect. The production of eDNA is known to play a crucial role in C. albicans biofilm formation (Martins et al., 2010; Sapaar et al., 2014) and in addition to the results presented here, eDNA production has been shown to enhance dual species biofilms of C. albicans and Staphylococcus epidermidis (Pammi et al., 2013). The deletion of comCDE from S. gordonii leads to dual species biofilms containing elevated amounts of eDNA. This is probably a response to the increased presence of C. albicans in the absence of CSP production. Thus, CSP can regulate composition of the dual species biofilm community by modulating EPS production and C. albicans retention.
Acknowledgements
We thank Alan Leard and Katy Jepson (Wolfson Bioimaging Facility, University of Bristol) for assistance with microscopy, Steven Bates and Mark Ramsdale for provision of strains, and Nick Jakubovics for helpful discussions. We thank Richard Silverman for help with biofilm development, and Lindsay Dutton and Jane Brittan for excellent technical assistance. This work was supported by the National Institutes of Health (NIDCR), Bethesda, USA (grant no. R01 DE016690).
Abbreviations:
- CSP
competence stimulating peptide
- eDNA
extracellular DNA
- EPS
extracellular polymeric substance
- QS
quorum sensing
- RFP
red fluorescent protein
- sCSP
scrambled CSP
- Sp
spectinomycin
Footnotes
Three supplementary figures and two supplementary tables are available with the online Supplementary Material.
References
- Aas J. A., Paster B. J., Stokes L. N., Olsen I., Dewhirst F. E. (2005). Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43, 5721–5732. 10.1128/JCM.43.11.5721-5732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford C. V., d’Mello A., Nobbs A. H., Dutton L. C., Vickerman M. M., Jenkinson H. F. (2009). Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77, 3696–3704. 10.1128/IAI.00438-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittan J. L., Buckeridge T. J., Finn A., Kadioglu A., Jenkinson H. F. (2012). Pneumococcal neuraminidase A: an essential upper airway colonization factor for Streptococcus pneumoniae. Mol Oral Microbiol 27, 270–283. 10.1111/j.2041-1014.2012.00658.x [DOI] [PubMed] [Google Scholar]
- Clayton Y. M., Noble W. C. (1966). Observations on the epidemiology of Candida albicans. J Clin Pathol 19, 76–78. 10.1136/jcp.19.1.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M. R., Graham C. E., Gagliano B. C., Lorenz M. C., Garsin D. A. (2013). Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun 81, 189–200. 10.1128/IAI.00914-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E., Chen T., Izard J., Paster B. J., Tanner A. C. R., Yu W.-H., Lakshmanan A., Wade W. G. (2010). The human oral microbiome. J Bacteriol 192, 5002–5017. 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton L. C., Nobbs A. H., Jepson K., Jepson M. A., Vickerman M. M., Aqeel Alawfi S., Munro C. A., Lamont R. J., Jenkinson H. F. (2014). O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. MBio 5, e00911. 10.1128/mBio.00911-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta M. L., Klein M. I., Colonne P. M., Scott-Anne K., Gregoire S., Pai C. H., Gonzalez-Begne M., Watson G., Krysan D. J. & other authors (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun 82, 1968–1981. 10.1128/IAI.00087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat Rev Microbiol 8, 623–633. [DOI] [PubMed] [Google Scholar]
- Foster J. S., Kolenbrander P. E. (2004). Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl Environ Microbiol 70, 4340–4348. 10.1128/AEM.70.7.4340-4348.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haisman R. J., Jenkinson H. F. (1991). Mutants of Streptococcus gordonii Challis over-producing glucosyltransferase. J Gen Microbiol 137, 483–489. 10.1099/00221287-137-3-483 [DOI] [PubMed] [Google Scholar]
- Håvarstein L. S., Gaustad P., Nes I. F., Morrison D. A. (1996). Identification of the streptococcal competence-pheromone receptor. Mol Microbiol 21, 863–869. 10.1046/j.1365-2958.1996.521416.x [DOI] [PubMed] [Google Scholar]
- Heng N. C., Tagg J. R., Tompkins G. R. (2006). Identification and characterization of the loci encoding the competence-associated alternative sigma factor of Streptococcus gordonii. FEMS Microbiol Lett 259, 27–34. 10.1111/j.1574-6968.2006.00238.x [DOI] [PubMed] [Google Scholar]
- Hogan D. A., Vik A., Kolter R. (2004). A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 54, 1212–1223. 10.1111/j.1365-2958.2004.04349.x [DOI] [PubMed] [Google Scholar]
- Jarosz L. M., Deng D. M., van der Mei H. C., Crielaard W., Krom B. P. (2009). Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell 8, 1658–1664. 10.1128/EC.00070-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasi E., Dewhirst F. E., Chalmers N. I., Kent R., Jr, Moore A., Hughes C. V., Pradhan N., Loo C. Y., Tanner A. C. R. (2010). Clonal analysis of the microbiota of severe early childhood caries. Caries Res 44, 485–497. 10.1159/000320158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont R. J., El-Sabaeny A., Park Y., Cook G. S., Costerton J. W., Demuth D. R. (2002). Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148, 1627–1636. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Morrison D. A. (1999). Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181, 5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembke C., Podbielski A., Hidalgo-Grass C., Jonas L., Hanski E., Kreikemeyer B. (2006). Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl Environ Microbiol 72, 2864–2875. 10.1128/AEM.72.4.2864-2875.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. H., Tang N., Aspiras M. B., Lau P. C., Lee J. H., Ellen R. P., Cvitkovitch D. G. (2002). A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184, 2699–2708. 10.1128/JB.184.10.2699-2708.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Klein M. I., Heim K. P., Fan Y., Bitoun J. P., Ahn S. J., Burne R. A., Koo H., Brady L. J., Wen Z. T. (2014). Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196, 2355–2366. 10.1128/JB.01493-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Burne R. A. (2011). The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J Bacteriol 193, 2826–2837. 10.1128/JB.00056-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ramsey M. M., Chen X., Koley D., Whiteley M., Bard A. J. (2011). Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc Natl Acad Sci U S A 108, 2668–2673. 10.1073/pnas.1018391108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Su C., Unoje O., Liu H. (2014). Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc Natl Acad Sci U S A 111, 1975–1980. 10.1073/pnas.1318690111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M., Uppuluri P., Thomas D. P., Cleary I. A., Henriques M., Lopez-Ribot J. L., Oliveira R. (2010). Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 169, 323–331. 10.1007/s11046-009-9264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L., Morrison D. A., Federle M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78, 589–606. 10.1111/j.1365-2958.2010.07361.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab R., Ford S. K., El-Sabaeny A., Barbieri B., Cook G. S., Lamont R. J. (2003). LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol 185, 274–284. 10.1128/JB.185.1.274-284.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J., Qi F. (2012). The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 27, 57–69. 10.1111/j.2041-1014.2011.00634.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne S. W., Cheetham J., Lloyd D., Aves S., Bates S. (2011). Cassettes for PCR-mediated gene tagging in Candida albicans utilizing nourseothricin resistance. Yeast 28, 833–841. 10.1002/yea.1910 [DOI] [PubMed] [Google Scholar]
- Naseem S., Gunasekera A., Araya E., Konopka J. B. (2011). N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J Biol Chem 286, 28671–28680. 10.1074/jbc.M111.249854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs A. H., Vickerman M. M., Jenkinson H. F. (2010). Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell 9, 1622–1634. 10.1128/EC.00103-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad B., Kilian M. (1990). Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res 24, 267–272. 10.1159/000261281 [DOI] [PubMed] [Google Scholar]
- Okshevsky M., Meyer R. L. (2013). The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 1–11. 10.3109/1040841X.2013.841639 [DOI] [PubMed] [Google Scholar]
- Palmer R. J., Jr, Gordon S. M., Cisar J. O., Kolenbrander P. E. (2003). Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J Bacteriol 185, 3400–3409. 10.1128/JB.185.11.3400-3409.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammi M., Liang R., Hicks J., Mistretta T. A., Versalovic J. (2013). Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol 13, 257. 10.1186/1471-2180-13-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHE (2013). Voluntary surveillance of candidaemia in England, Wales and Northern Ireland: 2012. Health Protection Report 7. HCAI.
- Piotrowski A., Luo P., Morrison D. A. (2009). Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW. J Bacteriol 191, 3359–3366. 10.1128/JB.01750-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A., Spellerberg B., Woischnik M., Pohl B., Lütticken R. (1996). Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177, 137–147. 10.1016/0378-1119(96)84178-3 [DOI] [PubMed] [Google Scholar]
- Ricker A., Vickerman M., Dongari-Bagtzoglou A. (2014). Streptococcus gordonii glucosyltransferase promotes biofilm interactions with Candida albicans. J Oral Microbiol 6, 23419. 10.3402/jom.v6.23419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapaar B., Nur A., Hirota K., Yumoto H., Murakami K., Amoh T., Matsuo T., Ichikawa T., Miyake Y. (2014). Effects of extracellular DNA from Candida albicans and pneumonia-related pathogens on Candida biofilm formation and hyphal transformation. J Appl Microbiol 116, 1531–1542. 10.1111/jam.12483 [DOI] [PubMed] [Google Scholar]
- Shak J. R., Vidal J. E., Klugman K. P. (2013). Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol 21, 129–135. 10.1016/j.tim.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R. J., Nobbs A. H., Vickerman M. M., Barbour M. E., Jenkinson H. F. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78, 4644–4652. 10.1128/IAI.00685-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M., Ahn S.-J., Guo Q., Burne R. A., Hagen S. J. (2012). Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86, 258–272. 10.1111/j.1365-2958.2012.08187.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztajer H., Szafranski S. P., Tomasch J., Reck M., Nimtz M., Rohde M., Wagner-Döbler I. (2014). Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J 8, 2256–2271. 10.1038/ismej.2014.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff H. T., Mitchell K. F., Edward J. A., Andes D. R. (2013). Mechanisms of Candida biofilm drug resistance. Future Microbiol 8, 1325–1337. 10.2217/fmb.13.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman M. M., Iobst S., Jesionowski A. M., Gill S. R. (2007). Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol 189, 7799–7807. 10.1128/JB.01023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Håvarstein L. S. (2012). Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl Environ Microbiol 78, 5897–5905. 10.1128/AEM.01343-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore S. E., Lamont R. J. (2011). The pathogenic persona of community-associated oral streptococci. Mol Microbiol 81, 305–314. 10.1111/j.1365-2958.2011.07707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39, 309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- Wright C. J., Burns L. H., Jack A. A., Back C. R., Dutton L. C., Nobbs A. H., Lamont R. J., Jenkinson H. F. (2013). Microbial interactions in building of communities. Mol Oral Microbiol 28, 83–101. 10.1111/omi.12012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Thompson A., Sobue T., Kashleva H., Xu H., Vasilakos J., Dongari-Bagtzoglou A. (2012). Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J Infect Dis 206, 1936–1945. 10.1093/infdis/jis607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Kreth J. (2013). Role of LytF and AtlS in eDNA release by Streptococcus gordonii. PLoS ONE 8, e62339. 10.1371/journal.pone.0062339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Jenkinson H. F., Dongari-Bagtzoglou A. (2014a). Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol 29, 99–116. 10.1111/omi.12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sobue T., Thompson A., Xie Z., Poon K., Ricker A., Cervantes J., Diaz P. I., Dongari-Bagtzoglou A. (2014b). Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol 16, 214–231. 10.1111/cmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnowski R., Westler W. M., Lacmbouh G. A., Marita J. M., Bothe J. R., Bernhardt J., Lounes-Hadj Sahraoui A., Fontaine J., Sanchez H. & other authors (2014). Novel entries in a fungal biofilm matrix encyclopedia. MBio 5, e01333–e14. 10.1128/mBio.01333-14 [DOI] [PMC free article] [PubMed] [Google Scholar]