Abstract
It has long been evident that cancer is a heterogeneous disease. But only relatively recently have we come to realize the extent of the heterogeneity. No single therapy is effective for every patient with tumors having the same histology. A clinical strategy based on a single-therapy approach results in overtreatment for the majority of patients. Biomarkers are knives slicing the disease ever more finely. The future of clinical research will be based on learning whether particular therapies are more appropriate for some biomarker-defined subsets than are other therapies. Therapies will eventually be tailored to narrow biomarker subsets. Determining which therapies are appropriate for which patients requires both biological science and empirical evidence from clinical trials. Neither aspect is easy. In this essay we describe some nascent approaches to designing clinical trials that are biomarker-based and adaptive. Our focus is on adaptive trials that address many questions at once. In a way these clinical experiments are themselves part of a much larger experiment: learning whether and how it is possible to design experiments that match patients in small subsets of disease with a therapies that are especially effective and possibly even curative for them.
Keywords: Adaptive clinical trials, Biomarkers in cancer trials, Imaging as auxiliary end point, I-SPY 2 TRIAL, BATTLE trial
Introduction
Despite a burgeoning number of cancer drugs in development, there are fewer new cancer drugs being submitted for market approval to the U.S. Food and Drug Administration (FDA). Moreover, the proportion of successful phase 3 clinical trials in oncology is the lowest among all therapeutic areas [1].
Recognizing the need to build a better foundation for drug development, the FDA initiated its Critical Path Initiative in 2004. Its goal was to accelerate the translation of biomedical discoveries into therapies. Among other recommendations this Initiative encouraged the use of innovative trial designs, including adaptive designs. Updating the Critical Path Initiative in 2006 the FDA indicated that they had “uncovered a consensus that the two most important areas for improving medical product development are biomarker development and streamlining clinical trials” [2]. These two areas are the principal focuses of the present article.
Other groups have also encouraged improving clinical trial design strategies. In concert with the FDA’s Critical Path Initiative, in 2005 statisticians at pharmaceutical companies formed an Adaptive Design Working Group under the auspices of Pharmaceutical Research and Manufacturers of America (PhRMA). “The objectives of the group were to foster and facilitate wider usage and regulatory acceptance of adaptive designs to enhance clinical development” [3]. In 2007 the European Medicines Agency (EMA) issued a “Reflection Paper on Methodological Issues in Confirmatory Clinical Trials Planned with an Adaptive Design” [4]. In 2010 the FDA released its draft guidance “Adaptive Design Clinical Trials for Drugs and Biologics” [5].
In a similar vein, in 2010 the Institute of Medicine (IOM) responded to a request from the National Cancer Institute (NCI) by publishing a review of the NCI’s Cooperative Group Program of clinical trials [6]. One conclusion of the IOM committee was that “Better Phase 2 trial designs are needed to more accurately assess which patients benefit from a particular therapy, and thus guide the decisions about whether to move into Phase 3 trials. Improved designs for Phase 3 trials … could lead to faster more accurate conclusions about new therapeutics and in the process reduce costs and conserve resources.”
These initiatives reflect wide recognition that traditional approaches to drug development too often fail and they too often fail in late phases, leading to excessive development costs and duration. Some of the failures are due to ineffective drugs, which should have been discovered sooner in more informative early phase clinical trials. Other reasons failures are effective drugs that were poorly developed. Moreover, some drugs that are eventually successful spend too much time (and resources) in clinical trials. Strategies to improve drug development should consider adaptive designs and using biomarkers to help guide trials having those designs, in “personalized therapy trials.” The goal of such trials is to identify which therapies are best for which patients while preserving the scientific integrity of the trial. The statistical issues are substantial, as are the logistics and timeliness of biomarker assessment and data flow [3, 7].
In this article we describe two personalized therapy trials, I-SPY 2 and BATTLE, including lessons learned in designing and running these trials. Both have prospective Bayesian designs. The Bayesian approach is ideally suited for building adaptive trials because its basic inferential measures—posterior probabilities of unknown parameters and predictive probabilities of future observations—can be updated (using Bayes rule) as information accrues in the trial [8-11].
Our description of personalized therapy trials is not comprehensive either with respect to adaptive or Bayesian approaches or using biomarkers. Generic descriptions have been published elsewhere. [3, 11-13] The two examples in the present article represent a very special kind of adaptive design—special in several ways, perhaps most noticeably in that they compare many therapies within the same trial, including those involving experimental drugs from different pharmaceutical companies. Quite generally, the adaptive Bayesian approach is most useful in trials that address many questions, including identifying which of many possible therapies are better for which patients.
I-SPY 2
I-SPY 2 (Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis 2) is a randomized phase 2 screening process that evaluates experimental agents in combination with standard neoadjuvant chemotherapy for patients with high-risk primary breast cancer, those with tumors at least 2.5 cm [14-15]. Pharmaceutical companies submit drugs to the trial’s Agent Selection Committee. Experimental arms can be added to the trial at any time, assuming adequate phase 1 safety information and assuming that the overall trial’s accrual rate is sufficient to accommodate additional treatment arms.
The primary endpoint is pathologic complete response (pCR) at the time of surgery, which is a potential path to accelerated marketing approval [16, Page 21]. An agent is evaluated for its effect on pCR and can be graduated from the trial at any time, together with its “biomarker signature.” This is one of 10 prospectively defined subsets of disease that make biological sense and have marketing interest as a consequence of their prevalence. Graduation requires having at least an 85% (Bayesian) predictive probability of success in a randomized 300-patient phase 3 trial having the same control arm as I-SPY2 and pCR as the end point. Experimental arms can be dropped from the trial at any time for lack of effect on pCR in any subset of the disease, less than 10% predictive probability of phase 3 success in all 10 biomarker signatures.
Patient screening includes an MRI to establish tumor size at baseline and a biopsy to identify the tumor’s hormone-receptor status (HR), HER-2neu status (HER2), and an NKI 70-gene profile (NKI) [17]. Patients within the HR/HER2/NKI strata are assigned to therapy in an adaptively randomized fashion. The randomization probabilities depend on the performance of the various therapies within the trial, in comparison with control (which has a fixed randomization probability of 20%), and in particular for patients in the same stratum as the patient being randomized. Therapies that have a high rate of pCR for such patients have greater randomization probabilities, thus moving better performing therapies through the process more rapidly.
Figure 1 illustrates the design of I-SPY 2. The patient population in Panel A is shown as being heterogeneous. It shows 5 experimental arms. (The ongoing trial has 3 experimental arms, with the 3 drugs from different companies, with additional drugs under consideration.) The adaptive randomization is within the patient subsets, as indicated above. Panel B shows the hypothetical possibility that experimental arm 2 graduates with a particular biomarker signature, indicated schematically by the subset of patient population symbols from Panel A. Panel C shows the setting where experimental arms 2, 3, and 5 have moved on from the trial and have been replaced by experimental arms 6 and 7.
Figure 1.
Panels A-D
Panel D of Figure 1 shows a configuration of arms that is possible but has not yet been used in I-SPY 2. The panel also suggests other settings and diseases by replacing standard therapy with SOC (standard of care) and indicating progression-free survival (PFS), overall survival (OS), along with pCR as possible endpoints. The bottom 4 arms constitute a factorial design in which agents C and D plus SOC are compared with SOC alone and combined. The trial could proceed just as when the arms are independent, but the analysis would exploit the benefits of the factorial design as a “subtrial” within the bigger trial. The randomization probabilities for the single-arm arms would be down weighted within subsets of the disease if the combination C+D is shown to be better than both alone within those subsets. This approach could be used in an effort to increase the efficiency of early studies of new therapeutics, where separate trials are often used to explore the efficacy of monotherapy and combinations with SOC, sometimes including separate biomarker-defined cohorts.
The “standard therapy” in I-SPY 2 consists of 12 weekly cycles of paclitaxel followed by 4 cycles of doxorubicin/cyclophosphamide. Experimental arms have experimental agents added to standard therapy during the paclitaxel phase of treatment. MRIs to assess change in tumor volume from baseline are conducted at weeks 3 and 12 of the paclitaxel phase. Consistent with the Bayesian approach, randomization and phase 3 success probabilities are based on all available data, including MRI volume measurements for all patients. Week 3 and week 12 measurements for those patients having surgery are used to inform a longitudinal statistical model for predicting pCR. This model is used to (multiply) impute pCR results for those patients who have not yet had surgery but who have had at least one post-randomization MRI. Longitudinal MRI volume measurements are predictive of outcomes at surgery [18-19]. The predictions are not perfect, but interim MRI measurements are informative and improve the performance of the adaptive design algorithm.
I-SPY 2 is sponsored by The Biomarkers Consortium of the Foundation for the National Institutes of Health (FNIH) [20], a public-private partnership that includes the FDA, the NIH, and major pharmaceutical companies, and QuantumLeap Healthcare [21] (Figure 1-2).
BATTLE-1
Multiple signaling pathways have been implicated in the development and progression of NSCLC. Important differences in signaling pathway alterations between chemo-naive and -resistant tumor tissues in patients with advanced NSCLC necessitate molecular examination of the tumor at the time of therapy selection for these patients, rather than using data from the original diagnostic biopsy. In the Department of Defense-sponsored BATTLE-1 trial (Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination) [22] we developed a program to obtain fresh tissue biopsies from patients with refractory NSCLC. We employed real-time molecular analyses of those biopsies to guide treatment decisions while continuing to discover new pathways and markers relevant to this disease. These molecular assessments were performed in the thoracic research laboratory and were used to guide patient assignments, via a Bayesian adaptive randomization algorithm, to four corresponding targeted therapies: erlotinib (EGFR inhibitor), vandetanib (dual EGFR/VEGFR inhibitor), bexarotene + erlotinib (targeting cyclin D1/RXR pathways and EGFR, respectively), and sorafenib (RAF/VEGFR2/PDGFR inhibitor). Based on the most up-to-date patient-derived data elucidating the relationship between biomarker status and treatment outcomes, the adaptive randomization model assigned patients to more effective treatments with higher probability depending on the current results within each individual patient’s biomarker profile.
Following consent and enrollment, patients underwent a core biopsy of their lung tumor or metastasis for biomarker analysis of 11 prespecified biomarkers/marker groups: EGFR, KRAS, and BRAF gene mutation (PCR), EGFR and cyclin D1 copy number (FISH), and 6 proteins by IHC (VEGFR and RXR receptors/cycD1). Of critical importance, every patient’s identification and consent, tissue collection, biomarker analysis, and randomization all occurred within 14 days of enrollment. The primary endpoint was 8-week disease control rate (DCR), with patients being treated until disease progression or unacceptable toxicity.
Findings from the BATTLE-1 trial serve to underscore the significance of our proposed studies in lung cancer research. From November 30, 2006, to October 28, 2009, 341 patients were enrolled in BATTLE-1, of which 255 were randomized to one of the 4 treatments previously listed. The patients were heavily pretreated, and many had received multiple therapies for metastatic disease, including prior erlotinib therapy (116 pts; 45%). The mandated biopsies were shown to be feasible and safe, with 11.5% pneumothorax incidence in patients receiving lung biopsies and, of these, only 1 patient with a Grade 3 pneumothorax (no Grades 4 or 5). The first 97 patients (~40%) were equally randomized into the four treatments to acquire sufficient data to inform the statistical model. We then “switched” to the adaptive randomization phase for the remaining 60% of patients. Associations between tumor molecular profiles and treatment efficacy were calculated and continually updated during the trial, allowing us to increasingly randomize new patients to the most effective treatments for that profile. The overall 8-week DCR was 46%.
The major outcomes of this trial include the following:
More than 250 pts were biopsied and randomized to one of the 4 treatments in less than 3 years (an unprecedented accrual rate of >8 patients/month), with biomarker analysis completed in our Thoracic Molecular Pathology research laboratory within 2 weeks. The study achieved its primary endpoint (assessment of disease control rate) and demonstrated our ability to successfully complete a large, complex, biopsy-driven clinical trial with mandated fresh tumor biopsies in poor-prognosis NSCLC patients.
Findings that EGFR mutations were predictive for erlotinib benefit confirmed knowledge emerging at the time of BATTLE-1’s development (2005), and demonstrated the potential of biomarkers to predict patients’ outcomes after treatment with a targeted agent.
We observed an unexpected level of benefit in sorafenib-treated pts with both wt- and mut-KRAS; however, the biologic underpinnings of this activity are unknown. Given the lack of response in KRAS-mutated patients to any targeted agent to date, including sorafenib, these results warrant further study of sorafenib’s clinical activity and potential markers to identify the specific patients most likely to benefit in a more durable fashion.
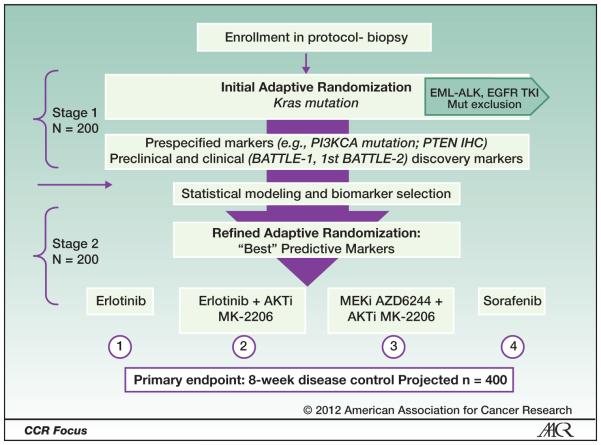
Building on lessons learned from the BATTLE-1 program, we have the unique opportunity and proven ability to study KRAS effects on response to inhibitors of its down-stream signaling pathways through our prospective, multi-arm, adaptively randomized trial titled “BATTLE-2 Program: A Biomarker-Integrated Targeted Therapy Study in Previously Treated Patients with Advanced Non-Small Cell Lung Cancer” (BATTLE-2). In particular, BATTLE-2 will identify new biomarkers that can effectively predict disease control for EGFR-wt patients treated with targeted agents. In this trial (see Figure 3), 400 patients with refractory NSCLC will undergo a mandated fresh biopsy prior to therapy and receive one of four treatments, including combinations targeting downstream markers of KRAS-activated pathways and Discovery of new Markers and Mutations in Patients with no known dominant pathway, as guided by Clinical Laboratory Improvement Amendments (CLIA)-certified molecular analyses of their tumor tissue:
Figure 3.
Battle-2 schema: advanced refractory NSCLC.
Erlotinib (EGFR inhibitor)
Erlotinib plus an AKT inhibitor (MK-2206)
MK-2206 plus a MEK inhibitor (AZD6244)
Sorafenib.
The laboratory component of this clinical trial will identify novel biomarkers for more effectively selecting patients who may benefit from these therapies. We will use high throughput sequencing technologies to identify gene mutations in BATTLE-1 and BATTLE-2 tumor tissues. These high-throughput technologies will include analysis of hot-spot mutation in 20 known NSCLC-related oncogenes (via Sequenom, Inc.), and the newly developed next-generation (nex-gen) sequencing platform, SOLiD (Life Technologies, Inc.), encompassing whole genome sequencing (DNA), full transcriptome sequencing (mRNA) and miRNA analysis.
We have evidence from studies using a panel of molecularly characterized NSCLC cell lines that different KRAS amino acid substitutions may have varying effects on KRAS-activated signaling. We have also identified new compounds that selectively inhibit proliferation of cells with mutant but not wild-type (wt) KRAS, and will explore their mechanism of action. We have fully annotated clinical data and biopsy samples from our BATTLE-1 NSCLC trial that fully support the feasibility and scientific strength of this approach and that can also be used for validation of our discoveries; thus, we will have available more than 400 tissue (core needle biopsy, CNB) and cytology (fine needle aspiration, FNA) specimens collected prospectively from BATTLE-2, as well as clinical and molecular data from both unique, biopsy-driven adaptive clinical trial programs — BATTLE-1 and BATTLE-2 — to further explore the efficacy of a personalized medicine approach to the treatment of NSCLC and to better understand and target this critical oncogenic pathway.
Limitations of I-SPY 2 and BATTLE and Alternatives
If a predictive biomarker is expected to identify a patient population that will respond to a new therapy with high confidence, the simplest path to development of both the drug and the companion diagnostic test (based on the predictive biomarker) is to restrict study enrollment to the selected population early in development. Notable recent successful examples of this strategy are the development of vemurafenib (BRAF-inhibitor) and crizotinib (ALK inhibitor), along with their companion diagnostic tests, in melanoma and non-small cell lung cancer, respectively. In both of these cases, enrollment was restricted beginning in late phase 1 studies.
However, a major risk with this strategy is the selection of a predictive biomarker that is predominantly based on preclinical studies. Preclinical models often do not fully recapitulate the clinical setting, and can suggest incorrect predictive biomarkers, as in the case of EGFR and IGFR1 protein expression for EGFR and IGFR-targeting antibodies, respectively [23-24]. With the increasing demands of efficiency in drug development, selection of the “wrong” biomarker in early studies in which enrollment is restricted can lead to an incorrect “no go” due to apparent lack of efficacy. Another issue with the restricted enrollment development strategy is that studies to determine the effect of the new drug in the “biomarker negative” population may be delayed or never done, leaving open the question of whether or not the drug would have efficacy in that population. An example is a study of trastuzumab in combination with chemotherapy in HER2-low breast cancer patients, which was initiated in 2011, more than 10 years after the initial approval of the drug (NCT01275677).
Conversely, using “all comers” approaches early in clinical drug development can also be risky in settings where there is an expectation for an early efficacy signal before investment in large phase 2 or 3 studies, especially when the prevalence of the responsive population is low among a histologically defined cancer type. For example, ~5% of patients with non-small cell lung cancer have cancers with an ALK translocation that is associated with responsiveness to crizotinib. Assuming that a 20-patient lung cancer phase 1b cohort is used as a screen for efficacy, the probability of enrolling 3 patients with an ALK translocation is 8%, and the probability of enrolling 2 such patients is 26%. Thus, assuming the drug has little activity in patients who do not have ALK-translocated tumors, the most likely outcome using an all comers approach in this scenario is that there would be 0 or 1 responses among these 20 patients, which could result in a “no go” for the development of the drug in lung cancer.
The advantages of BATTLE and I-SPY approaches over the “all comers” or “restricted enrollment” clinical trial strategies include the following: 1) single control arm used for multiple experimental drugs, 2) no “screen failures” for enrollment based on a specific diagnostic assay, 3) each drug is evaluated for efficacy among multiple biomarker-defined subgroups. These advantages have the potential to greatly improve the efficiency of the process of co-development of a new therapeutic with a matching diagnostic. However, there are limitations to the BATTLE and I-SPY approaches. First, since patients are assigned to treatments according to results of biomarker analyses, the biomarker assays must be chosen carefully. Similar to the restricted enrollment strategy, selection of “wrong” biomarkers may lead to an incorrect “no go” decision for a given compound.
A related limitation is the selection of the cutpoint for biomarkers that are measured by a continuous scale, which is used to classify patients as “positive” or “negative” with regard to the biomarker, and which must be decided before the study starts. Considering the limited clinical information that is typically available about the relationship between the biomarker and the efficacy of an investigational drug, it may be difficult to select this cutpoint. Setting the cutpoint too high could reduce the ability of adaptive randomization to discriminate among potential treatments for this subpopulation. Conversely, setting the cutpoint too low could dilute the effect size of an investigational treatment in the “biomarker positive” group and also decrease the discriminatory ability of the approach.
An alternative adaptive approach, which does not require selection of biomarkers or cutpoints before a study begins, has been proposed but not yet implemented in the clinic. Referred to as the adaptive signature design (or the related cross-validated adaptive signature design) [25-26], this is an adaptive but frequentist approach to testing multiple biomarkers in a relatively large study. The key aspect of this design is that a potential predictive biomarker is identified using a randomly selected “training” set of the enrolled patients, with the remaining patients used to validate the predictive biomarker. An attractive aspect of this approach is that whole genome, agnostic methods to identify a predictive biomarker can be used in the test set, which arguably decreases the risk of selection of the “wrong” biomarker. An additional important aspect of this approach is that analytically validated tests for potential predictive biomarkers are not needed until the time of the final analysis. If embraced by regulatory authorities and drug developers, similar to I-SPY and BATTLE, this approach has the potential to improve the efficiency of co-development of a new therapeutic with a matching diagnostic.
Conclusion
Adaptive clinical trials that use prospectively assessed biomarkers to assign therapy are feasible. They offer promise in shortening overall drug development time and in addressing more accurately which patients benefit from which therapies. We have described some example trials. For reasons we indicated, none is the final answer to personalizing cancer medicine. They are themselves part of an experiment. It is an essential experiment to help us understand how—or whether—we can build complicated and yet informative and efficient clinical trials that match biomarker subsets with therapies.
Figure 2.
Comparison of the I-SPY 2 trial with a standard approach. Figure adapted with permission from the AACR Cancer Progress Report (2011).
References
- 1. ( http://insidebioia.files.wordpress.com/2011/02/bio-ceo-biomedtracker-bio-study-handout-final-2-15-2011.pdf)
- 2. http://www.fda.gov/ScienceResearch/SpecialTopics/CriticalPathInitiative/default.htm.
- 3.Gallo P, Chuang-Stein C, Dragalin V, Gaydos B, Krams M, Pinheiro J. Adaptive designs in clinical drug development—An executive summary of the PhRMA Working Group. Journal of Biopharmaceutical Statistics. 2006;16:275–283. doi: 10.1080/10543400600614742. [ISSN: 1054-3406 print/1520-5711 online. DOI: 10.1080/10543400600614742] [DOI] [PubMed] [Google Scholar]
- 4. http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500003616.pdf.
- 5. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm201790.pdf.
- 6. http://www.iom.edu/Reports/2010/A-National-Cancer-Clinical-Trials-System-for-the-21st-Century-Reinvigorating-the-NCI-Cooperative.aspx.
- 7.Gaydos B, Anderson K, Berry D, Burnham N, Chuang-Stein C, Dudinak J, Fardipour P, Gallo P, Givens S, Maca J, Pinheiro J, Pritchett Y, Lewis R, Krams M. Good practices for adaptive clinical trials. Drug Information Journal. 2009;43539:556. [Google Scholar]
- 8.Berry DA. Statistics: A Bayesian Perspective. Duxbury Press; Belmont, California: 1996. [Google Scholar]
- 9.Berry DA. Bayesian clinical trials. Nature Reviews Drug Discovery. 2006;527:36. doi: 10.1038/nrd1927. [DOI] [PubMed] [Google Scholar]
- 10.Berry DA. Statistical Innovations in Cancer Research. In: Holland J, Frei T, editors. Cancer Medicine. BC Decker; London: 2009. pp. 446–463. e.8. Ch 35. et al. [Google Scholar]
- 11.Berry DA. Adaptive clinical trials in oncology. Nat Rev Clin Oncol. doi: 10.1038/nrclinonc.2011.165. advance online publication 8 November 2011; doi:10.1038/nrclinonc.2011.165. [DOI] [PubMed] [Google Scholar]
- 12.Biswas S, Liu DD, Lee JJ, Berry DA. Bayesian clinical trials at the University of Texas M. D. Anderson Cancer Center. Clinical Trials. 2009;6205:216. doi: 10.1177/1740774509104992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry DA. Adaptive clinical trials: The promise and the caution. Journal of Clinical Oncology. 2010;311423:1426. doi: 10.1200/JCO.2010.32.2685. [DOI] [PubMed] [Google Scholar]
- 14. http://ispy2.org/, http://clinicaltrials.gov/ct2/show/NCT01042379.
- 15.Barker AD, Sigman CC, Kelloff GJ, Hylton NM, Berry DA, Esserman LJ. I-SPY 2: An adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clinical Pharmacology & Therapeutics. 2009;8697:100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 16. http://www.fda.gov/AboutFDA/ReportsManualsForms/Reports/ucm274333.htm.
- 17.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. doi:10.1038/415530a. PMID 11823860. [DOI] [PubMed] [Google Scholar]
- 18.Esserman LJ, Berry DA, DeMichele et al. Pathologic Complete Response Predicts Recurrence-Free Survival More Effectively by Cancer Subtype: Results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) 2011 doi: 10.1200/JCO.2011.39.2779. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esserman LJ, Berry DA, Cheang MCU, et al. Chemotherapy Response and Recurrence-free Survival in Neoadjuvant Breast Cancer Depends on Biomarker Profiles: Results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) 2011 doi: 10.1007/s10549-011-1895-2. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. http://www.fnih.org/
- 21. http://www.quantumleaphealth.org/
- 22.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Tsao A, Stewart DJ, Hicks ME, Erasmus J, Gupta S, Alden CM, Liu S, Tang X, Khuri FR, Tran HT, Johnson BE, Heymach JV, Mao L, Fossella F, Kies MS, Papadimitrakopoulou V, Davis SE, Lippman SM, Hong WK. The BATTLE trial: Personalizing therapy for lung cancer. Cancer Discovery. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. doi:10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803–10. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 24.Gualberto A, Dolled-Filhart M, Gustavson M, et al. Molecular analysis of non-small cell lung cancer identifies subsets with different sensitivity to insulin-like growth factor I receptor inhibition. Clin Cancer Res. 2010;16:4654–65. doi: 10.1158/1078-0432.CCR-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Freidlin B, Simon R. Adaptive signature design: an adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clin Cancer Res. 2005;117872:8. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- 26.Freidlin B, Jiang W, Simon R. The cross-validated adaptive signature design. Clin Cancer Res. 2010;16691:8. doi: 10.1158/1078-0432.CCR-09-1357. [DOI] [PubMed] [Google Scholar]