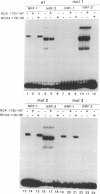
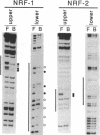
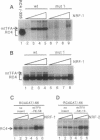
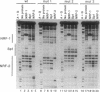
Abstract
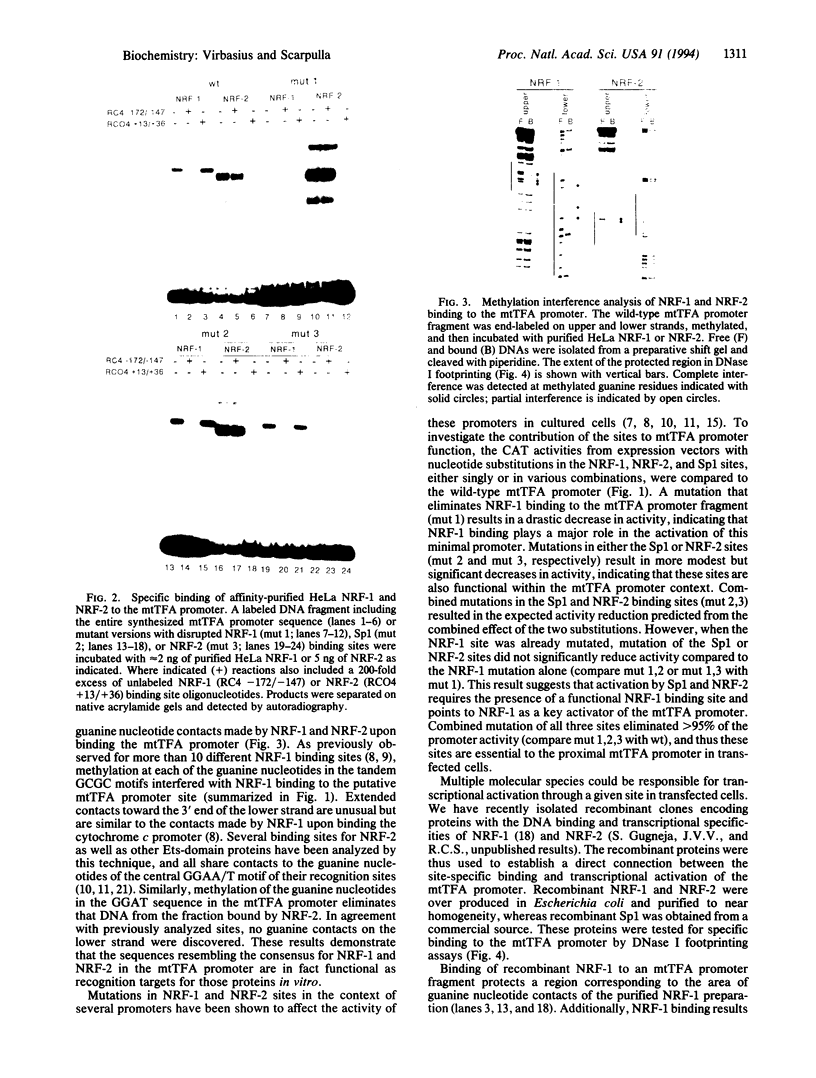
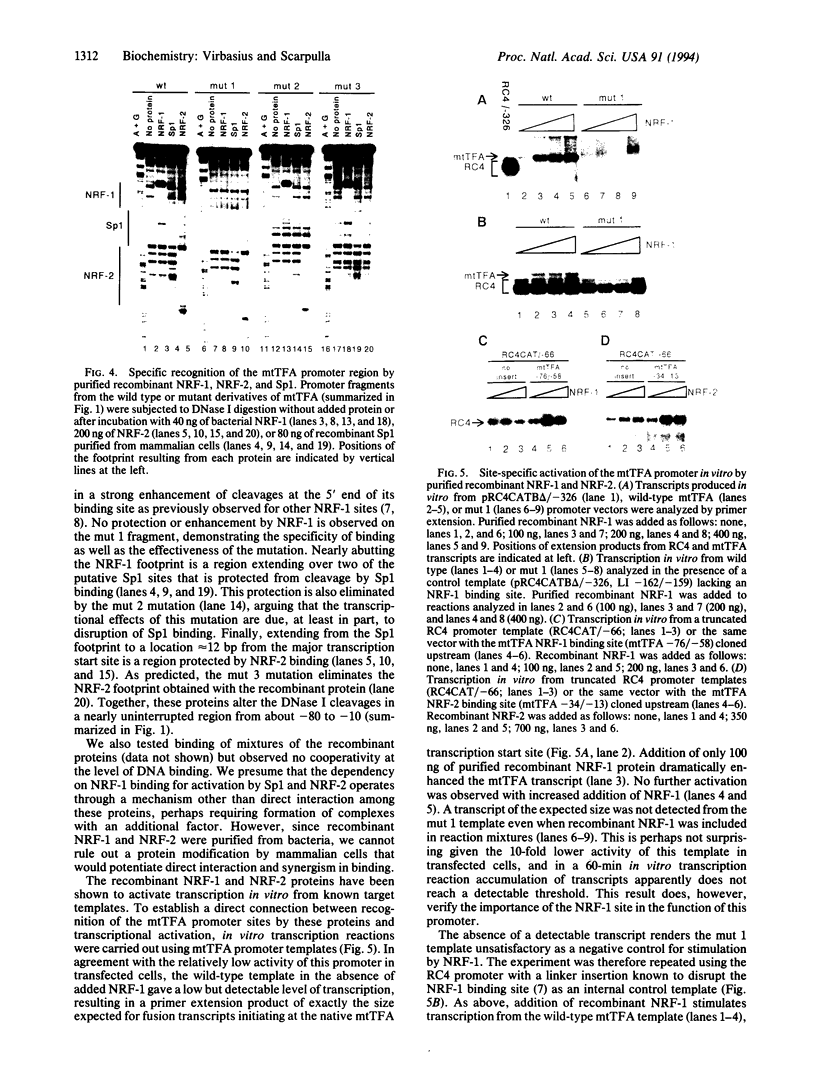
Mitochondrial transcription factor A (mtTFA), the product of a nuclear gene, stimulates transcription from the two divergent mitochondrial promoters and is likely the principal activator of mitochondrial gene expression in vertebrates. Here we establish that the proximal promoter of the human mtTFA gene is highly dependent upon recognition sites for the nuclear respiratory factors, NRF-1 and NRF-2, for activity. These factors have been previously implicated in the activation of numerous nuclear genes that contribute to mitochondrial respiratory function. The affinity-purified factors from HeLa cells specifically bind to the mtTFA NRF-1 and NRF-2 sites through guanine nucleotide contacts that are characteristic for each site. Mutations in these contacts eliminate NRF-1 and NRF-2 binding and also dramatically reduce promoter activity in transfected cells. Although both factors contribute, NRF-1 binding appears to be the major determinant of promoter function. This dependence on NRF-1 activation is confirmed by in vitro transcription using highly purified recombinant proteins that display the same binding specificities as the HeLa cell factors. The activation of the mtTFA promoter by both NRF-1 and NRF-2 therefore provides a link between the expression of nuclear and mitochondrial genes and suggests a mechanism for their coordinate regulation during organelle biogenesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braidotti G., Borthwick I. A., May B. K. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. J Biol Chem. 1993 Jan 15;268(2):1109–1117. [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987 Mar 6;235(4793):1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987 Feb;6(2):409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989 Jan 13;56(1):131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- Chau C. M., Evans M. J., Scarpulla R. C. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2 alpha, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem. 1992 Apr 5;267(10):6999–7006. [PubMed] [Google Scholar]
- Clayton D. A. Transcription and replication of animal mitochondrial DNAs. Int Rev Cytol. 1992;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J., Scarpulla R. C. Both upstream and intron sequence elements are required for elevated expression of the rat somatic cytochrome c gene in COS-1 cells. Mol Cell Biol. 1988 Jan;8(1):35–41. doi: 10.1128/mcb.8.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J., Scarpulla R. C. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem. 1989 Aug 25;264(24):14361–14368. [PubMed] [Google Scholar]
- Evans M. J., Scarpulla R. C. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990 Jun;4(6):1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Clayton D. A. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988 Aug;8(8):3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarco K., Thompson C. C., Byers B. P., Walton E. M., McKnight S. L. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991 Aug 16;253(5021):789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Mutvei A., Husman B., Andersson G., Nelson B. D. Thyroid hormone and not growth hormone is the principle regulator of mammalian mitochondrial biogenesis. Acta Endocrinol (Copenh) 1989 Aug;121(2):223–228. doi: 10.1530/acta.0.1210223. [DOI] [PubMed] [Google Scholar]
- Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992 Jun;6(6):975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Parisi M. A., Clayton D. A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991 May 17;252(5008):965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- Parisi M. A., Xu B., Clayton D. A. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol Cell Biol. 1993 Mar;13(3):1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. C., Brown T. A., McKnight S. L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991 Aug 16;253(5021):762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Tominaga K., Akiyama S., Kagawa Y., Ohta S. Upstream region of a genomic gene for human mitochondrial transcription factor 1. Biochim Biophys Acta. 1992 Jun 15;1131(2):217–219. doi: 10.1016/0167-4781(92)90082-b. [DOI] [PubMed] [Google Scholar]
- Tominaga K., Hayashi J., Kagawa Y., Ohta S. Smaller isoform of human mitochondrial transcription factor 1: its wide distribution and production by alternative splicing. Biochem Biophys Res Commun. 1993 Jul 15;194(1):544–551. doi: 10.1006/bbrc.1993.1854. [DOI] [PubMed] [Google Scholar]
- Virbasius C. A., Virbasius J. V., Scarpulla R. C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993 Dec;7(12A):2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- Virbasius J. V., Scarpulla R. C. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol Cell Biol. 1991 Nov;11(11):5631–5638. doi: 10.1128/mcb.11.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius J. V., Virbasius C. A., Scarpulla R. C. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 1993 Mar;7(3):380–392. doi: 10.1101/gad.7.3.380. [DOI] [PubMed] [Google Scholar]
- Xu B., Clayton D. A. Assignment of a yeast protein necessary for mitochondrial transcription initiation. Nucleic Acids Res. 1992 Mar 11;20(5):1053–1059. doi: 10.1093/nar/20.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]