Abstract
Previous experiments using enzyme inhibitors and RNAi in cell lysates and cultured cells have suggested that tripeptidyl peptidase II (TPPII) plays a role in creating and destroying MHC class I-presented peptides. However, its precise contribution to these processes has been controversial. To elucidate the importance of TPPII in MHC class I antigen presentation, we analyzed TPPII-deficient gene-trapped mice and cell lines from these animals. In these mice, the expression level of TPPII was reduced by >90% compared to wild-type mice. Thymocytes from TPPII gene-trapped mice displayed more MHC class I on the cell surface, suggesting that TPPII normally limits antigen presentation by destroying peptides overall. TPPII gene-trapped mice responded as well as did wild-type mice to four epitopes from lymphocytic choriomeningitis virus (LCMV). The processing and presentation of peptide precursors with long N-terminal extensions in TPPII gene-trapped embryonic fibroblasts was modestly reduced, but in vivo immunization with recombinant lentiviral or vaccinia virus vectors revealed that such peptide precursors induced an equivalent CD8 T cell response in wild type and TPPII-deficient mice. These data indicate while TPPII contributes to the trimming of peptides with very long N-terminal extensions, TPPII is not essential for generating most MHC class I-presented peptides or for stimulating CTL responses to several antigens in vivo.
Keywords: Processes-Antigen Presentation/Processing, Animals-Rodent, Molecules-MHC, Infections-Viral, Cells-T Cells, Cytotoxic
Introduction
Cytotoxic T lymphocytes (CTLs) survey the MHC class I molecules on the surface of cells searching for ones that contain immunogenic peptides (1, 2). The MHC class I molecules display peptides derived from the cell’s expressed genes. In most situations, these presented peptides are from normal cellular proteins and are ignored by CD8 T cells. However, if the abnormal peptides are present, e.g. ones containing mutations or from viral proteins, then CTLs will recognize these complexes and be stimulated to destroy the abnormal cell. This process protects the host against tumors and viral infections.
Peptides that bind to MHC class I are produced from intracellular proteins as a byproduct of normal protein catabolism (3–5). The major protease responsible for the initial cleavage of cellular proteins into oligopeptides is the proteasome, a large particle in the cytosol and nucleus of cells. The majority of peptides produced by proteasomes are very rapidly hydrolyzed into amino acids by the concerted action of aminopeptidases and endopeptidases in the cytosol (6). However, a small fraction of peptides escape destruction and are transported by TAP into the endoplasmic reticulum (ER) where ones of the right size and sequence bind to newly assembled class I molecules (7). MHC class I molecules bind peptides that are of a precise size, which depending on the specific MHC class I molecule are between 8 and 10 amino acids. Less than 5% of the peptides produced by the proteasome are actually of the proper size to stably bind to any particular class I molecule (8). Proteasomes more frequently generate peptides (~10–20%) that are too long to bind to MHC class I molecules, but can serve as potential antigenic precursors (8). These long precursors can be converted to MHC class I-binding peptides by aminopeptidases, especially ER aminopeptidase 1 (ERAP1; ERAAP) (9–14), or may be completely degraded to amino acids by aminopeptidases and endopeptidases.
Where examined, most aminopeptidases preferentially degrade relatively short peptides and in vitro have little or no activity on peptides that are longer than about 16 amino acids (15, 16). An exception to this rule is tripeptidyl peptidase II (TPPII). TPPII (EC 3.4.14.10) is an abundant cytosolic aminopeptidase that sequentially removes tripeptides from the amino terminus of peptides, and also has a poorly-understood endoproteolytic activity (17, 18). TPPII is capable of degrading quite long peptides (at least as long as 41 amino acids) (17), and in vitro is the major activity in cells that degrades peptides longer than 15 amino acids (16). However, since only about 10% of peptides produced by the proteasome are longer than 15 amino acids (8), the importance of this activity is not clear.
Several groups have reported a role for TPPII in MHC class I antigen presentation (16, 19–24). Most of these reports suggest a specialized role for TPPII in processing a limited number of presented peptides; however, one group suggested that in intact cells, proteasomes mainly generate very long peptides (in contrast to the behavior of purified proteasomes in vitro), and that TPPII is essential for processing these long peptides for antigen presentation (16). We have previously tested the role of TPPII in MHC class I antigen presentation in tissue culture, using siRNA to eliminate TPPII from human (HeLa) cells (21). We found that under these conditions, overall MHC class I antigen presentation was only slightly affected, even though presentation of peptides derived from very long model peptides (14 to 17 residues) was markedly reduced in the absence of TPPII. We concluded that (as with purified proteasomes in vitro) proteasomes in intact cells mainly generate relatively short peptides that can be degraded by many intracellular peptidases, and that TPPII is not essential for MHC class I antigen presentation in tissue culture. In this study, we investigated whether TPPII is also dispensable for antigen presentation in primary and cultured mouse cells and intact mice.
Materials and Methods
Mice and PCR
Mice containing a “gene-trap” cassette that abrogates TPPII expression were purchased from Lexicon Pharmaceutical (Woodlands, TX). These mice were produced by insertional mutagenesis of embryonic stem-cells using a retroviral cassette containing a splice acceptor and a poly-A signal. In these cells, the retroviral cassette was inserted between exons 2 and 3, so that a spliced TPPII containing exons 1 and 2 (98 out of 1262 amino acids) is expressed instead of full-length TPPII (Fig. 1A). The presence of the gene-trap cassette was tested by genomic PCR using primers specific for a wild-type (mTPPIIg-F1: 5’-AGAATAGCCCATGTGCCAAC-3’, mTPPIIg-R1: 5’-CAACGAAACTTGCCTTCACA-3’) and a gene-trapped allele (mTPPIIg-F1: 5’-AGAATAGCCCATGTGCCAAC-3’, Lex-LTR2: 5’-ATAAACCCTCTTGCAGTTGCATC-3’), respectively. Real-time PCR of TPPII was performed on spleens, kidneys, and embryonic fibroblasts to quantify the knockdown level of full-length TPPII. We first used a Taqman system (Applied Biosystems, Foster City, CA), in which the amplicon sequence was not provided by the manufacturer. Therefore, we also designed a set of defined primers that could be applied for a qPCR SYBR Green assay. The primers used were TPP2 ex11 forward: 5’-GTGCCTAACTGGACATTGAG-3’ and TPP2 ex12 reverse: 5’-CAACATTATTTGCTTTCAGCCCTG-3’ for amplification around exon 11 and 12. The real-time PCR was performed on a MyiQ machine (BioRad, Hercules, CA). We used the mice back-crossed with C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME) for 6 generations in all experiments except the one using mouse embryonic fibroblasts (MEFs), in which we used the mice back-crossed for 2 generations, using MEFs derived from littermates as controls. In all experiments, we used age- and sex-matched mice for analysis. All mice were housed under specific pathogen-free conditions in the animal facility in the University of Massachusetts Medical School. Handling of the mice was performed according to institutional guidelines in the University of Massachusetts Medical School.
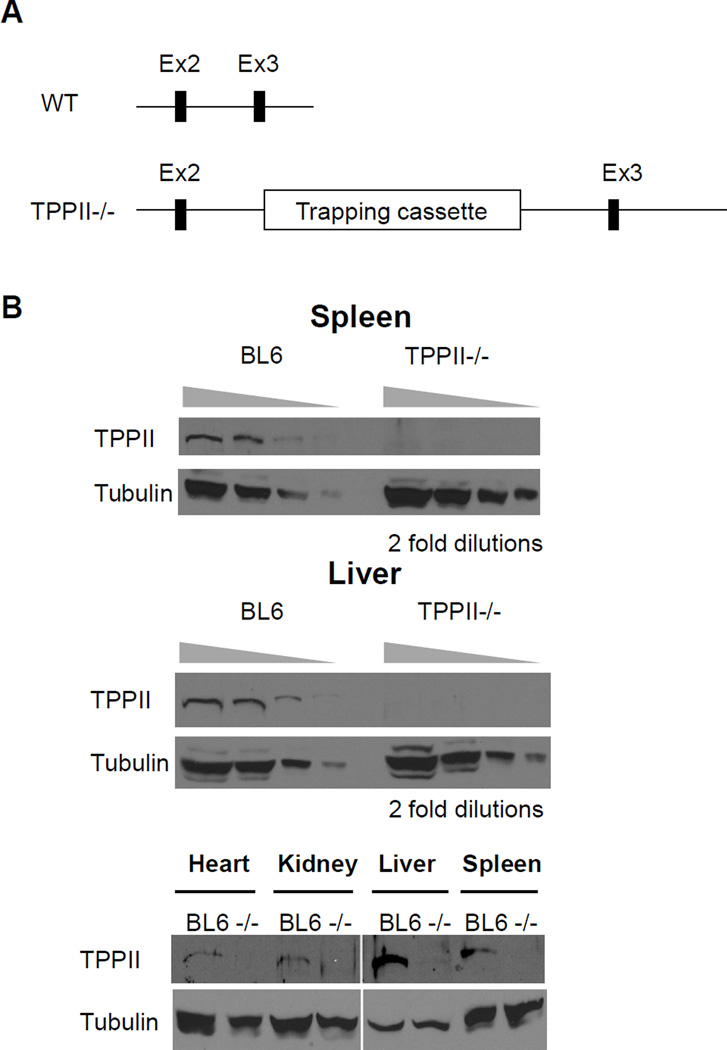
Figure 1. Construction and assessment of TPPII gene-trapped mice.
(A) Gene trap diagram. The TPPII gene-trapped mice were produced by insertional mutagenesis of embryonic stem-cells using a retroviral cassette containing a splice acceptor and a poly-A signal. In these cells, the retroviral cassette was inserted between exons 2 and 3, so that a spliced TPPII containing exons 1 and 2 (98 out of 1262 amino acids) is expressed instead of full-length TPPII. (B) Undetectable expression level of the TPPII protein in TPPII gene-trapped mice. Tissue extracts were subjected to Western blotting either with anti-TPPII antibody or with anti-α-tubulin antibody.
Cells
Mouse embryonic fibroblasts (MEFs) were produced from crosses of gene-trapped heterozygous mice as previously described (25). The genomic DNA extracted from the cells was analyzed by PCR to genotype each cell line. MHC-I antigen presentation was analyzed in homozygous gene-trapped, homozygous wild-type and heterozygous MEFs. MEFs were cultured in DMEM supplemented with 20% fetal bovine serum (FBS) in a 37 °C/10% CO2 incubator.
Mouse bone marrow-derived dendritic cells (DCs) were generated using standard protocols (26). Briefly, bone marrow cells were cultured in HCM media (RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% FBS, 2 mM L-glutamine, 10 mM HEPES, 50 µM 2-mercaptoethanol, penicillin/streptomycin and nonessential amino acids) in a 37 °C/5% CO2 incubator. On day 1, the cells were plated in the presence of 10 ng/ml mouse GM-CSF and 5 ng/ml mouse IL-4. On day 4, additional 10 ng/ml mouse GM-CSF and 5 ng/ml mouse IL-4 were added to the culture media.
293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS and 2 mM L-glutamine.
Construction of plasmids and recombinant lentiviral vectors
We tested presentation of a model peptide SIINFEKL (S8L), the H-2Kb-restricted immunodominant epitope from chicken ovalbumin (OVA). Construction of plasmids expressing N-extended S8L peptides has been previously described (21). Briefly, S8L preceded by N-terminal extensions of various lengths was inserted downstream of ubiquitin to generate a ubiquitin-peptide fusion protein. When expressed in cells, the N-terminal ubiquitin is rapidly cleaved by ubiquitin C-terminal hydrolases, yielding peptides with defined N-terminal residues (27). Green fluorescent protein (GFP) was expressed from the same transcript as the ubiquitin-peptide using an internal ribosome entry site.
Recombinant lentiviral vectors expressing full-length OVA, ubiquitin, ubiquitin fused with S8L, and ubiquitin fused with 9-amino acids N-extended S8L were constructed by inserting each gene and an IRES-GFP cassette at an EcoRI-BamHI site of FUGW (kindly provided by Dr. Eicke Latz, University of Massachusetts Medical School, Worcester, MA) (28).
Peptides used in these experiments are summarized in Table I.
Table I.
Peptides expressed by plasmids
| Name of peptide | Peptide sequence | Length |
|---|---|---|
| S8L | SIINFEKL | 8mer |
| N2S8L | LE SIINFEKL | 10mer |
| N4S8L | EQLE SIINFEKL | 12mer |
| N6S8L | GLEQLE SIINFEKL | 14mer |
| N8S8L | VSGLEQLE SIINFEKL | 16mer |
| N9S8L | EVSGLEQLE SIINFEKL | 17mer |
| N10S8L | DEVSGLEQLE SIINFEKL | 18mer |
Transfection and transduction in vitro
MEFs were transiently transfected using FuGENE6 (Roche) as described previously (12). In some cases, MEFs were treated with 50 U/ml of murine interferon-γ (IFN-γ) (BD Biosciences, San Jose, CA) 5 hours after transfection.
For production of lentivirus, 293T cells were transiently transfected with each recombinant lentiviral vector together with packaging construct delta-8.91 and a VSV-G expression plasmid (gifts from Dr. Eicke Latz, University of Massachusetts Medical School, Worcester, MA) using TransIT293 transfection reagent (Mirus Bio, Madison, WI). Supernatant containing recombinant lentivirus was collected after 2 and 3 days of transfection, and stored at −80 °C until just prior to use. For in vivo injection, the supernatant was further concentrated by ultracentrifugation (29). Lentiviral titers (transducing units (TU)) were determined using NIH3T3 cells based on the GFP-positive cell ratio on day 2.
Bone marrow cells were transduced with recombinant lentivirus in the presence of 4 µg/ml polybrene (Sigma, St Louis, MO) on day 2. Half of the media was replaced on day 3 to reduce a toxic effect of polybrene on the cells. On day 7, cells were stained and analyzed by flow cytometry.
Handling of viral vectors was performed according to the guidelines of BSL-2+ laboratories established by the recombinant DNA committee of University of Massachusetts Medical School.
Poly I:C treatment of mice
Mice were injected intraperitoneally (i.p.) with 200 µg of poly I:C (GE Healthcare, Piscataway, NJ) in a total volume of 100 µl of PBS. Spleens from the mice were harvested after 24 h and then stained for flow cytometric analysis.
Viral infection in vivo and peptide stimulation in vitro
Mice were injected i.p. with 5 × 104 pfu/mouse of lymphocytic choriomeningitis virus (LCMV) Armstrong (a gift from Dr. Ray Welsh, University of Massachusetts Medical School, Worcester, MA), or with 1.25 × 106 pfu/mouse of recombinant vaccinia virus expressing full-length OVA, ubiquitin, ubiquitin fused with S8L, or ubiquitin fused with 9-amino acids N-extended S8L. Mice were injected in the footpad with 3.3 × 105 TU/mouse of recombinant lentivirus or with 4.8 × 105 pfu/mouse of recombinant vaccinia virus. Eight (LCMV and recombinant lentivirus) or seven (recombinant vaccinia virus) days later, splenocytes (LCMV and recombinant vaccinia virus) or lymphocytes from a draining lymph node (recombinant lentivirus) were harvested and incubated for 5 hours with 5 µM of the appropriate peptide, or with 0.5 µg/ml anti-CD3ε (BD Biosciences), in the presence of GolgiPlug (BD Biosciences). Peptides that were used to stimulate IFN-γ production after LCMV infection were gp33 (KAVYNFATC), NP205 (YTVKYPNL), gp276 (SGVENPGGYCL), and NP396 (FQPQNGQFI). For recombinant vaccinia and lentivirus, S8L peptide was used. All peptides were synthesized by Anaspec, Inc. (San Jose, CA). Cells were stained for CD8, CD44 (BD Biosciences) and intracellular IFN-γ (eBioscience, San Diego, CA) using commercial antibodies, and analyzed by flow cytometry.
Antibodies and flow cytometry
The mAb 25.D1.16 (which recognizes S8L in combination with H-2Kb) (30), Y3 (anti-H-2Kb) (31), 28.14.8S (anti-H-2Db) (32) and H36.4.5 (anti-influenza hemagglutinin; for isotype control) (a gift of W. Gerhard, The Wistar Institute, University of Pennsylvania) (33) were used as primary antibodies in staining MEFs and lentivirus-transduced DCs for flow cytometry. After treatment with donkey anti-mouse F(ab’)2 fragments conjugated to Cy5 (Jackson Immunoresearch, West Grove, PA), flow cytometry was performed on a FACSCalibur apparatus (BD Biosciences), followed by analysis with FlowJo software (Tree Star, San Carlos, CA). Transfected cells were identified by gating on GFP fluorescence. In the case of staining of DCs, PE-conjugated anti-CD11c antibody (eBioscience) was also used as a secondary antibody.
For staining cells isolated from spleens, bone marrows and draining lymph nodes, AF6-88.5 (anti-H-2Kb), KH95 (anti-H-2Db) and AF6-120.1 (anti-I-Ab), BB7.2 (anti-HLA-A2; for isotype control) and anti-Gr1 antibodies conjugated to a fluorophore were used according to the manufacturer’s directions (BD Biosciences). Other fluorophore-conjugated antibodies against cell surface markers (CD4, B220, CD11c, CD86) were purchased from eBioscience. The cells were then analyzed by flow cytometry.
Western blotting
Tissue extracts were prepared in homogenization buffer (50 mM Tris (pH 7.4), 0.25 M Sucrose, 5 mM DTT, 5 mM MgCl2, 2 mM ATP, 10% glycerol) with complete protease inhibitor cocktail minitablets (Roche) using a dounce homogenizer on ice. The homogenate was spun at 325 g for 10 min, and the supernatant was further spun at 16,000 g for 20 min. The total amount of protein in the supernatant was quantified by BCA assay (Pierce). The supernatant was mixed with SDS/DTT sample buffer (New England Biolabs, Ipswich, MA), and was heated to 100°C for 5 min. The resulting lysate was resolved by SDS-PAGE and was transferred to a nitrocellulose membrane. After the membrane was blocked with 5% milk in PBS containing 0.1% Tween 20, the blot was probed with chicken anti-TPPII antibody (Cedarlane Labs, Burlington, NC) or mouse anti-α-tubulin antibody (Abcam, Cambridge, MA), followed by HRP-conjugated anti-chicken IgY (Promega, Madison, WI) or HRP-conjugated anti-mouse IgG (Jackson Immunoresearch, West Grove, PA), and detection was performed using the ECL system (Pierce Biotechnology, Rockford, IL).
Radiolabeling and immunoprecipitation
Thymocytes (25 × 106) were incubated in 700 microliters of RMPI 1640 without methionine (Sigma-Aldrich, Saint Louis, MO) at 37°C and 5% CO2 for 2.5 hours and then 500 uCi L-[35S]-Methionine L-[35S]-Cysteine mix (EasyTag express protein labeling mix, Perkin Elmer, Boston, MA) was added and the incubation continued for four hours. Cells were spun down, washed with PBS, and lysed on ice by addition of 700 microliters of ice-cold lysis buffer (Tris-buffered saline pH 8.0 1% (w/v) deoxycholic acid 0.5% (v/v) Nonidet P-40) with protease inhibitors (complete protease inhibitor cocktail tablets, Roche, Indianapolis, IN). The resulting lysates were cleared by centrifugation through a 0.22 micrometer pore cellulose acetate membrane (SpinX, Corning, Corning, NY).
For immunoprecipitation, protein A-conjugated magnetic beads (15 microliters/sample Dynabeads protein A, Invitrogen, Carlsbad, CA) were washed with lysis buffer (Tris-buffered saline pH 8.0 1% (w/v) deoxycholic acid 0.5% (v/v) Nonidet P-40) and incubated 2 hours at 4°C with rabbit anti-H-2Kb exon 8 (2 microliters/sample, a gift from Jonathan. Yewdell, NIH, Bethesda, MD) or rabbit anti-beta-2 microglobulin (1 microliter/sample, Dako, Carpinteria, CA) and then added to radiolabeled cell lysates. One third of each cell lysate volume was incubated with fifteen microliters of antibody-beads overnight at 4°C with shaking. The beads were then washed three times with lysis buffer using a magnet (DynaMag-2, Invitrogen, Carlsbad, CA), resuspended in 50 microliters/sample of sample buffer and reducing agent (XT sample buffer and reducing agent, Bio-Rad, Hercules, CA) and heated to 90°C for 10 minutes. 30 microliters/sample were run in a 12% 30/0.8 acrylamide/bisacrylamide (Duracryl, Proteomic solutions, Saint Marcel, France) 360 mM bis-Tris (Calbiochem, EMD Chemicals, Gibbstown, NJ) gel. The running buffer was 50 mM MOPS 50 mM Tris 1 mM EDTA 0.1% (w/v) SDS 5 mM sodium bisulfite. The gel was washed with distilled water, incubated for 1 hour in scintillation phosphor solution (Autofluor, National Diagnostics, Atlanta, GA), dried under vacuum, and autoradiography film was exposed to it at −80°C for one to five days. The films were developed and scanned, and the band intensities were quantified using ImageJ software (NIH, Bethesda, MD).
Results
TPPII gene-trapped mice are viable
Mice with a gene trap disrupting TPPII were obtained from Lexicon Pharmaceuticals. In the gene-trapped allele, the gene-trap cassette (containing a splice acceptor and a poly-A signal) was inserted between exons 2 and 3, leading to expression of a truncated TPPII (98 out of 1262 amino acids) and impairment of expression of full-length TPPII (Fig. 1A). In order to evaluate the reduction in TPPII, TPPII mRNA levels were analyzed in tissues and cells by real-time quantitative PCR. TPPII mRNA levels derived in spleens, kidneys and MEFs from gene-trapped mice were reduced down to 3.2%, 7.5% and 2.9% of wild-type mice, respectively. We also analyzed levels of TPPII mRNA by amplifying the active site region in exon 11 and found that mRNA levels in the gene-trapped MEFs were 4.9% of those in wild-type MEFs. These results indicate that the TPPII mRNA level is severely reduced (~95% reduction) in the gene-trapped mice; the residual mRNA level is possibly due to alternative splicing. To evaluate the effects of the reduction in mRNA levels on TPPII protein levels, we analyzed spleen, liver, heart, and kidney tissue from wild type and gene-trapped mice by semi-quantitative western blotting. TPPII protein was present in wild type mice but was undetectable in the gene-trapped animals, demonstrating that TPPII protein levels are reduced by at least 87.5% (Fig. 1B). These analyses indicated that the TPPII-gene trapped mice are markedly deficient in TPPII.
Mice with a gene-trap disrupting TPPII (Fig. 1A) were viable and fertile, and were normal in appearance and behavior. In a previous description of TPPII knockout mice, splenomegally due to extramedullary hematopoiesis and evidence of chronic inflammation were observed (34). However, in our TPPII-deficient gene-trapped mice, we did not observe any evidence of extramedullary hematopoiesis or inflammation, including splenomegaly, increased splenic myeloid cells (GR1+) cells (Fig. 2), or abnormal histology (data not shown) or evidence of systemic cytokine effects (described below).
Figure 2. TPPII gene-trapped mice do not show extramedullary hematopoiesis.
The bars represent averages with standard deviations (n=3). (A) Spleens of 5- to 6-months-old BL6 and TPPII gene-trapped mice. (B) Splenocytes were stained with anti-Gr1 antibody to identify splenic granulocytes. (C) Comparison of spleen weights of whole body weights. (D) Comparison of number of splenocytes.
Surface MHC class I levels are increased on a subset of TPPII-deficient cells
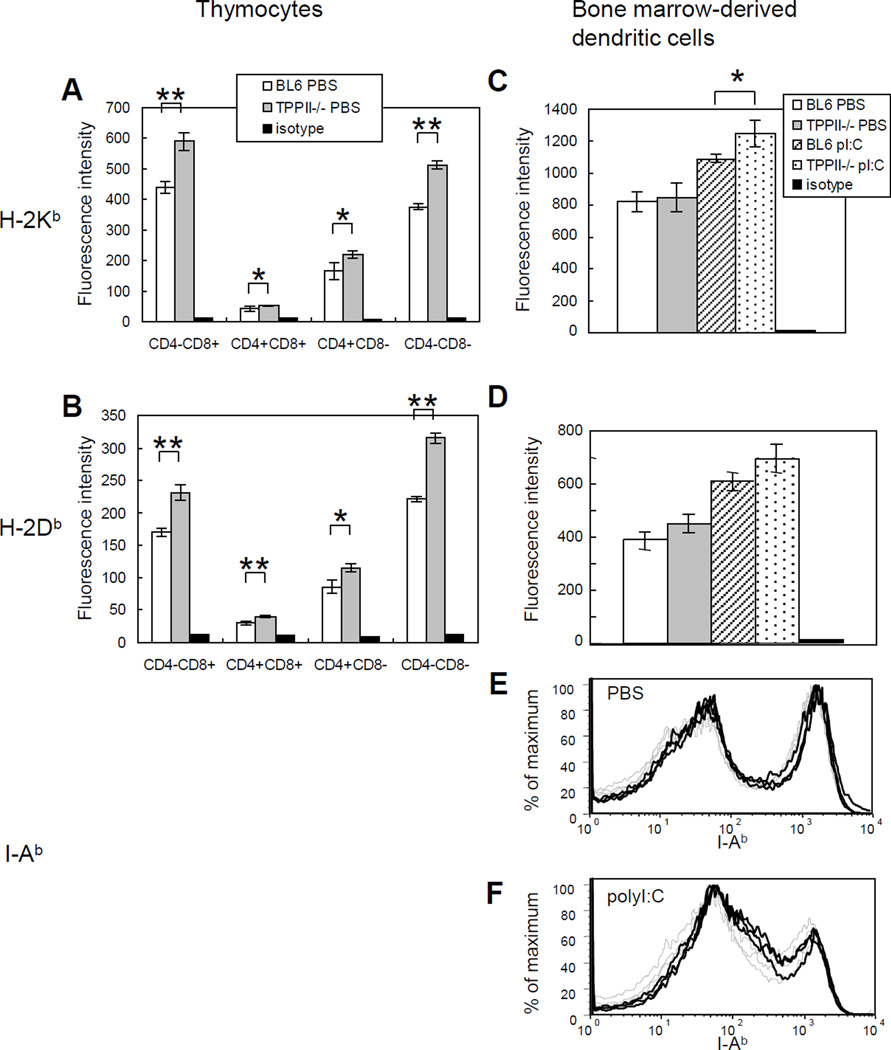
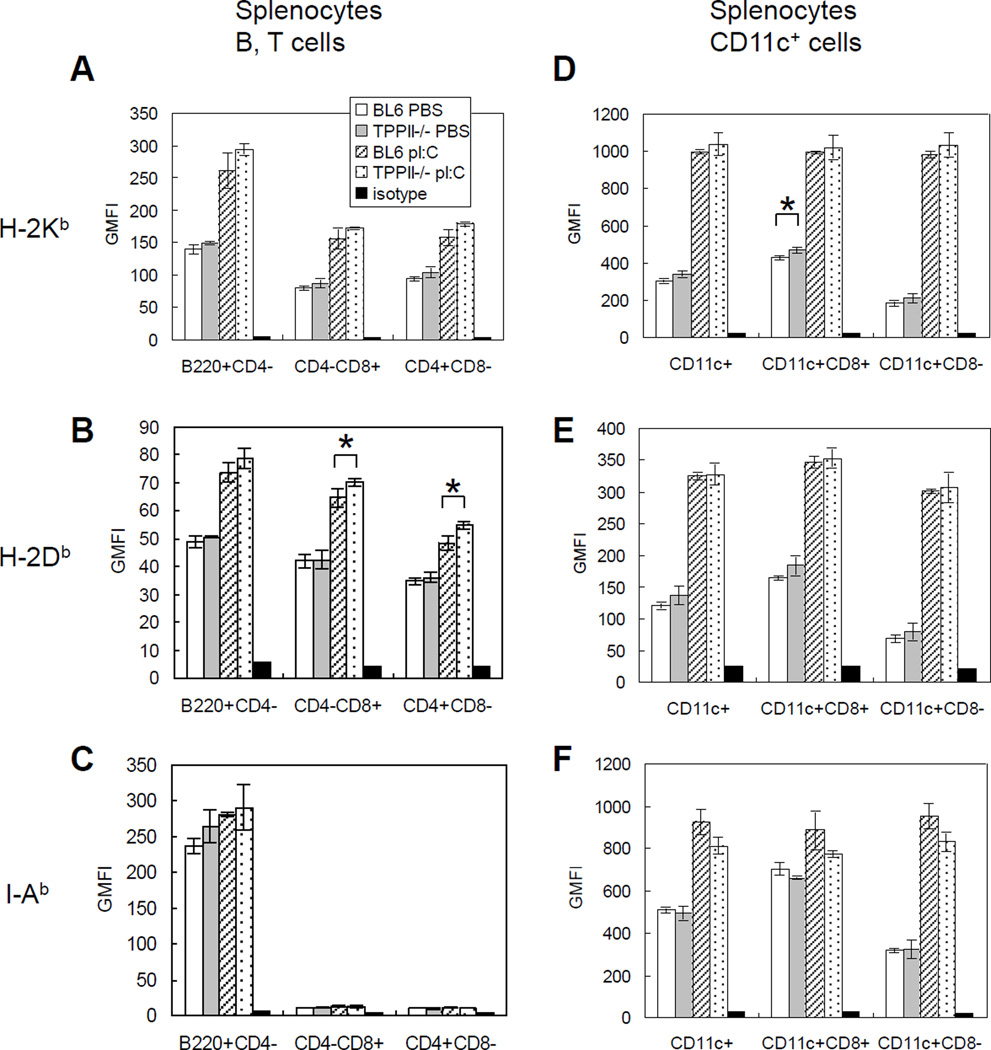
In order for MHC class I molecules to be transported to the cell surface, they must first bind a peptide in the ER. Therefore, surface expression of MHC class I molecules is an indirect measure of overall peptide supply. Elimination of TPPII might either reduce the peptide supply to MHC class I if TPPII contributes to the degradation of MHC class I-binding peptides, or increase peptide supply if TPPII helps produce such peptides. We therefore measured MHC class I surface expression on splenic B and T cells, CD11c+ splenic dendritic cells and thymocytes as well as on bone marrow-derived dendritic cells. The most striking finding in this analysis was an increase in MHC class I levels on thymocytes. H-2Kb and H-2Db on all thymic subsets from TPPII gene-trapped mice were 20–30% higher than those on thymocytes from wild-type C57BL/6 mice (Fig. 3A, 3B). There was also a trend for increased levels of MHC class I molecules on dendritic cells (Fig. 3C, 3D) and splenic lymphocytes (Fig. 4A, 4B, 4D, 4E) from TPPII-deficient mice, but in most cases this was not statistically significant. When mice were treated with the type I interferon inducer poly I:C, MHC class I levels increased on peripheral lymphocytes from TPPII-deficient versus wild type mice, but were not significantly different between the strains. I-Ab was similarly expressed on dendritic cells and splenic B cells from TPPII gene-trapped mice compared to wild-type animals (Fig. 3E, Fig. 4C, 4F).
Figure 3. TPPII gene-trapped and wild-type thymocytes show differences in MHC class I, but not in MHC class II, levels.
The mice were injected intraperitoneally with PBS. After 24 h, thymocytes were stained with appropriate antibodies and analyzed by FACS. Bone marrow cells were also harvested and cultured for 6 days, followed by addition of PBS or poly I:C (pI:C) and staining with appropriate antibodies on day 7 and analysis by FACS. Expression levels of H-2Kb ((A) and (C)), H-2Db ((B) and (D)) expressed as geometric mean fluorescence intensity. The Student t test was used to determine statistical significance (*, 0.01<p<0.05; **, p<0.01). Error bars represent standard deviations (n=3). Data are representative of three independent experiments. Flow cytometry traces of bone marrow-derived dendritic cells (gated on CD11chigh cells) stained for I-Ab after 6 days of culture, followed by 24 h of treatment with (E) PBS or (F) poly I:C. Bone marrow-derived dendritic cells from wild-type mice are indicated by dashed light traces. Bone marrow-derived dendritic cells from TPPII gene-trapped mice are indicated by solid dark traces.
Figure 4. TPPII gene-trapped and wild-type splenocytes show differences in MHC class I, but not in MHC class II, levels.
The mice were injected intraperitoneally with PBS or poly I:C (pI:C). After 24 h, splenocytes were stained with appropriate antibodies and analyzed by FACS. Each graph shows geometric mean fluorescence intensity (GMFI), which represents the expression levels of H-2Kb ((A) and (D)), H-2Db ((B) and (E)) or I-Ab ((C) and (F)). The Student t test was used to determine statistical significance (*, 0.01<p<0.05). Error bars represent standard deviations (n=3). Data are representative of three independent experiments.
TPPII gene-trapped mice respond normally to viral infection
To test the contribution of TPPII to immune responses to a viral infection, we examined the CTL responses to lymphocytic choriomeningitis virus (LCMV). C57BL/6 and TPPII gene-trapped mice were infected with LCMV. At the peak of the response on day 8 after infection, splenic lymphocytes were isolated, stimulated in vitro with individual LCMV epitopes, and stained for intracellular IFN-γ levels. The frequency of CTL producing IFN-γ in response to four LCMV epitopes in TPPII gene-trapped mice was not significantly different from those in C57BL/6 mice, suggesting that any contribution of TPPII to creating or destroying these presented peptides is not sufficient to affect immune responses in vivo (Fig. 5).
Figure 5. TPPII gene-trapped and wild-type mice respond similarly to LCMV infection in vivo.
Splenocytes from TPPII−/− and BL6 mice infected with lymphocytic choriomeningitis virus (LCMV) were harvested on day 8, and stimulated in vitro with peptides corresponding to LCMV epitopes for 5 h. Cells were then stained for intracellular interferon and analyzed by flow cytometry by gating on CD8+CD44+ cells. The graph represents average percentages of IFN-γ-positive cells with standard deviations as error bars (n=6). There was no significant difference between TPPII−/− and BL6 mice in their response to any of the four epitopes tested.
TPPII knockdown affects the trimming of long peptide precursors in some cell types
In our previous study using HeLa-Kb cells, we demonstrated that siRNA-mediated silencing of TPPII inhibited by about 50% the trimming of long peptide precursors (from 14 to 17 amino acids long) to generate MHC class I-presented peptides (21). To examine whether TPPII similarly contributes to trimming of long precursors in murine cells, we isolated multiple independent MEF lines from the progeny of TPPII+/− mouse crosses, thus generating homozygous gene-trapped, heterozygous, and wild-type MEFs. The homozygous TPPII gene-trapped and wild-type lines were indistinguishable in terms of their morphology and growth characteristics for at least 25 passages.
Since TPPII is a cytosolic peptidase, we examined the ability of the gene-trapped MEFs to generate presented peptides from precursors expressed in the cytosol. For this experiment, several independent MEF lines were transfected with plasmids expressing SIINFEKL (S8L) as ubiquitin fusion proteins, either as the mature epitope (SIINFEKL) or with N-terminal extensions of varying lengths as summarized in Table I. Cleavage by ubiquitin C-terminal hydrolases, which reside in the cytosol, releases the peptide of interest without an initiating methionine, thus generating peptides similar to those generated by the proteasome. Subsequent trimming of the N-terminal residues releases SIINFEKL (S8L), which if presented on H-2Kb, can be detected by staining with the monoclonal antibody 25.D1.16.
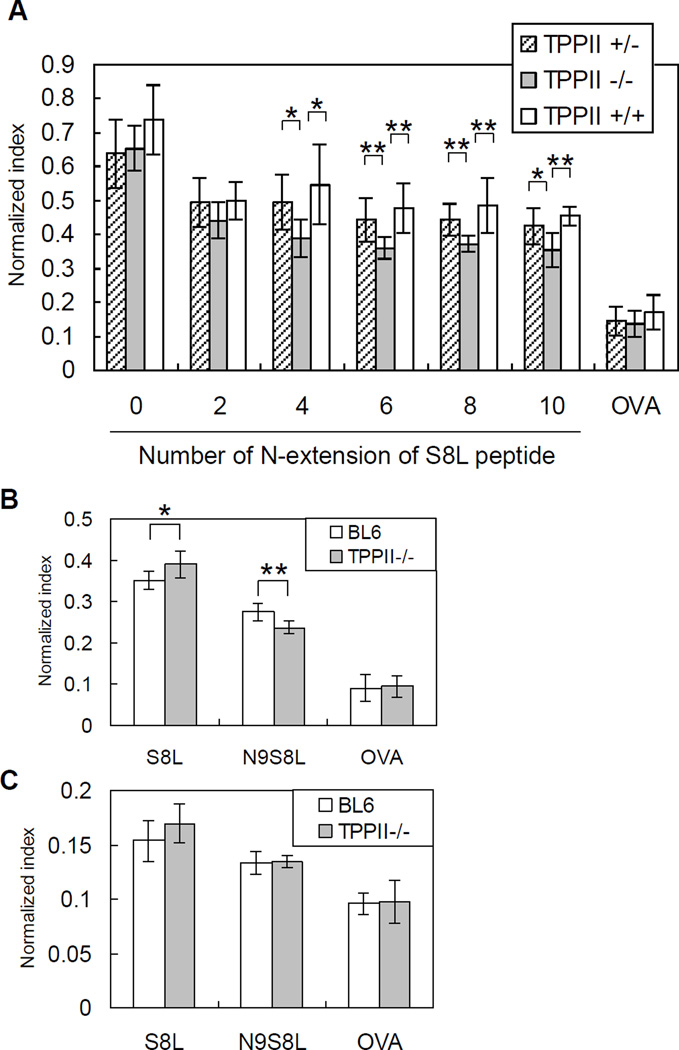
MEFs were transfected with various constructs and analyzed by flow cytometry by gating on cell populations expressing comparable amounts of GFP. Because absolute levels of surface H-2Kb were variable between different independent MEF lines, peptide presentation was normalized to the H-2Kb level of each cell. Presentations of mature S8L and of S8L with a 2-amino acid N-terminal extension were not statistically different between WT, heterozygous, and KO MEFs. In contrast, presentation of S8L with a 4 to 10-amino acid N-terminal extension was significantly reduced in the KO MEFs, compared to WT and heterozygous MEFs (Fig. 6A). These results are similar to (though less marked than) those previously observed in human HeLa cells (21) and indicate that TPPII contributes to the trimming of very long N-terminal extensions in some primary murine cells.
Figure 6. TPPII gene-trapped and wild-type embryonic fibroblasts, but not cultured dendritic cells, respond differently to N-extended epitope precursors.
(A) TPPII−/−, TPPII+/− or TPPII+/+ mouse embryonic fibroblasts (MEFs) were transfected with plasmids expressing, in the cytosol, the ovalbumin H-2Kb-restricted epitope SIINFEKL”S8L”) extended at the N-terminus by 0, 2, 4, 6, 8, or 10 amino acids as ubiquitin fusion constructs, or full-length ovalbumin (OVA), and co-expressing GFP on the same mRNA. After 24 h the MEFs were stained with 25.D1.16 antibody. Cells were gated on similar levels of GFP expression. Data are normalized to expression level of H-2Kb in each MEF line. The Student t test was used to determine statistical significance (*, 0.01<p<0.05; **, p<0.01). Error bars represent standard deviations (n=9 for TPPII+/−, n=7 for TPPII−/−, n=4 for TPPII+/+). (B) and (C), TPPII−/− or BL6 bone marrow-derived dendritic cells were transduced with recombinant lentiviral vectors encoding S8L or N9S8L as ubiquitin fusion constructs, or full-length OVA, and then stained with 25.D1.16 antibody at 5 days post-transduction. Cells were gated on (B) high or (C) low levels of GFP expression. Data are normalized by expression level of H-2Kb in each dendritic cell line. The Student t test was used to determine statistical significance. Error bars represent standard deviations (n=6).
To further test the role of TPPII in trimming N-extended peptides in another primary cell type, we transduced bone marrow-derived dendritic cells with lentiviruses expressing various S8L precursors. Lentivirus transduction does not affect DC maturation, or antigen presentation (35). We constructed recombinant lentiviral vectors encoding S8L as an ubiquitin fusion protein, S8L with a 9-residue N-terminal extension as a ubiquitin fusion protein (N9-S8L), ubiquitin alone (as a control), or full-length OVA. Bone marrow-derived DC progenitors from WT or KO mice were transduced with each lentivirus on day 2, and stained with H-2Kb-S8L-specific antibody 25.D1.16 on day 7 by gating on GFP+ CD11c+ cells. These cells presented the full length OVA and S8L similarly. However, TPPII-deficient dendritic cells also showed reduced presentation compared with wild type cells of the N9-S8L construct when it was expressed at high levels (GFPhigh cells) (Fig. 6B). Interestingly, the difference in N9-S8L presentation by WT and KO cells was not seen at more limiting antigen concentrations (GFPlow cells) (Fig 6C), presumably because the alternate trimming mechanisms for long peptides only become limiting at the higher antigen dose.
Analysis of the role of TPPII in trimming long precursors in vivo
Since TPPII has some effect on the trimming of long precursor peptides in some cell types, we sought to examine its role in this process in animals. He et al reported that subcutaneous injection of lentivirus resulted in direct transduction of skin-derived DCs and potent and prolonged antigen presentation (36). We injected recombinant lentiviruses into footpads of TPPII gene-trapped mice and C57BL/6 mice as described (36, 37). We used recombinant lentivirus harboring genes for the various S8L-derived peptides described above and full-length OVA. In this system the magnitude of the CD8 T cell response was dependent on the amount of virus injected, and therefore we used limiting doses of viruses so as to be on a sensitive portion of the dose response curve. On day 8 after infection, lymphocytes from the draining popliteal lymph node were stimulated in vitro with the S8L peptide, followed by staining for intracellular IFN-γ levels. The frequency of IFN-γ-positive CTLs to S8L and to the N-extended S8L precursor was not different between WT and TPPII-deficient mice (Fig. 7A), even at limiting doses of the virus (where responses were not maximal).
Figure 7. TPPII gene-trapped and wild-type mice respond similarly to N-extended epitope precursors in vivo.
(A) Mice were injected in the footpad with recombinant lentivirus encoding full-length OVA, ubiquitin alone (-), ubiquitin fused with SIINFEKL (S8L), or ubiquitin fused with S8L extended by 9 amino acids at the N terminus (N9S8L). (B) Mice were injected in the footpad with recombinant vaccinia virus encoding the same constructs. (C) Mice were injected i.p. with 1.25 × 106 pfu/mouse of recombinant vaccinia virus encoding the same constructs. Eight days later (A) or seven days later (B, C), lymphocytes from a draining lymph node (A) or splenocytes (B and C) were harvested and stimulated for 5 h with 5 µM of the appropriate epitope. Cells were then stained for intracellular IFN-γ and analyzed by flow cytometry by gating on CD8+CD44+ cells. The graphs represent average percentages of IFN-γ-positive cells with standard deviations as error bars (n=5). Data are representative of two independent experiments. Student’s t test was used to determine statistical significance (*, 0.01<p<0.05). The difference in response to i.p. vaccinia-N9S8L seen between TPPII knockout and wild-type mice in (C) (p=0.03) was not seen in the repeat experiment (p=0.59).
We next immunized mice with recombinant vaccinia viral vectors encoding the same set of genes as recombinant lentiviral vectors described above. We infected wild-type and gene-trapped mice either by footpad injection or i.p. injection with the various recombinant vaccinia viruses. On day 7, splenocytes were similarly stimulated in vitro with the S8L peptide, followed by staining for intracellular IFN-γ levels. No consistent, statistically significant difference in response to any of the constructs was seen, although a statistically significant decrease of IFN-γ-positive CTLs to N9S8L was observed in one of two i.p-injected experiments (Fig.7B and 7C).
Discussion
Immune surveillance for virally infected cells and tumors is dependent on cells processing their proteins into fragments and displaying a fraction of these peptides on MHC class I molecules on the cell surface. The magnitude of the CTL response to a particular peptide and the immunodominance hierarchy of responses are at least partly dependent on the numbers of MHC class I-peptide complexes at the cell surface. This in turn is influenced by several factors, such as the affinity of binding between the peptide and MHC class I allele, the number of protein precursors, and the frequency of which the peptide is generated or destroyed by the antigen processing pathway. In order to understand and predict the specificity of responses, therefore, it is important to identify proteases that generate or destroy peptides that bind, or could bind, MHC class I molecules.
Most intracellular proteins are degraded by proteasomes, and proteasomes are essential for generating most MHC class I-presented epitopes (38). Experiments with model peptides and proteasome inhibitors have shown that the proteasome is generally the only activity in cells that can make the proper cleavage to generate the C-terminus of an MHC class I epitope (39–41). In contrast, peptides that have the correct C-terminal residue for binding MHC class I, but that are too long at the N-terminus, can be trimmed by aminopeptidases into mature epitopes. Epitopes can also be destroyed if aminopeptidases or endopeptidases trim them below the minimum size needed for MHC class I binding (6). Therefore, several groups have pursued the question of whether trimming by aminopeptidases is normally important in antigen presentation, and if so which aminopeptidases contribute to this process (16, 42–44).
A number of aminopeptidases have been proposed to play a role in MHC class I antigen presentation, based on biochemical observations, experiments in tissue culture, or knockout mice. ER aminopeptidase 1 (ERAP1) has been shown to play an important role trimming peptides in the ER for antigen presentation and in the generation of CD8 T cell responses in mice in ways that increase or decrease responses to certain peptides (9–14). There is also strong evidence that peptide trimming by aminopeptidases in the cytosol contributes to antigen presentation (10, 39, 42, 43, 45). Although biochemical experiments with various aminopeptidases (including leucine aminopeptidase, bleomycin hydrolase, and puromycin-sensitive aminopeptidase) suggested they may have important functions in antigen processing (42, 43), mice lacking these aminopeptidases have shown at most very limited effects on antigen presentation and immune responses (25, 46, 47). It is therefore important to fully understand the contribution of various peptidases to antigen presentation it is important to generate and analyze deficient mice.
Tripeptidyl peptidase II (TPPII) is an abundant intracellular peptidase that cleaves triplets of amino acids from the N-terminus of peptides and has been reported to also act as an endoprotease (19). The physiological functions of TPPII have been unclear. Early studies suggested that TPPII might be able to compensate for the loss of proteasomes (48), although subsequent experiments did not support this possibility (49). In cell extracts, TPPII has been shown to help generate a few MHC class I–presented peptides and studies with inhibitors have suggested a similar role in vivo (16, 19, 48).
A recent paper suggested that in fact TPPII may be essential for almost all MHC class I antigen presentation, and proposed a model in which proteasomes in intact cells normally produce only long peptides (more than 16 amino acids long) and TPPII was needed to trim these long precursors; in this model TPPII was an essential bridge between proteasome-generated peptides, and degradation to amino acids by conventional peptidases (16). We have previously tested this model in cultured human cells by using siRNA to knock down TPPII levels (21). We found that, while TPPII was indeed important (though not essential) for converting long peptides to shorter forms, TPPII was not required for MHC class I antigen presentation in cultured cells: presentation of peptides from full-length proteins required proteasomes, but TPPII knockdown did not reduce (and if anything slightly increased) MHC class I peptide production. We tentatively concluded that relatively few very long precursor peptides are normally generated in vivo and therefore TPPII is not required for most antigen presentation. However, the possibility remained that TPPII is required for a natural immune response in intact animals (for example, if the HeLa cells we tested are not representative of normal cells, or if the initiation of an immune response has different requirements for antigen presentation).
Mice containing a gene-trap between exons 2 and 3 have a >90% reduction in the expression of TPPII mRNA and at least >87.5% reduction in the expression of protein. They are viable, fertile, and are grossly normal in appearance and behavior; in particular, and in contrast to observations in TPPII knockout mice (34), the gene-trapped TPPII-deficient mice did not show extramedullary hematopoiesis or splenitis. These differences may be because of differences in housing or in mouse background, or may be because of very low (though undetectable in our assays) levels of functional TPPII in our mice. The lack of chronic inflammation in these gene-trapped mice offers the ability to compare antigen presentation in the presence and near-absence of TPPII, without the confounding effects of inflammation on MHC class I antigen presentation.
Some cells (particularly thymocytes) from TPPII gene-trapped mice express a higher level of surface MHC class I than do those from C57BL/6 mice (Fig. 2). In contrast, the synthesis of heavy chains (measured by immunoprecipitation of metabolically labeled H-2Kb) and light chains (ß2-microglobulin) was not significantly different in thymocytes from wild type versus TPPII-deficient mice (data not shown). Since MHC class I can only reach the cell surface when it is bound to an appropriate peptide, this suggests that the overall supply of presented peptides may be reduced by TPPII. In other words, the net effect of TPPII may be to destroy more peptides than it helps produce. It is possible that residual TPPII in the gene trap mice is contributing some function and a more pronounced phenotype would be observed in the complete absence of this peptidase. However, our findings are similar to observations with TPPII knock-out mice (24).
In previous studies, the most important contribution of TPPII to antigen presentation was its ability to trim very long precursor peptides (16, 21). However, this activity has only been examined in cultured human tumor cells, and not in primary cells or cells of murine origin. Therefore we made minigene constructs to test the trimming of full length protein, short and long antigenic precursors, and mature epitope in primary cells from gene-trapped mice. For these studies, we produced embryonic fibroblast cell lines (MEFs), transfected these cells with plasmids expressing various precursors of the H-2Kb-binding peptide SIINFEKL, and quantified presentation of H-2Kb-SIINFEKL as a measure of antigen processing. Presentation of SIINFEKL from short precursors tested was similar in wild-type, heterozygous and homozygous gene-trapped cell lines, while presentation of SIINFEKL from precursors with 4 to 10-amino acid N-terminal extensions was significantly reduced by TPPII-deficiency. A similar defect in processing a long precursor (N9-S8L) was observed in TPPII-deficient bone marrow-derived dendritic cells under conditions of high antigen expression. These results are consistent with our previous findings with HeLa-Kb cells (21), although the extent of reduction in the MEFs was less marked than in the previous experiments, perhaps because of the variability between independent MEF cell lines, different species (human vs. mouse), expression level of TPPII, and/or presence of other aminopeptidases in the different cell types.
We also tested the effect of TPPII knockdown on the generation of an authentic antiviral immune response to viral proteins and antigenic precursors. We infected TPPII gene-trapped mice and C57BL/6 mice with lymphocytic choriomeningitis virus (LCMV), and after 8 days we analyzed the frequency of CD8+ T cell responses to four LCMV MHC class I-restricted epitopes. Consistent with the recent findings (23, 24), we found no difference in the responses to any of the epitopes (Fig. 5). In addition, we specifically tested the importance of TPPII on generation of MHC class I epitopes from precursors with long N-terminal extensions, using recombinant lentiviruses and vaccinia viruses expressing ubiquitin-fused minigenes. TPPII-gene-trapped mice generated similar levels of CTL to these precursors as did wild-type mice, demonstrating that TPPII is not essential for generating immune responses to long N-extended precursors in vivo. We presume that this is because the reduction in presentation we observe in TPPII-deficient dendritic cells is too small to affect responses in vivo. Alternatively, since we only see a defect in presentation at high antigen concentrations it is possible that such high amounts of antigen expression are not obtained in vivo. Finally, it is also possible that the variability in responses between individual mice could obscure the detection of small differences.
Taken together, we conclude that TPPII is not required for MHC class I antigen presentation in vivo. This conclusion is consistent with recent papers using cultured cells (23) or TPPII knock-out mice (24) and adds to the weight of evidence against the model proposed by Reits et al (16). This could in principle be because TPPII is not required for trimming precursors with long N-terminal extensions, and/or because such precursors are not generated very often in cells.
A substantial amount of trimming of antigen precursor peptides occurs in the cytosol (10). Although a number of cytosolic aminopeptidase (LAP, BH, PSA and TPPII) can trim such precursors in cell extracts, their elimination from cells and in knockout mice does not inhibit peptide trimming in vivo ((25, 46, 47) and this paper). This raises the possibility that there may be other cytosolic aminopeptidases that are important to antigen presentation and remaining to be identified. Alternatively since the known cytoplasmic aminopeptidases generally have broad substrate specificity, the enzymes’ function may be sufficiently redundant so that the lack of one peptidase could be readily compensated by other ones. Crossing of several kinds of knock-out mouse strains will help to define the functional redundancy or a unique role of aminopeptidases.
Acknowledgments
We thank Dr. Raymond Welsh for providing LCMV, and Dr. Eicke Latz for providing lentiviral vectors.
Abbreviations used in this paper
- DC
dendritic cell
- KO
knock out
- LCMV
lymphocytic choriomeningitis virus
- MEF
mouse embryonic fibroblast
- TPPII
tripeptidyl peptidase II
- WT
wild type
Footnotes
This work was supported by grants from the National Institutes of Health (to K.L.R.). Core resources supported by the Diabetes Endocrinology Research Grant DK42520 were also used. M. K. was supported by JSPS Postdoctoral Fellowships for Research Abroad.
References
- 1.Rock KL, York IA, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat. Immunol. 2004;5:670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- 2.Purcell AW, Elliott T. Molecular machinations of the MHC-I peptide loading complex. Curr. Opin. Immunol. 2008;20:75–81. doi: 10.1016/j.coi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 3.York IA, Goldberg AL, Mo XY, Rock KL. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol. Rev. 1999;172:49–66. doi: 10.1111/j.1600-065x.1999.tb01355.x. [DOI] [PubMed] [Google Scholar]
- 4.Shastri N, Schwab S, Serwold T. Producing nature's gene-chips: the generation of peptides for display by MHC class I molecules. Annu. Rev. Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 5.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 6.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 7.Momburg F, Roelse J, Hammerling GJ, Neefjes JJ. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J. Exp. Med. 1994;179:1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 9.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 10.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat. Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 11.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg AL. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 12.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc. Natl. Acad. Sci. USA. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat. Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 14.Firat E, Saveanu L, Aichele P, Staeheli P, Huai J, Gaedicke S, Nil A, Besin G, Kanzler B, van Endert P, Niedermann G. The role of endoplasmic reticulum-associated aminopeptidase 1 in immunity to infection and in cross-presentation. J. Immunol. 2007;178:2241–2248. doi: 10.4049/jimmunol.178.4.2241. [DOI] [PubMed] [Google Scholar]
- 15.Levy F, Burri L, Morel S, Peitrequin AL, Levy N, Bachi A, Hellman U, Van den Eynde BJ, Servis C. The final N-terminal trimming of a subaminoterminal proline-containing HLA class I-restricted antigenic peptide in the cytosol is mediated by two peptidases. J. Immunol. 2002;169:4161–4171. doi: 10.4049/jimmunol.169.8.4161. [DOI] [PubMed] [Google Scholar]
- 16.Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout J, Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- 17.Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G. A giant protease with potential to substitute for some functions of the proteasome. Science. 1999;283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 18.Tomkinson B, Lindas A. Tripeptidyl-peptidase II: A multi-purpose peptidase. Int. J. Biochem. Cell Biol. 2005;37:1933–1937. doi: 10.1016/j.biocel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Seifert U, Maranon C, Shmueli A, Desoutter J, Wesoloski L, Janek K, Henklein P, Diescher S, Andrieu M, de la Salle H, Weinschenk T, Schild H, Laderach D, Galy A, Haas G, Kloetzel P, Reiss Y, Hosmalin A. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat. Immunol. 2003;4:375–379. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 20.Wherry EJ, Golovina TN, Morrison SE, Sinnathamby G, McElhaugh MJ, Shockey DC, Eisenlohr LC. Re-evaluating the generation of a "proteasome-independent" MHC class I-restricted CD8 T cell epitope. J. Immunol. 2006;176:2249–2261. doi: 10.4049/jimmunol.176.4.2249. [DOI] [PubMed] [Google Scholar]
- 21.York I, Bhutani N, Zendzian S, Goldberg A, Rock K. Tripeptidyl peptidase II is the major peptidase needed to trim long antigenic precursors, but is not required for most MHC class I antigen presentation. J. Immunol. 2006;177:1434–1443. doi: 10.4049/jimmunol.177.3.1434. [DOI] [PubMed] [Google Scholar]
- 22.Guil S, Rodriguez-Castro M, Aguilar F, Villasevil EM, Anton LC, Del Val M. Need for tripeptidyl-peptidase II in major histocompatibility complex class I viral antigen processing when proteasomes are detrimental. J. Biol. Chem. 2006;281:39925–39934. doi: 10.1074/jbc.M608522200. [DOI] [PubMed] [Google Scholar]
- 23.Basler M, Groettrup M. No essential role for tripeptidyl peptidase II for the processing of LCMV-derived T cell epitopes. Eur. J. Immunol. 2007;37:896–904. doi: 10.1002/eji.200636372. [DOI] [PubMed] [Google Scholar]
- 24.Firat E, Huai J, Saveanu L, Gaedicke S, Aichele P, Eichmann K, van Endert P, Niedermann G. Analysis of direct and cross-presentation of antigens in TPPII knockout mice. J. Immunol. 2007;179:8137–8145. doi: 10.4049/jimmunol.179.12.8137. [DOI] [PubMed] [Google Scholar]
- 25.Towne CF, York IA, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J. Immunol. 2008;180:1704–1712. doi: 10.4049/jimmunol.180.3.1704. [DOI] [PubMed] [Google Scholar]
- 26.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 28.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 29.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat. Protocols. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 30.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 31.Albert F, Boyer C, Leserman LD, Schmitt-Verhulst AM. Immunopurification and insertion into liposomes of native and mutant H-2Kb: quantification by solid phase radioimmunoassay. Mol. Immunol. 1983;20:655–667. doi: 10.1016/0161-5890(83)90010-x. [DOI] [PubMed] [Google Scholar]
- 32.Ozato K, Hansen TH, Sachs DH. Monoclonal antibodies to mouse MHC antigens. II. Antibodies to the H-2Ld antigen, the products of a third polymorphic locus of the mouse major histocompatibility complex. J. Immunol. 1980;125:2473–2477. [PubMed] [Google Scholar]
- 33.Staudt LM, Gerhard W. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. J. Exp. Med. 1983;157:687–704. doi: 10.1084/jem.157.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huai J, Firat E, Nil A, Million D, Gaedicke S, Kanzler B, Freudenberg M, van Endert P, Kohler G, Pahl H, Aichelel P, Eichmann K, Niedermann G. Activation of cellular death programs associated with immunosenescence-like phenotype in TPPII knockout mice. Proc. Natl. Acad. Sci. USA. 2008;105:5177–5182. doi: 10.1073/pnas.0801413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y, Zhang J, Mi Z, Robbins P, Falo LD., Jr Immunization with lentiviral vector-transduced dendritic cells induces strong and long-lasting T cell responses and therapeutic immunity. J. Immunol. 2005;174:3808–3817. doi: 10.4049/jimmunol.174.6.3808. [DOI] [PubMed] [Google Scholar]
- 36.He Y, Zhang J, Donahue C, Falo LD., Jr Skin-derived dendritic cells induce potent CD8(+) T cell immunity in recombinant lentivector-mediated genetic immunization. Immunity. 2006;24:643–656. doi: 10.1016/j.immuni.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, Majumder N, Lin H, Watkins S, Falo LD, Jr, You Z. Induction of therapeutic antitumor immunity by in vivo administration of a lentiviral vaccine. Hum. Gene Ther. 2005;16:1255–1266. doi: 10.1089/hum.2005.16.1255. [DOI] [PubMed] [Google Scholar]
- 38.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 39.Craiu A, Akopian T, Goldberg A, Rock KL. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc. Natl. Acad. Sci. USA. 1997;94:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mo XY, Cascio P, Lemerise K, Goldberg AL, Rock K. Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J. Immunol. 1999;163:5851–5859. [PubMed] [Google Scholar]
- 41.Stoltze L, Dick TP, Deeg M, Pommerl B, Rammensee HG, Schild H. Generation of the vesicular stomatitis virus nucleoprotein cytotoxic T lymphocyte epitope requires proteasome-dependent and -independent proteolytic activities. Eur. J. Immunol. 1998;28:4029–4036. doi: 10.1002/(SICI)1521-4141(199812)28:12<4029::AID-IMMU4029>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 42.Beninga J, Rock KL, Goldberg AL. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 43.Stoltze L, Schirle M, Schwarz G, Schroter C, Thompson MW, Hersh LB, Kalbacher H, Stevanovic S, Rammensee HG, Schild H. Two new proteases in the MHC class I processing pathway. Nat. Immunol. 2000;1:413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 44.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 45.Del-Val M, Lopez D. Multiple proteases process viral antigens for presentation by MHC class I molecules to CD8(+) T lymphocytes. Mol. Immunol. 2002;39:235–247. doi: 10.1016/s0161-5890(02)00104-9. [DOI] [PubMed] [Google Scholar]
- 46.Towne CF, York IA, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Leucine aminopeptidase is not essential for trimming peptides in the cytosol or generating epitopes for MHC class I antigen presentation. J. Immunol. 2005;175:6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 47.Towne CF, York IA, Watkin LB, Lazo JS, Rock KL. Analysis of the role of bleomycin hydrolase in antigen presentation and the generation of CD8 T cell responses. J. Immunol. 2007;178:6923–6930. doi: 10.4049/jimmunol.178.11.6923. [DOI] [PubMed] [Google Scholar]
- 48.Wang E, Kessler B, Borodovsky A, Cravatt B, Bogyo M, Ploegh H, Glas R. Integration of the ubiquitin-proteasome pathway with a cytosolic oligopeptidase activity. Proc. Natl. Acad. Sci. USA. 2000;97:9990–9995. doi: 10.1073/pnas.180328897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Princiotta MF, Schubert U, Chen W, Bennink JR, Myung J, Crews CM, Yewdell JW. Cells adapted to the proteasome inhibitor 4-hydroxy- 5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc. Natl. Acad. Sci. USA. 2001;98:513–518. doi: 10.1073/pnas.021132398. [DOI] [PMC free article] [PubMed] [Google Scholar]