Abstract
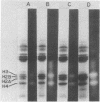
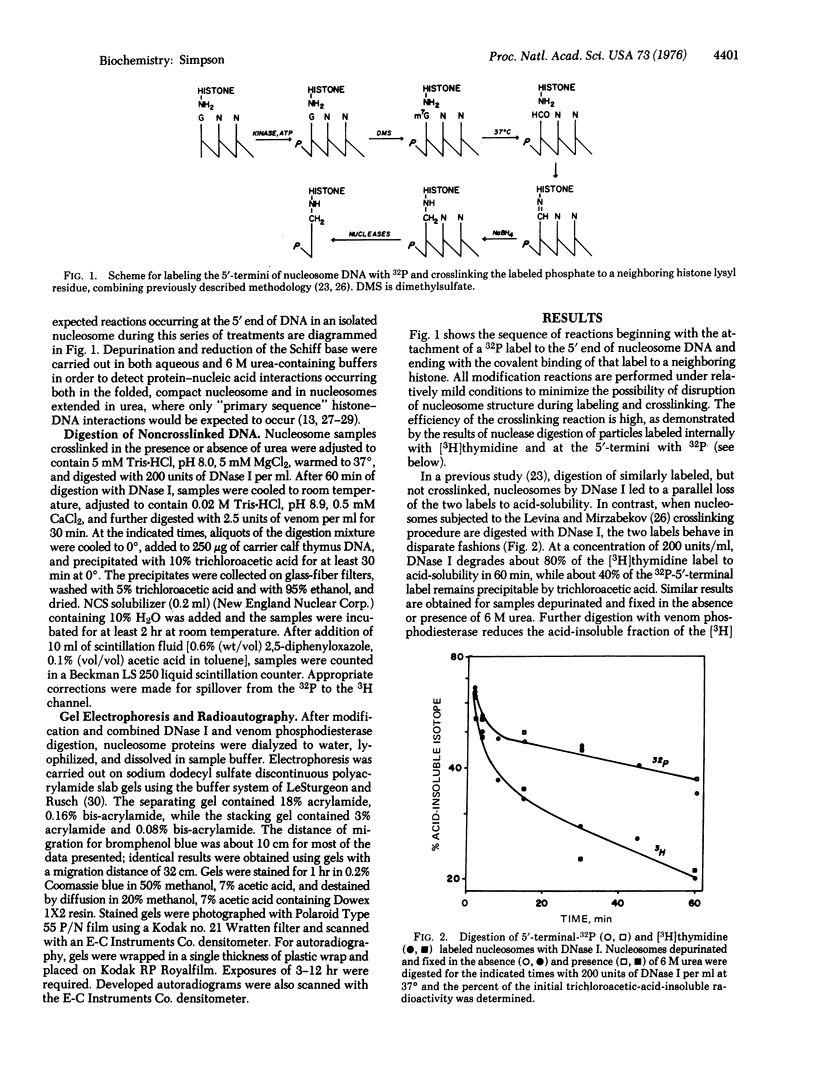
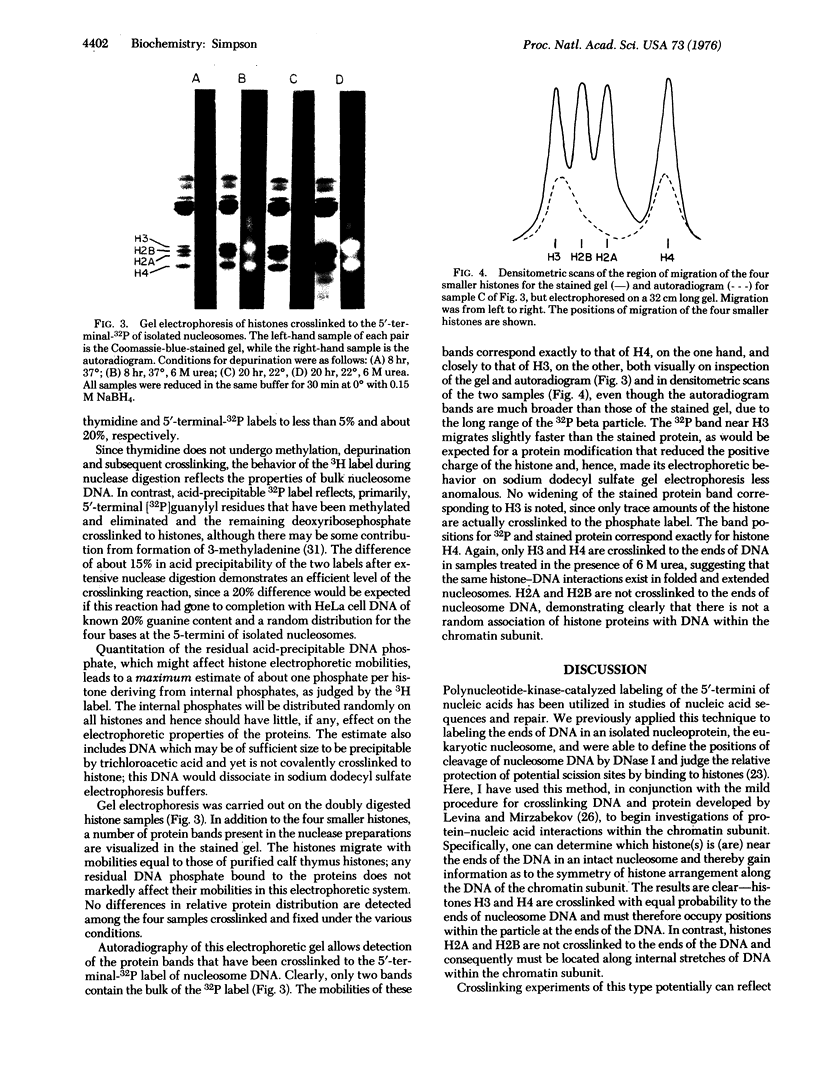
Isolated HeLa cell nucleosomes (core particles) were labeled at the 5'-termini of their DNA with 32P using [gamma-32P]ATP and polynucleotide kne by sequential methylation, depurination, Schiff base formation, and reduction with sodium borhydride. After digestion of the noncrosslinked DNA by DNase I and venom phosphodiesterase, histones were separated by gel electrophoresis and those crosslinked to the 5'-termini were identified by 32P-autoradiography. Histones H3 and H4 occur with equeal frequency as the nearest protein neighbors to the end of the DNA in nucleosomes. Histone arrangements within the core particle compatible with these results are discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Bartley J., Chalkley R. An approach to the structure of native nucleohistone. Biochemistry. 1973 Jan 30;12(3):468–474. doi: 10.1021/bi00727a017. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Pollard H. B. The presence of F3-F2a1 dimers and F1 oligomers in chromatin. Biochem Biophys Res Commun. 1975 May 5;64(1):282–288. doi: 10.1016/0006-291x(75)90250-8. [DOI] [PubMed] [Google Scholar]
- Carlson R. D., Olins A. L., Olins D. E. Urea denaturation of chromatin periodic structure. Biochemistry. 1975 Jul 15;14(14):3122–3125. doi: 10.1021/bi00685a013. [DOI] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hardison R. C., Eichner M. E., Chalkley R. An approach to histone nearest neighbours in extended chromatin. Nucleic Acids Res. 1975 Oct;2(10):1751–1770. doi: 10.1093/nar/2.10.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hyde J. E., Walker I. O. A model for chromatin sub-structure incorporating symmetry considerations of histone oligomers. Nucleic Acids Res. 1975 Mar;2(3):405–421. doi: 10.1093/nar/2.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- LeStourgeon W. M., Rusch H. P. Localization of nucleolar and chromatin residual acidic protein changes during differentiation in Physarum polycephalum. Arch Biochem Biophys. 1973 Mar;155(1):144–158. doi: 10.1016/s0003-9861(73)80017-7. [DOI] [PubMed] [Google Scholar]
- Levina E. S., Mirzabekov A. D. Kovalentnoe sviazyvanie belkov s DNK v sostave khromatina. Dokl Akad Nauk SSSR. 1975 Apr 11;(5):1222–1225. [PubMed] [Google Scholar]
- Lewis P. N. On the native structure of the histone H3-H4 complex. Biochem Biophys Res Commun. 1976 Jan 26;68(2):329–335. doi: 10.1016/0006-291x(76)91147-5. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., McCarthy B. J. Histone-histone associations within chromatin. Cross-linking studies using tetranitromethane. Biochemistry. 1975 Mar 11;14(5):1073–1078. doi: 10.1021/bi00676a030. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., Shetlar M. D., McCarthy B. J. Histone-histone interactions within chromatin. Crosslinking studies using ultraviolet light. Biochemistry. 1976 May 4;15(9):2002–2007. doi: 10.1021/bi00654a030. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Melnikova A. F. Localization of chromatin proteins within DNA grooves by methylation of chromatin with dimethyl sulphate. Mol Biol Rep. 1974 Sep;1(7):379–384. doi: 10.1007/BF00385669. [DOI] [PubMed] [Google Scholar]
- Moss T., Cary P. D., Crane-Robinson C., Bradbury E. M. Physical studies on the H3/H4 histone tetramer. Biochemistry. 1976 Jun 1;15(11):2261–2267. doi: 10.1021/bi00656a003. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Roark D. E., Geoghegan T. E., Keller G. H. A two-subunit histone complex from calf thymus. Biochem Biophys Res Commun. 1974 Jul 24;59(2):542–547. doi: 10.1016/s0006-291x(74)80014-8. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Bustin M. Histone compostion of chromatin subunits studied by immunosedimentation. Biochemistry. 1976 Sep 21;15(19):4305–4312. doi: 10.1021/bi00664a026. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Sperling R., Bustin M. Histone dimers: a fundamental unit in histone assembly. Nucleic Acids Res. 1976 May;3(5):1263–1275. doi: 10.1093/nar/3.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. Cleavable cross-links in the analysis of histone-histone associations. FEBS Lett. 1975 Oct 15;58(1):353–358. doi: 10.1016/0014-5793(75)80296-1. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Van Lente F. Dissection of chromosome structure with trypsin and nucleases. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4249–4253. doi: 10.1073/pnas.71.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Preparation and physical characterization of a homogeneous population of monomeric nucleosomes from HeLa cells. Nucleic Acids Res. 1976 Sep;3(9):2255–2266. doi: 10.1093/nar/3.9.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Safer J. P., Stanchfield J. E. Structural repeating units in chromatin. I. Evidence for their general occurrence. Exp Cell Res. 1976 Jan;97:101–110. doi: 10.1016/0014-4827(76)90659-5. [DOI] [PubMed] [Google Scholar]