Abstract
Pancreatic carcinoma is currently considered as a rapidly progressive and fatal disease, and is typically diagnosed late in its natural course. It is characterized by a poor diagnosis and lack of response to conventional therapy. Recent studies have suggested that disulfiram (DSF), a member of the dithiocarbamate family, may have antitumor activity. This study aimed to evaluate the in vitro effect of DSF on apoptosis in human pancreatic cancerous cell line (PANC-1). PANC-1 cells were cultured and treated with DSF at doses of 5, 10, 13 μM for 24 h and apoptosis was measured. Methylation specific PCR (MS-PCR) and real-time quantitative PCR were carried out to detect the methylation pattern and to estimate the mRNA expression levels of RASSF1A, p21 and Bax. MS-PCR analysis demonstrated that no unmethylated band was apeared in PANC-1 cell line after DSF treatments. The real-time quantitative PCR results showed no significant mRNA expression for RASSF1A (p>0.05); whereas p21 and Bax expression were significantly (p<0.01) enhanced after treatment with DSF. The results of the current study indicated that DSF can induce appoptosis in PANC-1 through p21 and Bax pathway but not through RASSF1A.
Keywords: Disulfiram, Human pancreatic cancerous cell line (PANC-1), Appotosis, RASSF1A, p21, Bax
INTRODUCTION
Pancreatic cancer is the fourth cause of cancer death all over the world (1). It is sometimes called a “silent killer” since it is not often diagnosed until it is progressd.
Patients with pancreatic adenocarcinoma have a poor prognosis and less than 5% have more than 5 years survival rate (2,3). Nowadays, there are many efforts to cure this disease.
Development of new curative approaches to enhance the therapeutic effect will be of great significance in the clinical management of pancreatic cancer. It is suggested that disulfiram (DSF) (Fig. 1) has anti-neoplastic effects in various cancerous cell lines such as melanoma, leukemia, small cell line carcinoma, osteosarcoma, cervical adenocarcinoma and colorectal adeno-carcinoma (4,5). DSF has a long history of alcoholic abuse treatment.
Fig. 1.
Chemical structure of disulfiram.
It contains strong thiol-reaction functional group which attacks the thiol group of cytosine and limits the catalytic mechanism of DNA methyltransferase inhibitor (DNMTi) on the C6 position of cytosine (6). DSF may induce appoptosis in these cell lines through p21, Bax and RASSF1A pathway. Fortunately RASSF1A re-expression is sufficient to reverse the tumorogenecity of human cancer. In this regard, different studies have demonstrated that several DNMT inhibitors have been developed and evaluated RASSF1A promoter demethylation in pre-clinical models and clinical trials (4,7,8,9,10,11).
Somatic epigenetic alterations are critical phenomenon in human cancer development (12,13,14). The epigenetic alterations may stay through cell division for the whole time of cell's life and also even last for multiple generations. Promoter CpG islands methylation is critical point of epigenetic mechanism that leads to gene silencing (15,16). Different studies have detected high frequencies of RASSF1A methylation in several human cancers (13).
RASSF1A has various roles in the cells such as cell cycle arrest, apoptosis, microtubule and genetic stability (17). RASSF1A is a key cell cycle gene at the G-S checkpoint by modulation of cycline D1 levels and inhibition of JNK pathway (10). Furthermore, it can upgrade CDKI (cycline-dependent kinase inhibitor) P21 which is mediated by P53 when DNA damage occurs in order to arrest the cell cycle (18). Cyclin-dependent kinase inhibitor 1 (P21 Cip1/Waf1) is a production of Ras/Raf pathway. Its expression occurs at the transcriptional and post-transcriptional levels making cell cycle arrest (19).
Bax is a member of the Bcl2 family and an important component of the apoptotic machinery. RASSF1A directly binds to the modulator of apoptosis-1 (MOAP1), a Bax-binding protein. This interaction is improved by the activated K-Ras, and knocking down RASSF1A reduces the ability of oncogenic K-Ras to activate Bax (7,10,11,20).
Since little is known about DSF anti-tumor effect and its mechanism of action is not well defined, this study aimed to evaluate the apoptotic effects of DSF on human pancreatic cancer cell line (PANC-1) in vitro and to underestand the mechanism of this effect through p21, Bax and RASSF1A pathway.
MATERIALS AND METHODS
Cell line and cell culture
Human PANC-1 cell line was purchased from the national cell bank of Pasteur Institute of Iran. The cells were cultured in high glucose Dulbecco's Modified Eagle Medium (DMEM) (Sigma USA) supplemented with 10% fetal bovine serum (FBS) (Sigma USA) and 1% penicillin-streptomycin (Sigma USA) at 37 °C and in humified atmosphere containing 5% CO2. After 80% confluence in T25 culture flask, 104 cells were transferred to 24 well cultue plate. Drug treatment were done one day after seeding. DSF was purchased from Sigma (USA). 3-(4,5-dimethythiaziazol-2yl)-2,5-diphenyl tetrazoliumbromide (MTT) was procured from Merck (Germany).
Disulfiram treatment and IC50 assay
DSF stock solution (50 mM) was prepared by dissolving DSF in DMSO and storing at -20 °C. The final concentration of DSF was determined by the IC50 assay. 104 PANC-1 cells were transferred in each well of 24 well cell culture-microplate and treated with different concentrations of DSF (0, 0.5, 2.5, 5, 7.5, 10, 15, 20, 25, 30 μM) for 24 h (n=3). Then 3-(4,5-dimethythiaziazol-2yl)-2,5-diphenyl tetrazoliumbromide (MTT) survival assay was carried out for evaluating cell viability and dose-response plot was used to calculate the half maximal inhibitory concentration, IC50 (4).
MTT Assay
To examine the viability of the PANC-1 cells in the presence of DSF, MTT was dissolved in PBS at 5 mg/ml. The stock solution was added to the culture medium at a dilution of 1:10. The plates were incubated at 37 °C for 4 h. The medium was then aspirated and 400 μl DMSO (Sigma, USA) was added to extract the MTT formazan crystals and the absorbance of each well was detected by microplate Eliza-reader (Hiperion MPR 4, Germany) at 540 nm.
DNA extraction, bisulfite treatment and methylation-specific PCR
Genomic DNA was isolated by PrimePrep TM Genomic DNA Isolation Kit (GeNet Bio, Korea) according to its protocol. DNA was treated by sodium bisulfate using Epitect Bisulfate Kit (Qiagen, Germany), according to its instruction. The PCR reaction were carried out in thermal cycler via eppendorf mastercycler (EP)-gradient PCR (Ependorf, Germany) as follows: 94 °C for 5 min followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s and the final extension 72 °C for 5 min. PCR products were transferred to electrophoresis on a 2% agarose gel and visualized with staining by DNA green viewer. Methylated and unmethylated RASSF1A primers were M1 (forward) 5'-GTGTTAACGCGTTGCGTATC-3'; M (reverse) 5'AACCCCGCGAACTAAAA ACGA-3'; U1 (forward) 5'-TTTGGTTGG AGTGTGTTAATGTG-3'; U2 (reverse) 5'-CAAACCCCACAAACTAAAAACAA-3' (21).
RNA extraction and real-time PCR
Total RNA was isolated by RNeasy mini kit (Qiagen, Germany). RNA samples were treated by RNase free DNase (Qiagen, Germqny) to eliminate the genomic DNA. The RNA concentration was measured using a biophotometer (Eppendorf Germany). 5 μl of total RNA was reverse-transcribed to cDNA. The cDNA construction was performed by Revert Aid TM Kit (Fermentas EU) according to the protocols. Real-time RT-PCR was carried out by the Maxima SYBR Green Rox qPCR master mix kit (Fermentase EU). The real-time primers were designed using AlleleID software (Primer Biosoft) which generated the following sequences: RASSF1A (Forward –TCATCTGGGGCGTCGTG, reverse- CGTTCGTGTCCCGCTCC), P21 (forward- GA CCAGCATGACAGATTTCTACCA, reverse- A ACTGAGACTAAGGCAGAAGATG), Bax (forward- GACCAGCATGACAGATTTCTAC CA, reverse-AACTGAGACTAAGGCAGAA GATG) ACTB (forward- GTTGTCGACGACG AGCG, reverse- GCACAGAGCCTCGCCTT). Real-time PCR reactions were performed using the steponeplus quantitative real time PCR detection system (Applied Biosystem USA). The PCR amplification conditions consisted of 10 min at 95 °C followed by 40 cycles of denaturation step at 95 °C for 15 sec and annealing and extension for 1 min at 60 °C. As it was previously described, the expression level of each target gene was calculated as 2-ΔΔCt. The epression mRNA levels of RASSF1-A, p21 and Bax were briefly calculated by determining a ratio between their amount and endogenous control (ACT B). Melting curve analysis (60 °C → 95 °C increment at 0.3 °C/S) was applied to determine melting temperature of specific amplification products and primer. These experiments were carried out in triplicate and repeated independently at least three times (21,22).
Flowcytometric analysis
The apoptotic cell percentage was measured by annexinV fluous kit (Roche). About 106 cells for each sample were washed in PBS and re-suspended in 100 μL of annexine-V-FLUOS labeling solution. Then they were incubated for 10-15 min at room temperature. Finally, the apoptotic cells were counted by FAC scan flowcytometry (Becton Dickinson, Heidelberg, Germany). These experiments were carried out in triplicate and repeated independently at least three times (23).
Statistical analysis
All the quantitative data were presented as the mean ± standard deviations. Kolmogorov-Simonov test was used to assess normal distribution of variables. One-way-analysis of variance (ANOVA) with LSD post hoc test was performed to determine statistical significance among different groups using SPSS software package 16.0. Significance was accepted at P<0.05.
RESULTS
Half maximal inhibitory concentration
Using MTT assay, the dose response curve was generated for DSF identified from the initial screen to determine IC50 for PANC-1 line (Fig. 2). The results showed that the essential DSF concentration to achieve the IC50 in PANC-1 cells was 13 μmol/L for 24 h which was not cytotoxic. Furthermore, we tested 5 and 10 μM DSF concentration in PANC-1 (24) (Fig. 2).
Fig. 2.
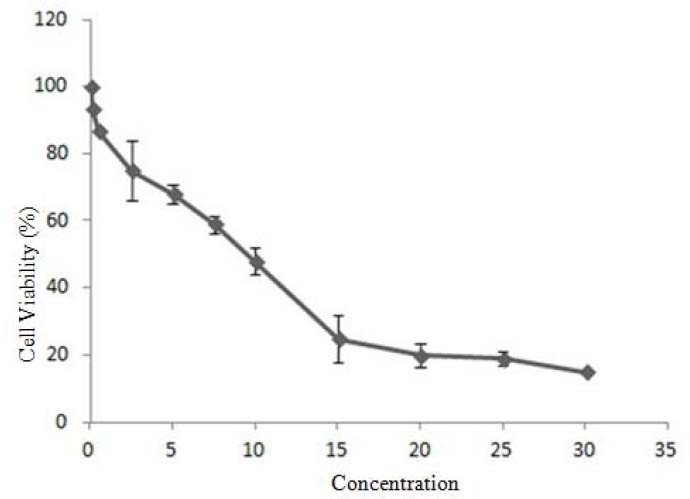
Cell viability of various disulfiram concentrations vs PANC-1 cancer cell viability which was used to calculate IC50 value. Cells were exposed to disulfiram at 0, 0.5, 2.5, 5, 7.5, 10, 15, 20, 25, 30 μM for 24 h (n=3) and cell viability was assessed by MTT assay.
Methylation specific PCR
The methylation specific PCR (MS-PCR) results in the untreated PANC-1 cells (without expousure to DSF) showed that the methylated DNA band was amplified while no corresponding DNA band was detected in treated cells. After treating cells with DSF, the results demonstrated no unmethylated band in MS-PCR while the methylated band was still intact. Taken together, these results showed that DSF at least at the tested concentration for 24 h could not demethylate the treated DNA (Fig. 3).
Fig. 3.
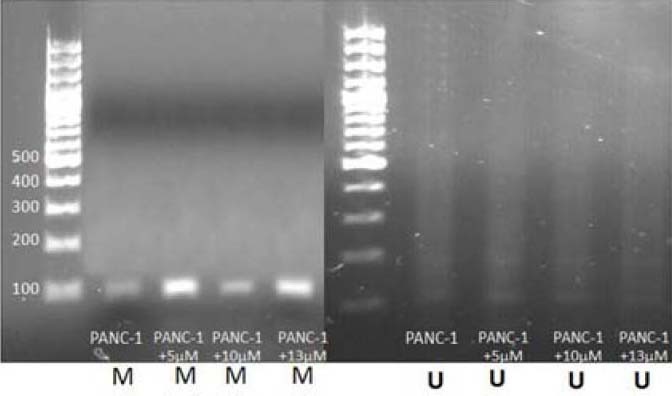
The methylation pattern detection in promoter region of RASSF1A by methylation specific PCR in PANC-1 before and after the treatment by disulfiram. A; Methylation band before and after the treatment. B; Unmethylated band before and after the treatment.
Real-time PCR
The result demonstrated no significant alteration (p>0.05) of RASSF1A expression after treating cells with DSF at the above-mentioned concentrations, whereas significant increase (p<0.01) of p21 and Bax expression was observed at these concentrations (Fig. 4).
Fig. 4.
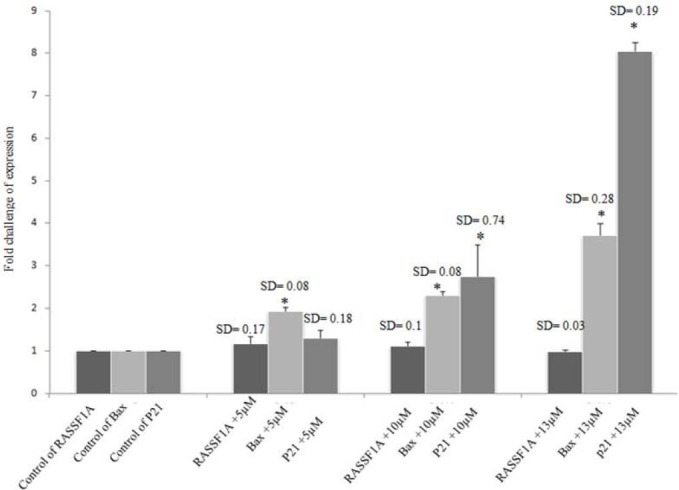
Disulfiram treatment in PANC-1, 5, 10 and 13 μM for 24 h. Disulfiram up-regulated p21 and Bax genes (p<0.01), but it did not have any effect on the expression of RASSF1A (p>0.05).
Flow cytometry
Simultaneous staining of cell surface phosphatidylserine with annexin-V-FITC and necrotic cell labeling with propidium iodide (PI) can discriminate between apoptotic and necrotic cells. During apoptosis, the phospholipid phosphatidylserine is exposed to the outer of the plasma membrane so annexin-V binds to it. Since positive PI result is a sign of cell necrosis, cells which are annexin-V positive, but PI negative, are generally defined as apoptotic cells. Analyses were performed on a FACSCalibur using the CellQuest Pro software. Given the definition, an apoptosis assessment was possible using the PI/annexin V dot plots as follows (Fig. 5): “Normal” includes cells that stain negatively for both annexin V and PI which are considered undamaged; “Apoptotic” includes cells stained with annexin V but are still PI negative and are considered apoptotic; and “Necrotic” includes cells that are both annexin V and PI positive and are necrotic. Compared to the control value (73.2%), the percentage of normal cells (annexin Vneg/PIneg) was reduced by 5, 10 and 13 μM DSF, (49%, 16% and 9%), respectively during 24 h, indicating that DSF treatments arrested PANC-1 cell proliferation (50%, > 80%, 90% of inhibition) within 24 h.
Fig. 5.
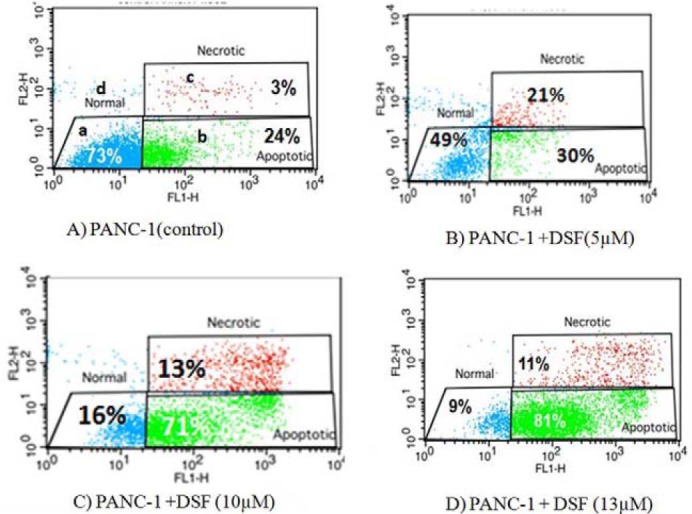
AnnexinV FITC and propidium iodide assay used to measure the percentage of apoptotic cells. The a quadrant contains viable non-apoptotic cells which are negative for annexin-V and exclude propidium iodide staining (A- /PI-). The b quadrant shows apoptotic cells which bind annexin-V but exclude propidium iodide (A+ /PI-). The c quadrant represents necrotic (late apoptotic cells binding annexin-V and propidium iodide (A+ /PI+). The d quadrant represent snecrotic cells which exclude annexin-V and binding propidium iodide (A- /PI+). A; Non-treated PANC-1. B; 5 μM disulfiram treated PANC-1. C; 10 μM disulfiram treated PANC-1. D; 13 μM disulfiram treated PANC-1
DISCUSSION
Disulfiram has widely been recognized antialcoholism drug used in the clinic and is approved by Food and Drug Administration (25). Our study revealed high cytotoxicity of DSF on PANC-1 cell lines. However, DSF IC50 concentration used in our study was different from that reported in previous study on PC3, DU145, C4-2B and CWR22Rv1 (5). This difference could be due to the different cell systems, different levels of cellular response, and/or different sensitivity of appoptosis activity assays. In proportion to previous reports by Wickstrom and coworkers, a monophasic cytotoxic effect of DSF was observed in PANC-1 cell lines similar to NCI-H69, H69AR, ACHN, Hela and hTERT RPE-1 (Fig. 2) (5). PANC-1 cells were killed at 5, 10 and 13 μM concentration (26).
To answer the question of whether DSF have demethylating effect on PANC-1 cells as it has on prostatic cancer cell line (4), we tested this effect in the current study. The result of our study is against those of Lin that expressed demethylating effect of DSF on RARB and APC genes on prostatic cancer cell line, but is consistent with Lyko findings that DSF like other known non-nucleoside DNMTi has significant lower DNA demethylation potency than nucleoside ones (4,27). The results of our study showed no un-methylated band using MS-PCR (Fig. 3). Although, DSF has been suggested as DNMTi, in the present study no epigenetic-reversion effect was observed. Furthermore the RT-PCR results demonstrated no RASSF1A mRNA expression (Fig. 4).
In line with previous studies (Lloyd, Roper and Sewing), our results showed up-regulation of p21 mRNA expression enhancment as illustrated by RT-PCR and MTT assay (28,29,30). Furthermore, Shivakumar showed that p21 can contribute to the negative regulation of CDK2 in the H1299 cells and induce cell cycle arrest (22). In this study, there was no correlation between RASSF1A and p21 which is controversial to Thalar who demonstrated cell death by the activation of Ras pathway and p21 up-regulation in-vitro and in-vivo due to the age (6). In PANC-1, p53 is mutated and thus p21 expression is blocked in this way.
As the cell death molecular mechanism of DSF is not defined, we examined the expression of p21 and Bax genes, as two important genes in cell cycle arrest and apoptosis, related to re-expression of RASSF1A (10).
However, there was no statistically significant correlation between RASSF1A and Bax expression in our study. It seems that although DSF demethylating effect is weaker than nucleoside analogous effect, it can make up regulation of p21 and Bax in mentioned concentrations in other non-epigenetic mechanisms. It is clear that growth and death signaling pathways are linked with each other and growth deregulation starts apoptotic pathway.
Wickstrom pronounced that the cell death mechanisms induced by DSF was relatively with typical signs of apoptosis, i.e. nuclear fragmentation/condensation and activation of caspase-3, occurring after 24 h. Membrane permeabilization and substrate detachment occurred at later time and necrosis was induced along with apoptosis after 48 h (5). Furthermore, other antineoplastic activities are proposed for DSF mechanisms by Lin; for instance, inhibition of NF-κB pathway and topoisomerase I and II catalytic activities, inhibition of invasion and angiogenesis and a direct inhibitory effect on matrix metallo-proteinases (MMP-2, MMP-9). Moreover it changes the intracellular superoxide levels and decreases mitochondrial membrane polarization, induces apoptosis and finally proteasome inhibition in a variety of cancer cell lines (4).
CONCLUSION
In conclusion, although DSF showed no DNA demethylation effect, its multi-potential antineoplastic effects and tumor-specific toxicity make it has less side effect in comparison to other compounds in cancer therapy. More investigation of possible mechanisms action of DSF regarding its apoptotic effect is needed.
ACKNOWLEDGMENTS
This study was supported by Isfahan University of Medical Sciences. We would like to appreciate Dr. Batool Hashemi Beni, Dr. Mohammad. Kazemi, and Mrs. Maryam Aliakbari for their sincere help. This study could not be performed without their contributions.
REFERENCES
- 1.Hariharan D, Saied A, Kocher H. Analysis of mortality rates for pancreatic cancer across the world. HPB. 2008;10:58–62. doi: 10.1080/13651820701883148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hruban RH, Brune K, Fukushima N, Maitra A. Pancreatic intraepithelial neoplasia. Pancreatic Cancer. 2008;378:41–51. [Google Scholar]
- 3.Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, et al. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene. 2003;22:3806–3812. doi: 10.1038/sj.onc.1206582. [DOI] [PubMed] [Google Scholar]
- 4.Lin J, Haffner MC, Zhang Y, Lee BH, Brennen WN, Britton J, et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. The Prostate. 2010;71:333–343. doi: 10.1002/pros.21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wickstrom M, Danielsson K, Rickardson L, Gullbo J, Nygren P, Isaksson A, et al. Pharmacological profiling of disulfiram using human tumor cell lines and human tumor cells from patients. Biochemical pharmacology. 2007;73:25–33. doi: 10.1016/j.bcp.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Veverka KA, Johnson KL, Mays DC, Lipsky JJ, Naylor S. Inhibition of aldehyde dehydrogenase by disulfiram and its metabolite methyl diethylthio-carbamoyl-sulfoxide. Biochemical pharmacology. 1997;53:511–518. doi: 10.1016/s0006-2952(96)00767-8. [DOI] [PubMed] [Google Scholar]
- 7.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 8.Brueckner B, Kuck D, Lyko F. DNA methyl transferase inhibitors for cancer therapy. The Cancer Invest. 2007;13:17–22. doi: 10.1097/PPO.0b013e31803c7245. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Epigenetics in cancer. New Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 10.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 11.Vos MD, Dallol A, Eckfeld K, Allen NPC, Donninger H, Hesson LB, et al. The RASSF1A tumor suppressor activates Bax via MOAP-1. J Biol Chem. 2006;281:4557–4563. doi: 10.1074/jbc.M512128200. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 13.Wang T, Liu H, Chen Y, Liu W, Yu J, Wu G. Methylation associated inactivation of RASSF1A and its synergistic effect with activated K-Ras in nasopharyngeal carcinoma. J Exp Clin Canc Res. 2009;28:1–11. doi: 10.1186/1756-9966-28-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gharagozloo M, Kazemi M, Esmaeil N, Roshankhah S, Golmohammadi R, Mobarakian M, et al. The apoptotic effects of sirtuin 1 inhibitor on the MCF-7 and MRC-5 cell lines. Res Pharm Sci. 2012;8:79–89. [PMC free article] [PubMed] [Google Scholar]
- 15.Bird A. Perceptions of epigenetics. NatureLondon. 2007;447:396–388. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 16.Gheibi A, Kazemi M, Baradaran A, Akbari M, Salehi M. Study of promoter methylation pattern of 14-3-3 sigma gene in normal and cancerous tissue of breast: A potential biomarker for detection of breast cancer in patients. Adv Biom Res. 2012;1:1–5. doi: 10.4103/2277-9175.102990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 18.Kierszenbaum ALM. 2nd edition. 2007. Histology and Cell Biology: An Introduction to Pathology published February; pp. 896–897. E-Book. [Google Scholar]
- 19.Thaler S, Hähnel PS, Schad A, Dammann R, Schuler M. RASSF1A mediates p21Cip1/Waf1-dependent cell cycle arrest and senescence through modulation of the Raf-MEK-ERK pathway and inhibition of Akt. Cancer Res. 2009;69:1748–1757. doi: 10.1158/0008-5472.CAN-08-1377. [DOI] [PubMed] [Google Scholar]
- 20.Arlt A, Müerköster SS, Schäfer H. Targeting apoptosis pathways in pancreatic cancer. Cancer Lett. 2010;10:1–13. doi: 10.1016/j.canlet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Avramouli A, Tsochas S, Mandala E, Katodritou E, Ioannou M, Ritis K, et al. Methylation status ofRASSF1A in patients with chronic myeloid leukemia. Leukemia Res. 2009;33:1130–1132. doi: 10.1016/j.leukres.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Kabiri A, Esfandiari E, Hashemibeni B, Kazemi M, Mardani M, Esmaeili A. Effects of FGF-2 on human adipose tissue derived adult stem cells morphology and chondrogenesis enhancement in Transwell culture. Biochem Bioph Res Co. 2012;424:234–238. doi: 10.1016/j.bbrc.2012.06.082. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Yashiro M, Ohira M, Ren J, Hirakawa K. Synergic antiproliferative effect of DNA methyltransferase inhibitor in combination with anticancer drugs in gastric carcinoma. Cancer Science. 2006;97:938–944. doi: 10.1111/j.1349-7006.2006.00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X, Xu B, Pandey S, Goessl E, Brown J, Armesilla AL, et al. Disulfiram/copper complex inhibiting NFκB activity and potentiating cytotoxic effect of gemcitabine on colon and breast cancer cell lines. Cancer Lett. 2010;290:104–113. doi: 10.1016/j.canlet.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Eneanya DI, Bianchine JR, Duran DO, Andresen BD. The actions and metabolic fate of disulfiram. Annu Rev Pharmacol. 1981;21:575–596. doi: 10.1146/annurev.pa.21.040181.003043. [DOI] [PubMed] [Google Scholar]
- 26.Chung KC, Park JH, Kim CH, Lee HW, Sato N, Uchiyama Y, et al. Novel biphasic effect of pyrrolidine dithiocarbamate on neuronal cell viability is mediated by the differential regulation of intracellular zinc and copper ion levels, NF‐κb, and MAP kinases. J Neurosci Res. 2000;59:117–125. [PubMed] [Google Scholar]
- 27.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. Journal of the National Cancer Institute. 2005;97:1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd AC, Obermüller F, Staddon S, Barth CF, McMahon M, Land H. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes & Development. 1997;11:663–677. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- 29.Roper E, Weinberg W, Watt FM, Land H. p19ARF-independent induction of p53 and cell cycle arrest by Raf in murine keratinocytes. EMBO reports. 2001;2:145–150. doi: 10.1093/embo-reports/kve020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]


