Abstract
Importance
As APOE locus variants contribute to both risk of late-onset Alzheimer disease and differences in age-at-onset, it is important to know if other established late-onset Alzheimer disease risk loci also affect age-at-onset in cases.
Objectives
To investigate the effects of known Alzheimer disease risk loci in modifying age-at-onset, and to estimate their cumulative effect on age-at-onset variation, using data from genome-wide association studies in the Alzheimer’s Disease Genetics Consortium (ADGC).
Design, Setting and Participants
The ADGC comprises 14 case-control, prospective, and family-based datasets with data on 9,162 Caucasian participants with Alzheimer’s occurring after age 60 who also had complete age-at-onset information, gathered between 1989 and 2011 at multiple sites by participating studies. Data on genotyped or imputed single nucleotide polymorphisms (SNPs) most significantly associated with risk at ten confirmed LOAD loci were examined in linear modeling of AAO, and individual dataset results were combined using a random effects, inverse variance-weighted meta-analysis approach to determine if they contribute to variation in age-at-onset. Aggregate effects of all risk loci on AAO were examined in a burden analysis using genotype scores weighted by risk effect sizes.
Main Outcomes and Measures
Age at disease onset abstracted from medical records among participants with late-onset Alzheimer disease diagnosed per standard criteria.
Results
Analysis confirmed association of APOE with age-at-onset (rs6857, P=3.30×10−96), with associations in CR1 (rs6701713, P=7.17×10−4), BIN1 (rs7561528, P=4.78×10−4), and PICALM (rs561655, P=2.23×10−3) reaching statistical significance (P<0.005). Risk alleles individually reduced age-at-onset by 3-6 months. Burden analyses demonstrated that APOE contributes to 3.9% of variation in age-at-onset (R2=0.220) over baseline (R2=0.189) whereas the other nine loci together contribute to 1.1% of variation (R2=0.198).
Conclusions and Relevance
We confirmed association of APOE variants with age-at-onset among late-onset Alzheimer disease cases and observed novel associations with age-at-onset in CR1, BIN1, and PICALM. In contrast to earlier hypothetical modeling, we show that the combined effects of Alzheimer disease risk variants on age-at-onset are on the scale of, but do not exceed, the APOE effect. While the aggregate effects of risk loci on age-at-onset may be significant, additional genetic contributions to age-at-onset are individually likely to be small.
Keywords: Alzheimer Disease, Alzheimer Disease Genetics, Alzheimer’s Disease - Pathophysiology, Genetics of Alzheimer Disease, Aging
INTRODUCTION
Alzheimer Disease (AD) [MIM 104300] affects more than 13% of individuals aged 65 years and older, and its prevalence increases with age, occurring in fewer than 1% of those age 65 years and younger and in as much as 40% of the population after age 90.1-4 While genetic studies of late-onset Alzheimer Disease (LOAD) confirmed at least ten loci contributing to risk of disease, including APOE, PICALM, CLU, CR1, BIN1, CD2AP, EPHA1, MS4A4A, CD33, and ABCA7,5,6 genes modifying age-at-onset (AAO) of LOAD have not been widely studied. Earlier linkage and candidate gene studies identified only a few loci possibly underlying variation of AAO (e.g., GSTO1),7 but only variation in the APOE region has been consistently confirmed.8-12
A multitude of studies have attempted to identify susceptibility genes for AAO in AD. The first study to identify a genetic association with AAO showed a lower mean AAO among cases for each additional copy of the ε4 allele at the APOE locus on chromosome 19q (0 copies: 84.3 years; 1 copy: 75.5; 2 copies: 68.4),13 a finding which has since been replicated.14 Subsequent genome-wide linkage scans examining AAO in AD patients and unaffected family members (using age at study entry) found suggestive evidence of linkage on chromosome 19 to APOE (LOD = 3.28),15 which was confirmed in later studies.16 Multiple studies identified other suggestive linkage signals on chromosomes 4q, 8q,16 1q, 6p,17 7q, 15, and 19p18 in Caucasian families, and chromosomes 5q, 7q, 14q, and 17q19 in Caribbean Hispanics, though the specific loci driving these linkage signals remain unknown. More recently, an AAO GWAS in 2,222 Caucasian AD cases confirmed association at APOE, and also found strong evidence of association (P=4.95×10−7) on chromosome 4q31.3 in the gene DCHS2.20
The lack of overlap in the regions identified across these studies may have resulted from differences in the approaches applied, such as varied strategies for censoring unaffected pedigree members and differences in covariates adjusted for in analyses. Reduced statistical power from the limited availability of extended families for analysis may also have contributed to the differences in findings between these early linkage and association studies. The high variability in approaches and findings highlights the need for a more comprehensive approach to identify genetic risk factors which may influence LOAD AAO as well as LOAD risk directly. Notably, variants in the ten confirmed LOAD risk loci have not been examined for their possible influence on AAO among LOAD cases.
Using data from 9,162 LOAD cases from a recent genome-wide association study (GWAS) of LOAD by the Alzheimer’s Disease Genetics Consortium,6 we examined whether variants most significantly associated with LOAD risk in 10 LOAD loci are also associated with differences in AAO among LOAD cases. Furthermore, we used a genetic burden analysis approach to determine the proportion of variation in AAO accounted for by variants in these established LOAD risk genes.
MATERIALS AND METHODS
Ascertainment and Collection of Genotypic and Phenotypic Data
A detailed description of ascertainment and the collection of genotype and phenotype data in the individual datasets of the ADGC is available elsewhere.6 Briefly, subjects in each dataset (eTable 1) were genotyped using either Illumina or Affymetrix commercially-available GWAS high-density SNP genotyping microarrays. All LOAD subjects met NINCDS/ADRDA criteria for definite, probable or possible LOAD,21 and age-at-onset (AAO) of LOAD, which was abstracted from medical records for most subjects, was defined as the age when LOAD-related symptoms manifested, as reported by the subject or an informant. Age-at-ascertainment (WU, ADNI) was substituted for datasets lacking AAO information (eTable 1). Unaffected subjects and LOAD cases lacking AAO information, cases with an age-at-onset or age-at-death less than 60 years, and non-Caucasians of European ancestry were excluded from the association analyses. Post hoc analyses revealed no significant differences in association findings between subjects with age-at-onset and those with age-at-ascertainment information (data not shown).
Quality Control
Subjects were excluded if Affymetrix chip genotypes were called for fewer than 95% of SNPs or if Illumina chip genotypes were called for fewer than 98% for SNPs. Additionally, samples were excluded if reported gender differed from genetic gender by X-chromosome analysis (PLINK software).22 Samples were dropped from family datasets if reported relationships differed from estimated relatedness from IBD using the program PREST.23 If samples were duplicated in different datasets, only one sample per duplicate pair was kept in analysis. After exclusions, data on 9,162 cases remained for subsequent analyses.
After sample quality control, genotyped SNPs were excluded from analysis if their minor allele frequencies (MAF) were less than 0.02 for Affymetrix chips or less than 0.01 for Illumina chips, or if the SNPs were observed to be out of Hardy-Weinberg equilibrium with P<10−6. Imputed SNPs were excluded if the quality score (“Info” from IMPUTE2)24 was less than 0.50. Genome-wide genotype imputation was performed in each cohort using IMPUTE2 software24 with 1000 Genomes (December 2010 release) CEPH Utah pedigree (CEU) reference haplotypes. Imputation quality was assessed using the ‘Info’ statistic, and only SNPs imputed with Info ≥0.50 were included in analysis. The ten SNPs examined here were among the common set of SNPs produced in imputation.
Statistical Analysis
We performed association analysis on individual datasets assuming an additive model on log-transformed age-at-onset with covariate adjustment for population substructure. For cases from case-control datasets, linear regression was performed in PLINK,22 while for analysis of cases from family datasets (used only in the primary analysis of risk variants), generalized estimating equations (GEE) with a linear model as implemented in the R statistical package25 were used. To account for the effects of population substructure, we performed a principal components (PC) analysis on cases within each dataset using EIGENSTRAT26 on a subset of 21,109 SNPs common to all genotyping platforms. The first three PCs from analysis were incorporated in our minimal model for covariate adjustment. We also performed analyses conditioning on the major AAO-modifying effects of APOE through an extended model of covariate adjustment which included sex and number of APOE ε4 alleles (0, 1, or 2). Results from individual datasets were combined in the meta-analysis using inverse-variance weighting as implemented in METAL,27 applying a genomic control to each dataset. With this set of 9,162 cases, we expected to have 80% power to detect loci with as little effect as 9 months difference in AAO per allelic copy for very common variants (MAF>0.34), with power to detect 1 year difference in AAO per allelic copy for variants of even modest frequency (MAF>0.14).28
We performed a discovery genome-wide association meta-analysis among 6,143 cases in 10 ADGC case-control datasets to determine whether SNPs with more modest LOAD risk associations may contribute to differences in AAO, and to assess genetic burden attributable to these variants. Methods, results, and a brief summary are provided in the eAppendix.
In addition to association meta-analysis, we performed genetic burden analyses to determine the percent contribution of LOAD susceptibility SNPs in ten LOAD candidate genes to variation in AAO. Risk-weighted genetic burden analyses of AAO linearly modeled locus-specific effects as the product of the meta-analysis-estimated effect size (across-study change in AAO for each copy of the minor allele) and the dosage of the minor allele (scale 0-2; estimated from genotype-specific imputation probabilities), and were implemented in analyses of risk variants. Additional covariate adjustment in the burden model included covariates for population substructure from principal components analysis and dataset-specific effects.
RESULTS
ADGC Data Characteristics
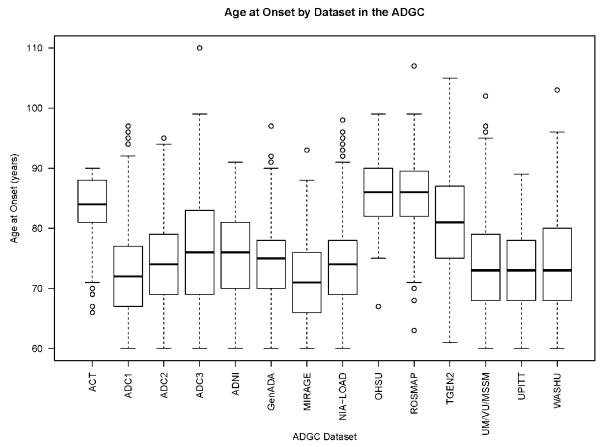
Descriptive characteristics of the individual ADGC datasets are shown in eTable 1. There were more female cases (N = 5,480; 60%) than males. The mean age-at-onset of was 74.3 years (y) (standard deviation = 7.64 y) for the entire group. Several datasets had later ages at onset; two of these were population-based cohorts of aging and memory loss, ROSMAP [mean AAO (SD) = 85.6 y (6.26 y)] and ACT [mean AAO (SD) = 83.9 y (4.76 y)], and one case-control dataset, OHSU, which intentionally ascertained individuals with later age-at-onset [mean AAO (SD) = 86.1 y (5.53 y)]. While data from these studies did not largely change the patterns of association observed (data not shown) in association testing, we performed several sub-analysis to assess their effect on the genetic burden analyses as described below.
LOAD Susceptibility Variant Associations with AAO
We confirmed the association of the APOE ε4 allele with lower AAO, with each additional copy of the ε4 allele reducing AAO by 2.45 years (β=−2.45; P=3.30×10−96). Examining the variants most strongly associated with LOAD in nine genomic regions with genome-wide statistically significant associations in our GWAS of LOAD risk (Table 1),6 we observed that several LOAD risk loci also demonstrated statistically significant associations (P<0.005) with AAO, including rs6701713 in CR1 (P=7.17×10−4), rs7561528 in BIN1 (P=4.78×10−4), rs561655 in PICALM (P=0.00223). Both rs6701713 in CR1 and rs7561528 in BIN1 demonstrated a reduced age-at-onset for each copy of the risk variant with each copy of the risk allele A at rs6701713 (MAF=0.24) advancing AAO by approximately five months [β (95% CI): −0.41 (−0.65, −0.17)], and each copy of the risk allele A at rs7561528 (MAF=0.37) advancing AAO by slightly less than four months [β (95% CI): −0.31 (−0.52, −0.09)]. In contrast, each copy of the more common risk allele (A; frequency=0.62) at rs561655 in the PICALM gene corresponded with earlier onset by approximately four months [β (95% CI): −0.33 (−0.55, 0.12)]. These patterns of association remained largely unchanged after adjustment for APOE ε4 allele dose and sex for the CR1 and BIN1 variants [rs6701713: −0.41 (−0.69, −0.12), P=0.00488; rs7561528: −0.32 (−0.57, −0.08), P=0.00985]. While the size and direction of the association remained the same as in the minimally adjusted model, the association of the PICALM variant demonstrated only marginal significance after this additional adjustment [rs561655: 0.32 (0.07, 0.57), P=0.0112]. Investigation of AAO associations in the vicinity of these AD risk variants revealed no substantially different associations among nearby variants. Directions of variant effects were concordant between AD risk and AAO; all variants that increase risk also lower AAO.
Table 1. Association with age-at-onset (AAO) of SNPs most significantly associated with LOAD in nine genomic regions and APOE.
SNPs presented demonstrated strongest associations within each of ten genomic regions with associations of genome-wide statistical significance (P ≤5.0×10−8) with LOAD risk. P-values for AAO associations exceeding the multiple hypothesis testing threshold (P<0.005) are shown in bold
| SNP | CH:MB | Nearest Gene |
MA | MAF | AAO (Minimal Adjustment Model) |
AAO (Extended Adjustment Model) |
LOAD Risk (from Naj et al.) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (95% CI) | P | Het P | β (95% CI) | P | Het P | OR (95% CI) | P | |||||
| rs6701713 | 1:207.8 | CR1 | A | 0.24 | −0.41 (−0.65, −0.17) | 7.17×10−4 | 0.405 | −0.41 (−0.69, −0.12) | 4.88×10−3 | 0.422 | 1.16 (1.11, 1.22) | 4.6×10−10 |
| rs7561528 | 2:127.9 | BIN1 | A | 0.37 | −0.31 (−0.52, −0.09) | 4.78×10−4 | 0.855 | −0.32 (−0.57, −0.08) | 9.85×10−3 | 0.684 | 1.17 (1.13, 1.22) | 4.2×10−14 |
| rs9349407 | 6:47.5 | CD2AP | C | 0.32 | −0.03 (−0.25, 0.19) | 0.765 | 0.266 | −0.14 (−0.40, 0.11) | 0.273 | 0.860 | 1.12 (1.07, 1.18) | 1.0×10−6 |
| rs11767557 | 7:143.1 | EPHA1 | C | 0.18 | 0.03 (−0.26, 0.32) | 0.830 | 0.861 | 0.07 (−0.24, 0.39) | 0.659 | 0.657 | 0.87 (0.83, 0.92) | 2.4×108−7 |
| rs1532278 | 8:27.5 | CLU | T | 0.37 | 0.05 (−0.18, 0.28) | 0.661 | 0.137 | 0.0038 (−0.26, 0.27) | 0.977 | 0.108 | 0.89 (0.85, 0.93) | 8.3×10−8 |
| rs4938933 | 11:60.0 | MS4A4A | C | 0.36 | 0.09 (−0.14, 0.31) | 0.448 | 0.454 | 0.018 (−0.23, 0.27) | 0.8874 | 0.584 | 0.88 (0.85, 0.92) | 1.7×10−9 |
| rs561655 | 11:85.8 | PICALM | G | 0.38 | 0.33 (−0.12, 0.55) | 2.23×10−3 | 0.915 | 0.32 (0.07, 0.57) | 0.0112 | 0.957 | 0.87 (0.84, 0.91) | 7.0×10−11 |
| rs3752246 | 19:1.1 | ABCA7 | G | 0.34 | −0.27 (−0.55, 0.02) | 0.0640 | 0.700 | −0.19 (−0.51, 0.13) | 0.242 | 0.748 | 1.15 (1.09, 1.21) | 5.8×10−7 |
| Haplotype (rs7412/ rs429358) |
19:45.4 | APOE | ε4 | 0.35 | −2.45 (−2.68, −2.21) | 3.30×10−96 | 0.0941 | −0.24 (−0.75, 0.27) | 0.360 | 0.874 | 3.02 (2.86, 3.20) | 2.18×10−320 |
| rs3865444 | 19:51.7 | CD33 | A | 0.20 | 0.10 (−0.13, 0.33) | 0.377 | 0.596 | 0.13 (−0.13, 0.38) | 0.338 | 0.872 | 0.89 (0.86, 0.93) | 1.1×10−7 |
CH:MB, chromosome:position (in mega base pairs, build 19); MA, minor allele; MAF, minor allele frequency; β, Beta coefficient for AAO from meta-analysis (# years difference in AAO per copy of the minor allele); OR, odds ratio; 95% CI, 95% Confidence Interval; P, P-value; Het P, P-value for heterogeneity across studies.
Genetic Burden Analysis of AAO with LOAD Risk Variants
We examined the genetic burden of APOE and the LOAD risk variants in the nine genomic regions on variation in AAO (Table 2) in the 14 ADGC datasets with complete AAO data. In our baseline model, 19% of the variation in AAO (R2=0.189) was accounted for by population substructure and study-specific effects. The independent contributions of dosage of the APOE ε4 allele to genetic burden was roughly 3.1% of AAO variation (R2=0.220) while the cumulative effect of the nine LOAD risk variants was 0.93% (R2=0.198), together accounting for approximately 4.1% of genetic variation in AAO (R2=0.229). Excluding study-specific effects, APOE accounts for 3.9% of the remaining variation, the nine LOAD risk variants account for another 1.1%, for a combined contribution of 5% of the variation of AAO. Variant effects in burden modeling were consistent with the association results for individual variants described above.
Table 2. Risk-weighted burden analysis results for APOE and 9 LOAD Candidate Genes.
Beta coefficients (β), 95% Confidence Intervals, and P-values are from four logistic regression models examining weighted scores for the peak SNP associations in APOE and 9 LOAD candidate genes. Scores are the product of the log-transformed odds ratio for each SNP association multiplied by minor allele dosage from imputed genotype probabilities.
| Model 1: Adjustment for PCs & Site |
Model 2: Adjustment for Model 1 + APOE |
Model 3: Adjustment for Model 1 + 9 LOAD Genes |
Model 4: Adjustment for Model 1 + APOE + 9 LOAD Genes |
|||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | |
| (Intercept) | 74.2 (73.5, 75.0) | <10-32 | 70.6 (69.8, 71.4) | <10−32 | 72.9 (71.6, 74.1) | <10−32 | 69.1 (67.9, 70.4) | <10−32 |
| CR1 Score | -- | -- | -- | -- | 1.01 (0.59, 1.43) | 2.66×10−6 | 1.01 (0.60, 1.43) | 1.51×10−6 |
| BIN1 Score | -- | -- | -- | -- | −0.97 (−1.45, −0.50) | 5.27×10−5 | −1.04 (−1.51, −0.58) | 9.99×10−6 |
| CD2AP Score | -- | -- | -- | -- | −0.51 (−1.00, −0.03) | 0.0381 | −0.43 (−0.90, 0.05) | 0.0764 |
| EPHA1 Score | -- | -- | -- | -- | −0.73 (−1.49, 0.03) | 0.0586 | −0.67 (−1.41, 0.07) | 0.0778 |
| CLU Score | -- | -- | -- | -- | 0.83 (0.36, 1.29) | 4.63×10−4 | 0.89 (0.44, 1.35) | 1.11×10−4 |
| MS4A4A Score | -- | -- | -- | -- | 0.87 (0.34, 1.40) | 0.00120 | 0.91 (0.39, 1.43) | 5.58×10−4 |
| PICALM Score | -- | -- | -- | -- | 1.11 (0.63, 1.59) | 6.78x10−6 | 1.03 (0.56, 1.51) | 1.83×10−5 |
| ABCA7 Score | -- | -- | -- | -- | 0.92 (0.35, 1.50) | 0.00164 | 0.97 (0.41, 1.53) | 7.35×10−4 |
| CD33 Score | -- | -- | -- | -- | −0.84 (−1.41, −0.28) | 0.00315 | −0.81 (−1.36, −0.26) | 0.00405 |
| APOE Score | -- | -- | −0.77 (−0.85, −0.70) | 1.03×10−79 | -- | -- | -0.78 (−0.86, −0.70) | 5.48×10−81 |
| ACT | 9.67 (8.62, 10.7) | 3.31×10−72 | 10.3 (9.30, 11.4) | 1.10×10−84 | 11.15 (9.64, 12.66) | 8.89×10−47 | 11.9 (10.42, 13.39) | 6.69×10−55 |
| ADC1 | −1.76 (−2.60, −0.93) | 3.66x10−5 | 0.54 (−0.32, 1.39) | 0.219 | −1.74 (−2.81, −0.67) | 0.00139 | 0.62 (−0.45, 1.70) | 0.256 |
| ADC2 | −i.03 (−1.96, −0.10) | 0.0294 | −1.47 (−2.38, −0.56) | 0.00157 | 0.33 (−1.20, 1.87) | 0.671 | −0.09 (−1.59, 1.42) | 0.909 |
| ADC3 | 0.19 (−0.74, 1.13) | 0.684 | 0.40 (−0.52, 1.32) | 0.393 | 1.17 (−0.68, 3.02) | 0.215 | 1.31 (−0.50, 3.13) | 0.155 |
| ADNI | −1.20 (−2.57, 0.18) | 0.0882 | −0.91 (−2.26, 0.44) | 0.187 | −0.26 (−1.90, 1.39) | 0.758 | 0.22 (−1.39, 1.84) | 0.787 |
| GenADA | 0.36 (−0.57, 1.28) | 0.449 | −1.42 (−2.34, −0.50) | 0.00258 | 2.46 (1.19, 3.72) | 1.39×10−4 | 0.79 (−0.46, 2.04) | 0.216 |
| LOAD | −0.67 (−1.49, 0.15) | 0.111 | 0.42 (−0.40, 1.23) | 0.316 | −0.37 (−1.55, 0.80) | 0.533 | 0.77 (−0.38, 1.93) | 0.190 |
| MIRAGE | −3.08 (−4.04, −2.11) | 4.66×10−10 | 0.37 (−0.64, 1.38) | 0.472 | −2.17 (−3.51, −0.83) | 0.00146 | 1.36 (0, 2.72) | 0.0504 |
| OHSU | 11.9 (10.5, 13.3) | 2.06×10−59 | 12.5 (11.1, 13.9) | 5.29×10−68 | 13.55 (11.72, 15.39) | 5.32×10−47 | 14.4 (12.6, 16.2) | 1.16×10−54 |
| ROSMAP | 11.4 (10.3, 12.5) | 6.52×10−90 | 9.21 (8.11, 10.3) | 4.63×10−60 | 13.26 (11.57, 14.95) | 1.34×10−52 | 11.22 (9.55, 12.9) | 3.11×10−39 |
| TGEN2 | 0.55 (−1.03, 2.13) | 0.494 | 0.69 (−0.86, 2.24) | 0.385 | 1.47 (−3.09, 6.03) | 0.528 | 1.55 (−2.92, 6.03) | 0.497 |
| UM/VU/MSSM | −0.33 (−1.20, 0.53) | 0.449 | −0.6 (−1.45, 0.25) | 0.165 | 1.18 (−0.10, 2.46) | 0.0704 | 0.92 (−0.33, 2.18) | 0.149 |
| UPITT | −1.33 (−2.18, −0.47) | 0.0233 | −0.61 (−1.45, 0.23) | 0.155 | 0.7 (−0.54, 1.94) | 0.267 | 1.38 (0.16, 2.59) | 0.0261 |
| PC1 | 6.76 (3.09, 10.4) | 3.10×10−4 | 6.16 (2.56, 9.76) | 8.01×10−4 | 7.04 (3.38, 10.7) | 1.61×10−4 | 6.44 (2.85, 10.02) | 4.32×10−4 |
| PC2 | 1.12 (−2.60, 4.84) | 0.554 | 1.99 (−1.66, 5.64) | 0.285 | 0.81 (−2.90, 4.51) | 0.670 | 1.65 (−1.98, 5.28) | 0.372 |
| PC3 | −0.89 (−4.54, 2.77) | 0.634 | −0.34 (−3.93, 3.24) | 0.852 | −1.07 (−4.71, 2.57) | 0.564 | −0.5 (−4.07, 3.06) | 0.782 |
|
| ||||||||
| F (df1, df2) | 164.2 (13, 9114) | 184.8 (14, 9113) | 103.3 (22, 9105) | 119.1 (23, 9104) | ||||
| P | <10−100 | <10−100 | <10−100 | <10−100 | ||||
| Multiple R2 | 0.1898 | 0.2211 | 0.1998 | 0.2313 | ||||
| Adjusted R2 | 0.1886 | 0.2199 | 0.1979 | 0.2294 | ||||
To determine whether ascertainment differences may have influenced the amount of variation in AAO attributable to LOAD risk variants, we examined the effects of the three datasets with much later average AAO (ACT, OHSU, and ROSMAP) and the two-family based datasets (NIA-LOAD and MIRAGE) on genetic burden analyses. In analyses excluding the datasets with later average AAO (eTable 2), we found that these datasets account for much of the dataset-specific AAO variation, reducing the effect of dataset on AAO variation from nearly 19% to 1.7% (R2=0.0165). In these analyses, after excluding dataset-specific effects, the percent variation attributable to APOE was slightly lower at 3.6% (R2=0.0523), the effect attributable to the nine LOAD risk variants was similar to before at 1.1% (R2=0.0270), and the combined contribution of both was observed to be 4.7% (R2=0.0631). Removal of the family datasets (eTable 3) did not appreciably change the variation attributable to study-specific effects (R2=0.221), nor did it substantially change the relative effects of APOE and the 9 LOAD risk variants on AAO variation.
DISCUSSION
Our analysis of more than 9,000 LOAD cases with age-at-onset information is the largest genetic study of LOAD AAO to date. Examining AAO associations at LOAD risk loci, we confirmed the association of APOE region variation with AAO and found additional strong associations with AAO among variants at three of the other nine established risk loci (CR1, BIN1, and PICALM). Burden analysis demonstrated that the cumulative variation explained by SNPs at nine LOAD risk loci was about a third as much as the percent variation in AAO from APOE, The smaller effect of APOE ε4 on differences in AAO here (3-4%) than in previous studies (7-9%)29 may be due to differences in study design; for instance, all previous estimates were made in multiplex pedigrees, whereas most cases examined here were unrelated (2,302 of 9,162 [25.1%] of cases were from family datasets). However, in addition to confirming the predominance of the effect of APOE on AAO, we showed that the cumulative effects of risk loci associated with AAO may have an effect of similar scale on AAO. In our secondary analysis of genome-wide association, cumulative effects on genetic burden of SNPs associated with AAO but with little or no effect on LOAD risk accounted for more variation in AAO compared to the non-APOE risk variants (2.2% vs. 1.1%), but were still dwarfed by the effects of APOE on variation in AAO (~4%).
Several previous studies have suggested potential associations of risk variants at these loci with AAO. A recent study using a small subset of the cases used in this study (ADC1-3; n=2,569) identified an association with a PICALM risk variant (rs3851179, P=0.008.6×10−3).10 A study of the expression of the 10 LOAD risk genes in parietal lobe neurons from an autopsy series of AD brains demonstrated nominally significant evidence of an association between reduced BIN1 expression levels and earlier AAO (P=0.041),30 as well as an association with a longer duration of disease. A study by Jones et al. (2013)31 among persons with Down Syndrome, which is typically associated with elevated AD risk at an earlier AAO, showed risk variants in APOE (P=0.014) and PICALM (P=0.011) to be correlated with lower AAO in AD patients with Down syndrome.
Daw et al analyzed families with a high burden of AD and later age of onset in a multiplex family data set29 and found evidence for at least four additional genes with major effects on variation in AAO as large as those of APOE. The lack of major AAO-modifying effects outside of APOE in our study is not consistent with the Daw et al study and may reflect genetic heterogeneity of age-at-onset genetics within late-onset AD or, more likely, indicate the existence of large effect modifiers enriched in families with multiple affected members. APOE-related survival effects may have further complicated the identification of AAO-modifying genes. Furthermore, other genetic mechanisms including the effects of rare variants, epigenetic modification, and gene-environment interactions, which have been reported to influence dementia risk and cognitive decline,32-37 may also contribute to variation in age at onset of AD. Identification of other genetic modifiers of age-at-onset through studies of larger samples of LOAD cases and studies using next-generation sequencing approaches which can more thoroughly interrogate the genome may yet yield additional genetic risk factors that influence age-at-onset and provide new insights into LOAD pathogenesis.
Supplementary Material
Figure 1. Boxplots for age-at-onset (AAO) by ADGC dataset.
Acknowledgements
The National Institutes of Health, National Institute on Aging (NIH-NIA) supported multiple aspects of this work through the following grants: design and conduct of the study; centralized collection, management, analysis, and interpretation of the data; and preparation, review, and approval of the manuscript by the ADGC were funded by U01 AG032984 and RC2 AG036528; preliminary data collection and management, preliminary analyses, and approval of the manuscript by the individual component studies and datasets were funded for each member group as follows: NACC, U01 AG016976; NCRAD, U24 AG021886; NIA LOAD, U24 AG026395, R01AG041797, U24 AG026390; Banner Sun Health Research Institute P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01 AG025259, R01AG33193; Columbia University, P50 AG008702, R37 AG015473; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, UO1 AG06781, UO1 HG004610; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, MO1RR00096, UL1 RR029893, 5R01AG012101, 5R01AG022374, 5R01AG013616, 1RC2AG036502, 1R01AG035137; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG30146; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582, UL1RR02777; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573, P50, P50 AG016575, P50 AG016576, P50 AG016577; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383, AG05144; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653, AG041718; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991. The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by NINDS grant # NS39764, NIMH MH60451 and by Glaxo Smith Kline. Genotyping of the TGEN2 cohort was supported by Kronos Science. The TGen series was also funded by NIA grant AG034504 to AJM, The Banner Alzheimer’s Foundation, The Johnnie B. Byrd Sr. Alzheimer’s Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council, local NHS trusts and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council),South West Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England (HEFCE), Alzheimer’s Research Trust (ART), BRACE as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), The Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, Brain Net Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, Universitat de Barcelona. ADNI Funding for ADNI is through the Northern California Institute for Research and Education by grants from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, the Dana Foundation, and by the National Institute of Biomedical Imaging and Bioengineering and NIA grants U01 AG024904, RC2 AG036535, K01 AG030514. We thank Drs. D. Stephen Snyder and Marilyn Miller from NIA who are ex-officio ADGC members. Support was also from the Alzheimer’s Association (LAF, IIRG-08-89720; MP-V, IIRG-05-14147) and the US Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. P.S.G.-H. is supported by Wellcome Trust, Howard Hughes Medical Institute, and the Canadian Institute of Health Research.
Alzheimer’s Disease Genetics Consortium Coauthors*
Marilyn S. Albert, Ph.D.1, Roger L. Albin, M.D.2-4, Liana G. Apostolova, M.D.5, Steven E. Arnold, M.D.6, Robert Barber, Ph.D.7, Lisa L. Barnes, Ph.D.8,9, Thomas G. Beach, M.D., Ph.D.10, James T. Becker, Ph.D.11, Duane Beekly, B.S.12, Eileen H. Bigio, M.D.13,14, James D. Bowen, M.D.15, Adam Boxer, M.D., Ph.D.16, James R. Burke, M.D., Ph.D.17, Nigel J. Cairns, Ph.D., FRCPath18, Laura B. Cantwell, MPH19, Chuanhai Cao, Ph.D.20, Chris S. Carlson, Ph.D.21, Regina M. Carney, M.D.22, Minerva M. Carrasquillo, Ph.D.23, Steven L. Carroll, M.D., Ph.D.24, Helena C. Chui, M.D.25, David G. Clark, M.D.26, Jason Corneveaux, B.S.27, David H. Cribbs, Ph.D.28, Elizabeth A. Crocco, M.D.22, Charles DeCarli, M.D.29, Steven T. DeKosky, M.D.30, Malcolm Dick, Ph.D.31, Dennis W. Dickson, M.D.23, Ranjan Duara, M.D.32, Kelley M. Faber, M.S.33, Kenneth B. Fallon, M.D.24, Martin R. Farlow, M.D.34, Steven Ferris, Ph.D.35, Matthew P. Frosch, M.D., Ph.D.36, Douglas R. Galasko, M.D.37, Mary Ganguli, M.D.38, Marla Gearing, Ph.D.39,40, Daniel H. Geschwind, M.D., Ph.D.41, Bernardino Ghetti, M.D.42, John R. Gilbert, Ph.D.43,44, Sid Gilman, M.D., FRCP2, Jonathan D. Glass, M.D.45, John H. Growdon, M.D.46, Ronald L. Hamilton, M.D.47, Lindy E. Harrell, M.D., Ph.D.26, Elizabeth Head, Ph.D.48, Lawrence S. Honig, M.D., Ph.D.49, Christine M. Hulette, M.D.50, Bradley T. Hyman, M.D., Ph.D.46, Gregory A. Jicha, M.D., Ph.D.51, Lee-Way Jin, M.D., Ph.D.52, Anna Karydas, B.A.16, Jeffrey A. Kaye, M.D.53,54, Ronald Kim, M.D.55, Edward H. Koo, M.D.37, Neil W. Kowall, M.D.56,57, Joel H. Kramer, PsyD58, Frank M. LaFerla, Ph.D.59, James J. Lah, M.D., Ph.D.45, James B. Leverenz, M.D.60, Allan I. Levey, M.D., Ph.D.45, Ge Li, M.D., Ph.D.61, Andrew P. Lieberman, M.D., Ph.D.62, Chiao-Feng Lin, Ph.D.19, Oscar L. Lopez, M.D.63, Constantine G. Lyketsos, M.D., MPH64, Wendy J. Mack, Ph.D.65, Daniel C. Marson, J.D., Ph.D.26, Frank Martiniuk, Ph.D.66, Deborah C. Mash, Ph.D.67, Eliezer Masliah, M.D.37,68, Wayne C. McCormick, M.D., MPH69, Susan M. McCurry, Ph.D.70, Andrew N. McDavid, B.A.21, Ann C. McKee, M.D.56,57, Marsel Mesulam, M.D.14,71, Bruce L. Miller, M.D.16, Carol A. Miller, M.D.72, Joshua W. Miller, Ph.D.52, Jill R. Murrell, Ph.D.33,42, John M. Olichney, M.D.29, Vernon S. Pankratz, Ph.D.73, Joseph E. Parisi, M.D.74,75, Elaine Peskind, M.D.61, Ronald C. Petersen, M.D., Ph.D.76, Aimee Pierce, M.D.28, Wayne W. Poon, Ph.D.31, Huntington Potter, Ph.D.20, Joseph F. Quinn, M.D.53, Ashok Raj, M.D.20, Murray Raskind, M.D.61, Barry Reisberg, M.D.35,77, John M. Ringman, M.D.5, Erik D. Roberson, M.D., Ph.D.26, Howard J. Rosen, M.D.16, Roger N. Rosenberg, M.D.78, Mary Sano, Ph.D.79, Lon S. Schneider, M.D.25,80, William W. Seeley, M.D.16, Amanda G. Smith, M.D.20, Joshua A. Sonnen, M.D.60, Salvatore Spina, M.D.42, Robert A. Stern, Ph.D.56, Rudolph E. Tanzi, Ph.D.46, Tricia A. Thornton-Wells, Ph.D.81, John Q. Trojanowski, M.D., Ph.D.19, Juan C. Troncoso, M.D.82, Otto Valladares, M.S.19, Vivianna M. Van Deerlin, M.D., Ph.D.19, Linda J. Van Eldik, Ph.D.83, Badri N. Vardarajan, Ph.D.84, Harry V. Vinters, M.D.5,85, Jean Paul Vonsattel, M.D.86, Sandra Weintraub, Ph.D.14,87, Kathleen A. Welsh-Bohmer, Ph.D.17,88, Jennifer Williamson, M.S.49, Sarah Wishnek, MPH43, Randall L. Woltjer, M.D., Ph.D.89, Clinton B. Wright, M.D., M.S.90, Steven G. Younkin, M.D., Ph.D.23, Chang-En Yu, Ph.D.69, Lei Yu, Ph.D.8
Alzheimer’s Disease Genetics Consortium Coauthor Affiliations
1Department of Neurology, Johns Hopkins University, Baltimore, Maryland; 2Department of Neurology, University of Michigan, Ann Arbor, Michigan; 3Geriatric Research, Education and Clinical Center (GRECC), VA Ann Arbor Healthcare System (VAAAHS), Ann Arbor, Michigan; 4Michigan Alzheimer Disease Center, Ann Arbor, Michigan; 5Department of Neurology, University of California Los Angeles, Los Angeles, California; 6Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania; 7Department of Pharmacology and Neuroscience, University of North Texas Health Science Center, Fort Worth, Texas; 8Department of Neurological Sciences, Rush University Medical Center, Chicago, Illinois; 9Department of Behavioral Sciences, Rush University Medical Center, Chicago, Illinois; 10Civin Laboratory for Neuropathology, Banner Sun Health Research Institute, Phoenix, Arizona; 11Departments of Psychiatry, Neurology, and Psychology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania; 12National Alzheimer’s Coordinating Center, University of Washington, Seattle, Washington;13Department of Pathology, Northwestern University Feinberg School of Medicine, Chicago, Illinois; 14Cognitive Neurology and Alzheimer’s Disease Center, Northwestern University, Chicago, Illinois; 15Swedish Medical Center, Seattle, Washington; 16Department of Neurology, University of California San Francisco, San Francisco, California; 17Department of Medicine, Duke University, Durham, North Carolina; 18Department of Pathology and Immunology, Washington University, St. Louis, Missouri; 19Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania; 20USF Health Byrd Alzheimer’s Institute, University of South Florida, Tampa, Florida; 21Fred Hutchinson Cancer Research Center, Seattle, Washington; 22Department of Psychiatry and Behavioral Sciences, Miller School of Medicine, University of Miami, Miami, Florida; 23Department of Neuroscience, Mayo Clinic, Jacksonville, Florida; 24Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama; 25Department of Neurology, University of Southern California, Los Angeles, California; 26Department of Neurology, University of Alabama at Birmingham, Birmingham, Alabama; 27Neurogenomics Division, Translational Genomics Research Institute, Phoenix, Arizona; 28Department of Neurology, University of California Irvine, Irvine, California; 29Department of Neurology, University of California Davis, Sacramento, California; 30University of Virginia School of Medicine, Charlottesville, Virginia; 31Institute for Memory Impairments and Neurological Disorders, University of California Irvine, Irvine, California; 32Wien Center for Alzheimer’s Disease and Memory Disorders, Mount Sinai Medical Center, Miami Beach, Florida; 33Department of Medical and Molecular Genetics, Indiana University, Indianapolis, Indiana; 34Department of Neurology, Indiana University, Indianapolis, Indiana; 35Department of Psychiatry, New York University, New York, New York; 36C.S. Kubik Laboratory for Neuropathology, Massachusetts General Hospital, Charlestown, Massachusetts; 37Department of Neurosciences, University of California San Diego, La Jolla, California; 38Department of Psychiatry, University of Pittsburgh, Pittsburgh, Pennsylvania; 39Department of Pathology and Laboratory Medicine, Emory University, Atlanta, Georgia; 40Emory Alzheimer’s Disease Center, Emory University, Atlanta, Georgia; 41Neurogenetics Program, University of California Los Angeles, Los Angeles, California; 42Department of Pathology and Laboratory Medicine, Indiana University, Indianapolis, Indiana; 43The John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida; 44Dr. John T. Macdonald Foundation Department of Human Genetics, University of Miami, Miami, Florida; 45Department of Neurology, Emory University, Atlanta, Georgia; 46Department of Neurology, Massachusetts General Hospital/Harvard Medical School, Boston, Massachusetts; 47Department of Pathology (Neuropathology), University of Pittsburgh, Pittsburgh, Pennsylvania; 48Sanders-Brown Center on Aging, Department of Molecular and Biomedical Pharmacology, University of Kentucky, Lexington, Kentucky; 49Taub Institute on Alzheimer’s Disease and the Aging Brain, Department of Neurology, Columbia University, New York, New York; 50Department of Pathology, Duke University, Durham, North Carolina; 51Sanders-Brown Center on Aging, Department Neurology, University of Kentucky, Lexington, Kentucky; 52Department of Pathology and Laboratory Medicine, University of California Davis, Sacramento, California; 53Department of Neurology, Oregon Health & Science University, Portland, Oregon; 54Department of Neurology, Portland Veterans Affairs Medical Center, Portland, Oregon; 55Department of Pathology and Laboratory Medicine, University of California Irvine, Irvine, California; 56Department of Neurology, Boston University, Boston, Massachusetts; 57Department of Pathology, Boston University, Boston, Massachusetts; 58Department of Neuropsychology, University of California San Francisco, San Francisco, California; 59Department of Neurobiology and Behavior, University of California Irvine, Irvine, California; 60Department of Pathology, University of Washington, Seattle, Washington; 61Department of Psychiatry and Behavioral Sciences, University of Washington School of Medicine, Seattle, Washington; 62Department of Pathology, University of Michigan, Ann Arbor, Michigan; 63University of Pittsburgh Alheimer’s Disease Research Center, Pittsburgh, Pennsylvania; 64Department of Psychiatry, Johns Hopkins University, Baltimore, Maryland; 65Department of Preventive Medicine, University of Southern California, Los Angeles, California; 66Department of Medicine - Pulmonary, New York University, New York, New York; 67Department of Neurology, University of Miami, Miami, Florida; 68Department of Pathology, University of California San Diego, La Jolla, California; 69Department of Medicine, University of Washington, Seattle, Washington; 70School of Nursing Northwest Research Group on Aging, University of Washington, Seattle, Washington; 71Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, Illinois; 72Department of Pathology, University of Southern California, Los Angeles, California; 73Department of Biostatistics, Mayo Clinic, Rochester, Minnesota; 74Department of Anatomic Pathology, Mayo Clinic, Rochester, Minnesota; 75Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota; 76Department of Neurology, Mayo Clinic, Rochester, Minnesota; 77Alzheimer’s Disease Center, New York University, New York, New York; 78Department of Neurology, University of Texas Southwestern, Dallas, Texas; 79Department of Psychiatry, Mount Sinai School of Medicine, New York, New York; 80Department of Psychiatry, University of Southern California, Los Angeles, California; 81Center for Human Genetics and Research, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, Tennessee; 82Department of Pathology, Johns Hopkins University, Baltimore, Maryland; 83Sanders-Brown Center on Aging, Department of Anatomy and Neurobiology, University of Kentucky, Lexington, Kentucky; 84Department of Neurology, Columbia University, New York, New York; 85Department of Pathology & Laboratory Medicine, University of California Los Angeles, Los Angeles, California; 86Taub Institute on Alzheimer’s Disease and the Aging Brain, Department of Pathology, Columbia University, New York, New York; 87Department of Psychiatry, Northwestern University Feinberg School of Medicine, Chicago, Illinois; 88Department of Psychiatry & Behavioral Sciences, Duke University, Durham, North Carolina; 89Department of Pathology, Oregon Health & Science University, Portland, Oregon; 90Evelyn F. McKnight Brain Institute, Department of Neurology, Miller School of Medicine, University of Miami, Miami, Florida
Author Contributions
Sample collection, phenotyping, and data management: J.D.Buxbaum, G.P.J., P.K.C., E.B.L., T.D.B., B.F.B., N.R.G., P.L.D., D.E., J.A.Schneider, M.M.C., N.E., S.G.Y., C.C., J.S.K.K., P.N., P.K., J.H., M.J.H., A.J.M., M.M.B., F.Y.D., C.T.B., R.C.G., E.R., P.S.G.-H., S.E.A., R.B., T.B., E.H.B., J.D.Bowen, A.B., J.R.B., N.J.C., C.S.C., S.L.C., H.C.C., D.G.C., J.C., C.W.C., J.L.C., C.D., S.T.D., R.D.-A., M.D., D.W.D., W.G.E., K.M.F., K.B.F., M.R.F., S.F., M.P.F., D.R.G., M.Ganguli, M.Gearing, D.H.G., B.Ghetti, J.R.G., S.G., B.Giordani, J.G., J.H.G., R.L.H., L.E.H., E.H., L.S.H., C.M.H., B.T.H., G.A.J., L.-W.J., N.J., J.K., A.K., J.A.K., R.K., E.H.K., N.W.K., J.J.L., A.I.L., A.P.L., O.L.L., W.J.M., D.C.Marson, F.M., D.C.Mash, E.M., W.C.M., S.M.M., A.N.M., A.C.M., M.M., B.L.M., C.A.M., J.W.M., J.E.P., D.P.P., E.P., R.C.P., W.W.P., J.F.Q., M.R., B.R., J.M.R., E.D.R., R.N.R., M.S., L.S.S., W.S., M.L.S., M.A.S., C.D.S., J.A.Sonnen, S.S., R.A.S., R.E.T., J.Q.T., J.C.T., V.M.V., H.V.V., J.P.V., S.W., K.A.W., J.W., R.L.., L.B.C., B.A.D., D.Beekly, M.I.K., A.J.S., E.M.R., D.A.B., A.M.G., W.A.K., T.M.F., J.L.H., R.M., M.A.P., L.A.F. Study management and coordination: L.B.C., D.Beekly, D.A.B., J.C.M., T.J.M., A.M.G., D.Blacker, D.W.T., H.H., W.A.K., T.M.F., J.L.H., R.M., M.A.P., L.A.F., G.D.S. Statistical methods and analysis: A.C.N., G.J., G.W.B., L.-S.W., B.N.V., J.B., P.J.G., R.M.C., R.A.R., M.A.S., K.L.L., E.R.M., J.L.H., M.A.P., L.A.F. Interpretation of results: A.C.N., G.J., G.W.B., L.-S.W., B.N.V., J.B., P.J.G., R.A.R., M.A.S., K.L.L., E.R.M., M.I.K., A.J.S., E.M.R., D.A.B., J.C.M., T.J.M., A.M.G., D.Blacker, D.W.T., H.H., W.A.K., T.M.F., J.L.H., R.M., M.A.P., L.A.F., G.D.S. Manuscript writing group: A.C.N., G.J., G.W.B., L.-S.W., B.N.V., J.B., P.J.G., J.L.H., R.M., M.A.P., L.A.F., G.D.S. Study design: D.A.B., J.C.M., T.J.M., A.M.G., D.Blacker, D.W.T., H.H., W.A.K., T.M.F., J.L.H., R.M., M.A.P.-V., L.A.F., G.D.S.
Competing Financial Interests
T.D.B. received licensing fees from and is on the speaker’s bureau of Athena Diagnostics, Inc. M.R.F. receives research funding from BristolMyersSquibb Company, Danone Research, Elan Pharmaceuticals, Inc., Eli Lilly and Company, Novartis Pharmaceuticals Corporation, OctaPharma AG, Pfizer Inc., and Sonexa Therapeutics, Inc; Receives honoraria as scientific consultant from Accera, Inc., Astellas Pharma US Inc., Baxter, Bayer Pharmaceuticals Corporation, BristolMyersSquibb, Eisai Medical Research, Inc., GE Healthcare, Medavante, Medivation, Inc., Merck & Co., Inc., Novartis Pharmaceuticals Corp., Pfizer, Inc., Prana Biotechnology Ltd., QR Pharma., Inc., The sanofi-aventis Group, and Toyama Chemical Co., Ltd.; and is speaker for Eisai Medical Research, Inc., Forest Laboratories, Pfizer Inc. and Novartis Pharmaceuticals Corporation. A.M.G. has research funding from AstraZeneca, Pfizer and Genentech, and has received remuneration for giving talks at Pfizer and Genentech. R.C.P. is on the Safety Monitory Committee of Pfizer, Inc. (Wyeth) and a consultant to the Safety Monitoring Committee at Janssen Alzheimer’s Immunotherapy Program (Elan), to Elan Pharmaceuticals, and to GE Healthcare. R.E.T. is a consultant to Eisai, Japan in the area of Alzheimer’s genetics and a shareholder in, and consultant to Pathway Genomics, Inc, San Diego, CA.
References
- 1.Bachman DL, Wolf PA, Linn R, et al. Prevalence of dementia and probable senile dementia of the Alzheimer type in the Framingham Study. Neurology. 1992 Jan;42(1):115–119. doi: 10.1212/wnl.42.1.115. [DOI] [PubMed] [Google Scholar]
- 2.Evans DA, Funkenstein HH, Albert MS, et al. Prevalence of Alzheimer’s disease in a community population of older persons. Higher than previously reported. Jama. 1989 Nov 10;262(18):2551–2556. [PubMed] [Google Scholar]
- 3.Canadian Study of Health and Aging Working Group Canadian study of health and aging: study methods and prevalence of dementia. CMAJ. 1994 Mar 15;150(6):899–913. [PMC free article] [PubMed] [Google Scholar]
- 4.Fratiglioni L, De Ronchi D, Aguero-Torres H. Worldwide prevalence and incidence of dementia. Drugs Aging. 1999 Nov;15(5):365–375. doi: 10.2165/00002512-199915050-00004. [DOI] [PubMed] [Google Scholar]
- 5.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011 May;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011 May;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YJ, Oliveira SA, Xu P, et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003 Dec 15;12(24):3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- 8.Myers RH, Schaefer EJ, Wilson PW, et al. Apolipoprotein E epsilon4 association with dementia in a population-based study: The Framingham study. Neurology. 1996 Mar;46(3):673–677. doi: 10.1212/wnl.46.3.673. [DOI] [PubMed] [Google Scholar]
- 9.Khachaturian AS, Corcoran CD, Mayer LS, Zandi PP, Breitner JC. Cache County Study I. Apolipoprotein E epsilon4 count affects age at onset of Alzheimer disease, but not lifetime susceptibility: The Cache County Study. Arch Gen Psychiatry. 2004 May;61(5):518–524. doi: 10.1001/archpsyc.61.5.518. [DOI] [PubMed] [Google Scholar]
- 10.Thambisetty M, An Y, Tanaka T. Alzheimer’s disease risk genes and the age-at-onset phenotype. Neurobiol Aging. 2013 Nov;34(11):2696–e2691-2695. doi: 10.1016/j.neurobiolaging.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JT, Li L, Yuan J, Zhang ZX. Correlation between Apolipoprotein E polymorphism and age at onset of Alzheimer’s disease in a Chinese Han population] Zhonghua yi xue za zhi. 2013 Jan 15;93(3):182–186. [PubMed] [Google Scholar]
- 12.Kwon OD, Khaleeq A, Chan W, Pavlik VN, Doody RS. Apolipoprotein E polymorphism and age at onset of Alzheimer’s disease in a quadriethnic sample. Dement Geriatr Cogn Disord. 2010;30(6):486–491. doi: 10.1159/000322368. [DOI] [PubMed] [Google Scholar]
- 13.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993 Aug 13;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 14.Blacker D, Haines JL, Rodes L, et al. ApoE-4 and age at onset of Alzheimer’s disease: the NIMH genetics initiative. Neurology. 1997 Jan;48(1):139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 15.Li YJ, Scott WK, Hedges DJ, et al. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002 Apr;70(4):985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi Y, Marchani EE, Bird TD, Steinbart EJ, Blacker D, Wijsman EM. Genome scan of age-at-onset in the NIMH Alzheimer disease sample uncovers multiple loci, along with evidence of both genetic and sample heterogeneity. Am J Med Genet B Neuropsychiatr Genet. 2011 Dec;156B(7):785–798. doi: 10.1002/ajmg.b.31220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmans P, Hamshere M, Hollingworth P, et al. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2005 May 5;135B(1):24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- 18.Dickson MR, Li J, Wiener HW, et al. A genomic scan for age at onset of Alzheimer’s disease in 437 families from the NIMH Genetic Initiative. Am J Med Genet B Neuropsychiatr Genet. 2008 Sep 5;147B(6):784–792. doi: 10.1002/ajmg.b.30689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Barral S, Cheng R, et al. Age-at-onset linkage analysis in Caribbean Hispanics with familial late-onset Alzheimer’s disease. Neurogenetics. 2008 Feb;9(1):51–60. doi: 10.1007/s10048-007-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamboh MI, Barmada MM, Demirci FY, et al. Genome-wide association analysis of age-at-onset in Alzheimer’s disease. Mol Psychiatry. 2012 Oct 18; doi: 10.1038/mp.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984 Jul;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000 Mar;66(3):1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010 Jul;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 25.R: a language and environment for statistical computing. computer program. [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010 Sep 1;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 computer program. [Google Scholar]
- 29.Daw EW, Payami H, Nemens EJ, et al. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000 Jan;66(1):196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7(11):e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones EL, Mok K, Hanney M, et al. Evidence that PICALM affects age at onset of Alzheimer’s dementia in Down syndrome. Neurobiol Aging. 2013 Oct;34(10):2441 e2441–2445. doi: 10.1016/j.neurobiolaging.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Migliore L, Coppede F. Genetics, environmental factors and the emerging role of epigenetics in neurodegenerative diseases. Mutation research. 2009 Jul 10;667(1-2):82–97. doi: 10.1016/j.mrfmmm.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Guerreiro R, Wojtas A, Bras J, et al. TREM2 Variants in Alzheimer’s Disease. N Engl J Med. 2012 Nov 14; doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonsson T, Stefansson H, Ph DS, et al. Variant of TREM2 Associated with the Risk of Alzheimer’s Disease. N Engl J Med. 2012 Nov 14; doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010 May 7;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 36.Castellano JF, Fletcher BR, Kelley-Bell B, Kim DH, Gallagher M, Rapp PR. Age-related memory impairment is associated with disrupted multivariate epigenetic coordination in the hippocampus. PLoS One. 2012;7(3):e33249. doi: 10.1371/journal.pone.0033249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayeux R, Ottman R, Maestre G, et al. Synergistic effects of traumatic head injury and apolipoprotein-epsilon 4 in patients with Alzheimer’s disease. Neurology. 1995 Mar;45(3 Pt 1):555–557. doi: 10.1212/wnl.45.3.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.