Abstract
Enterohemorrhagic E. coli (EHEC) is a food-borne pathogen that causes disease ranging from uncomplicated diarrhea to life-threatening hemolytic uremic syndrome (HUS) and nervous system complications. Shiga toxin 2 (Stx2) is the major virulence factor of EHEC and is critical for development of HUS. The genes encoding Stx2 are carried by lambdoid bacteriophages and the toxin production is tightly linked to the production of phages during lytic cycle. It has previously been suggested that commensal E. coli could amplify the production of Stx2-phages and contribute to the severity of disease. In this study we examined the susceptibility of commensal E. coli strains to the Stx2-converting phage ϕ734, isolated from a highly virulent EHEC O103:H25 (NIPH-11060424). Among 38 commensal E. coli strains from healthy children below 5 years, 15 were lysogenized by the ϕ734 phage, whereas lytic infection was not observed. Three of the commensal E. coli ϕ734 lysogens were tested for stability, and appeared stable and retained the phage for at least 10 cultural passages. When induced to enter lytic cycle by H2O2 treatment, 8 out of 13 commensal lysogens produced more ϕ734 phages than NIPH-11060424. Strikingly, five of them even spontaneously (non-induced) produced higher levels of phage than the H2O2 induced NIPH-11060424. An especially high frequency of HUS (60%) was seen among children infected by NIPH-11060424 during the outbreak in 2006. Based on our findings, a high Stx2 production by commensal E. coli lysogens cannot be ruled out as a contributor to the high frequency of HUS during this outbreak.
Keywords: EHEC, Stx2, bacteriophage lambda, lysogen, commensal E. coli
Introduction
Enterohemorrhagic Escherichia coli (EHEC) causes disease with manifestations ranging from mild diarrhea to severe illness comprising hemorrhagic colitis (HC) (Riley et al., 1983) and hemolytic uremic syndrome (HUS) (Karmali et al., 1983, 1985). The E. coli serotype O157:H7, which caused the first described EHEC outbreak in 1982 (Riley et al., 1983) is so far the best described EHEC serotype. However, non-O157:H7 serotype strains have been implicated in a number of outbreaks and sporadic cases of HC and HUS (Luna-Gierke et al., 2014). In Europe, E. coli serotypes such as O103:H25 and O104:H4 have caused severe outbreaks (Schimmer et al., 2008; Beutin and Martin, 2012).
EHEC strains possess a range of colonization and virulence factors that facilitate infection and contribute to development of disease. Shiga toxin (Stx) is recognized as one of the main virulence factors in enterohemorrhagic disease caused by E. coli and all EHEC strains produce one or both of the Shiga toxins Stx1 and Stx2 (Scotland et al., 1985). Stx2 has been shown to be far more potent (as quantified by LD50 in mice) than Stx1 and patients infected with the latter are much less likely to develop serious illness than those infected by the former (Tesh et al., 1993; Friedrich et al., 2002). After being produced in the large intestine the Stx toxin passes through the epithelial cells and is disseminated via the blood stream to the target organs. Stx binds specifically and with high-affinity to the glycosphingolipid receptor globotriaosylceramide (Gb3) which is highly expressed in kidney cells (Jacewicz et al., 1986; Hughes et al., 2000; Okuda et al., 2006; Shimizu et al., 2007; Shin et al., 2009). After binding to the receptor, Stx is translocated into the cytosol where it causes cell damage by inhibiting protein synthesis (Sandvig and van Deurs, 2000). The Stx induced cell damage appears to be central in the pathogenic events leading to HUS and occasionally chronic kidney disease (Obrig, 2010; Obrig and Karpman, 2012).
About 15 years ago, several E. coli and Shigella strains were lysogenized with labeled Stx2 phages in vitro (Schmidt et al., 1999; James et al., 2001), and successful in vivo transduction experiments with Stx derivative phages have also been reported (Acheson et al., 1998; Toth et al., 2003). When a bacterial cell is infected by an Stx-encoding phage, two different pathways are possible (Allison, 2007). During lytic infection, the phage DNA exists as a separate molecule within the cell and utilizes the host machinery to express its genes and to produce large amounts of new phage particles until the host cell bursts. The other outcome is lysogenic infection, where the phage genome is integrated into the chromosome as a prophage, and is replicated along with the host genome. The phage can remain in the lysogenic state as long as the phage genes are repressed. It has been shown in several studies that the stx2 genes are controlled by the phage late gene promoter, and that phage production is tightly linked to production of Stx toxin (Neely and Friedman, 1998; Unkmeir and Schmidt, 2000; Zhang et al., 2000; Wagner et al., 2002). Upon induction, the prophage can switch from the lysogenic state to the lytic cycle, accompanied by production of Stx and new phage particles (Herold et al., 2004; Waldor and Friedman, 2005). Several physical and chemical agents may act as prophage-inducing agents and all share the ability to activate the bacterial SOS response, mainly due to DNA damage (Kimmitt et al., 2000; Erill et al., 2007). Mitomycin C has often been used as prophage-inducing agent in studies of EHEC, however, H2O2 has been shown to be an effective prophage-inducer (Loś et al., 2009, 2010) and its presence in the gut may also increase Stx production (Wagner et al., 2001).
It has been reported that phages present in the gastrointestinal tract tend to enter the lysogenic pathway more often than the lytic pathway (Reyes et al., 2012). Factors like the number of infecting phages per bacterial cell and cell size prior to infection have been shown to influence whether the host will lyse or become lysogenic (St-Pierre and Endy, 2008). However, the mechanisms that determine the cell fate following phage-infection are complex and not fully understood.
Previous studies have shown that Stx-phages display a diverse host range, and also infect commensal E. coli (Wagner et al., 1999; Muniesa et al., 2003; Gamage et al., 2004). Gamage et al. (2004) demonstrated that commensal E. coli infected with Stx2 phages from E. coli O157:H7 were able to produce Stx2 and possibly increase the pathogenic potential of EHEC during infection. The contribution of commensal E. coli flora to Stx production was also demonstrated in a mouse model infected with E. coli O157:H7, where Stx was more commonly detected in mice colonized with E. coli sensitive to the Stx-phage than mice colonized with E. coli resistant to the Stx phage (Gamage et al., 2006). Children are particularly susceptible to EHEC infections and development of HUS (Tarr et al., 2005; Gyles, 2007). In 2006, Norway experienced a foodborne EHEC outbreak comprising 17 cases where all patients, except one (an adult aged 18), were children. The outbreak had an HUS frequency of 60%, which is extremely high, and all HUS patients were less than 9 years old (Schimmer et al., 2008). Due to the high HUS frequency, the causative strain, E.coli O103:H25 (NIPH-11060424), was considered to be particularly virulent (Schimmer et al., 2008). The strain was later shown to be closely related to the E. coli O104:H4 strain causing a large outbreak in Germany in 2011 (L' Abée-Lund et al., 2012). However, the genetic and phenotypic features underlying the extraordinary high virulence of the Norwegian outbreak strain are not yet known.
In this study, we examine the susceptibility of commensal E. coli isolates from young children to the Stx2-converting phage (ϕ734) from the 2006 Norwegian outbreak strain. We address the commensal E. coli strains sensitivity for lytic and lysogen infection and their ability to contribute to ϕ734 phage production and thereby Stx2 production.
Materials and methods
Bacterial strains and phages
The bacterial strains used in this study are listed in Table 1 and Supplementary Table 1. The commensal E. coli strains were isolated from fecal samples from healthy Norwegian children below 5 years of age in the years 2009–2014. All strains tested negative against Test Serum Anti-Coli O 103:K- and Anti-Coli O157:K- in agglutination tests (SIFIN, Germany). EHEC O103:H25 NIPH-11060424 is a highly virulent strain which caused a severe outbreak in Norway in 2006 (Schimmer et al., 2008; L' Abée-Lund et al., 2012). The phage infection experiments in this study were performed using the Stx2-converting phage ϕ734 from NIPH-11060424 (L' Abée-Lund et al., 2012) or the recombinant version of this phage (Table 1). The recombinant phage ϕ734 Cm in which stx2A is replaced by the chloramphenicol resistance gene (cat) was constructed by Dr. Muniesa, University of Barcelona, Spain, as described by Serra-Moreno et al. (2006). E. coli DH5α was used as a propagating strain for determination of phage concentration. A stable lysogen of the laboratory strain E. coli C600 carrying ϕ734 (C600:ϕ734) was created by infecting E. coli C600 with ϕ734. The lysogen was identified by PCR using the stx2 primers listed in Table 2.
Table 1.
E. coli strains and bacteriophages used in the study.
| Bacterial strains and phage | Characteristics | References |
|---|---|---|
| E. coli LABORATORY STRAINS | ||
| C600 | K-12 derivate | Appleyard, 1954 |
| DH5α | K-12 derivate | Hanahan, 1985 |
| EHEC STRAINS | ||
| NIPH-11060424 | Human isolate, Norwegian outbreak strain 2006, O103:H25. Possesses the Stx2-phage ϕ734 and the phi-like phage TL-2011b | Schimmer et al., 2008; L' Abée-Lund et al., 2012 |
| COMMENSAL E. coli ISOLATES | ||
| NVH-1034-NVH-1042, NVH-1064-NVH-1094 | Child isolates (n = 38) non-O103/O157 | This study |
| L1034-L1042, L1064-L1094 | ϕ734 Cm lysogens of the commensal E. coli isolates with corresponding numbering | |
| RECOMBINANT E. coli STRAINS | ||
| NIPH-11060424:ϕ734 Cm | ϕ734 Cm lysogen in NIPH-11060424a | This study |
| C600:ϕ734 Cm | ϕ734 Cm lysogen in C600a | This study |
| C600:ϕ734 | ϕ734 lysogen in C600a | This study |
| BACTERIOPHAGES | ||
| ϕ734 | Stx2-converting phage from NIPH-11060424 (GenBank acc no JQ011318.1) Synonyme name is TL-2011c | L' Abée-Lund et al., 2012 |
| ϕ734 Cm | ϕ734ΔstxA::cat | This study |
The strains were stable.
Table 2.
PCR primers used in the study.
| Primers | Sequence (5′-3′) | References |
|---|---|---|
| stx2 forward | GCGTTTTGACCATCTTCGT | Muniesa and Jofre, 1998 |
| stx2 reverse | ACAGGAGCAGTTTCAGACAG | Muniesa and Jofre, 1998 |
| cat forward | GGGCGAAGAAGTTGTCCATA | This study |
| cat reverse | TACACCGTTTTCCATGAGCA | This study |
| phi-phage forward | GCGGTCATGAAAACAAACCT | This study |
| phi-phage reverse | AGGCGGCAGGATTTATCAAG | This study |
Preparation of phage filtrates for phage infection experiments
E. coli strains carrying either ϕ734 or ϕ734 Cm were grown in Lysogeny broth (LB) to mid-exponential growth phase (OD600 = 0.3–0.5) and then left non-induced or induced by addition of either Mitomycin C (MMC) (0.5 μg/ml) or H2O2 (1.5 mM). The cultures were then further incubated overnight at 37°C followed by centrifugation for 10 min at 4500 rpm and sterile-filtrated using 0.22 μm filters (Millex-GP, Millipore, Bedford, MA). The phage concentration in the bacteria-free filtrate was determined by plaque assay using E. coli DH5α as a propagating strain. In order to remove any colicins, Trypsin (Sigma) was added to the phage-filtrate to a final concentration of 0.1 mg/ml followed by 1 h incubation at 37°C (Gordon and O'Brien, 2006).
Plaque assay
A plaque assay was used to determine the concentration of infective phage particles in the phage filtrates. A volume of 100 μl of phage filtrate was mixed with 900 μl of E. coli DH5α culture (OD600 = 0.3) containing 10 mM CaCl2, and then further incubated without agitation for 30 min. After incubation, the samples were mixed with 2.5 ml 0.7% LB agar and poured onto LB agar plates containing 10 mM CaCl2. The plates were incubated overnight at 37°C and plaques were counted. The phage concentration is given as plaque forming units/ml (PFU/ml).
Hybridization of plaques in E. coli DH5α lawn was performed to confirm that E. coli DH5α was susceptible to ϕ734 and ϕ734 Cm (see the Materials and Methods below). The results showed that 100% of the plaques were positive for the corresponding probe, either stx2A or cat. Thus, E. coli DH5α was used to quantify the number of phages in the phage filtrates.
Plaque hybridization
Plaque hybridization was performed according to a standard procedure (Datz et al., 1996; Sambrook and Russell, 2001) using Hybond-N+ membranes (Amersham Pharmacia Biotech). The membranes were hybridized against a DIG labeled PCR amplified probe (primers shown in Table 2). Labeling of probe and hybridization were performed using the DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instruction. The hybridization temperature used for all experiments was 56°C.
Lytic phage infection
To test for susceptibility to lytic infection, 5 μl of ϕ734 phage-filtrate was spotted on LB soft agar plates with 10 mM CaCl2, containing commensal E. coli strains, E. coli C600 or E. coli DH5α. The concentrations of the ϕ734 phage-filtrates used in the spot assay were 106 PFU/ml when propagated on NIPH-11060424 and 109 PFU/ml when propagated on E. coli C600. The LB plates were incubated overnight at 37°C. The susceptibility to lytic infection among the commensal E. coli strains were additionally tested using the plaque assay where the recipient culture had a cell density of 1 × 108 CFU/ml (OD600 = 0.3) and the ϕ734 phage concentration was either 1 × 108 PFU/ml or 1 × 105 PFU/ml, giving a multiplicity of infection (MOI) of 1 and 0.001, respectively.
Lysogen infection
The recombinant ϕ734 Cm phage was used to test commensal E. coli strains for susceptibility to lysogenic infection as described previously (Schmidt et al., 1999). The commensal E. coli strains were tested for Cm sensitivity prior to the experiment, and all strains were found sensitive. Colonies growing on LB plates containing 25 μg/ml of chloramphenicol were considered to be lysogens. Lysogens from each commensal strain were named by adding the prefix L to their wildtype number. Phage filtrate from 13 lysogens (Table 3) was prepared to examine their phage production by plaque assay using DH5α as a recipient strain. The stability of the ϕ734 Cm phage containing lysogens was tested by culturing lysogens in LB without antibiotic selection for 10 passages. After each passage, dilutions of the cultures were spread onto LB plates with chloramphenicol to examining the level of bacteria carrying ϕ734 Cm.
Table 3.
Susceptibility of 38 commensal E. coli strains to lysogenic infection by the ϕ734 Cm phage.
| E. coli isolates | ϕ734 Cm from NIPH-11060424: ϕ734 Cm MOI 0.005 | ϕ734 Cm from C600: ϕ734 Cm MOI 0.005 | ϕ734 Cm from C600: ϕ734 Cm MOI 0.5a | ϕ734 Cm from NVH-1090: ϕ734 Cm MOI 0.5 |
|---|---|---|---|---|
| NVH-1034 | −/− | −/− | −/− | −/− |
| NVH-1036 | −/− | −/− | −/− | −/− |
| NVH-1037 | −/− | −/− | 40/20 | 30/50 |
| NVH-1038 | −/− | −/− | −/− | −/− |
| NVH-1039 | −/− | −/− | −/− | −/− |
| NVH-1040 | −/− | −/− | −/− | −/− |
| NVH-1041 | −/− | −/− | −/− | −/− |
| NVH-1042 | −/− | −/− | −/− | −/− |
| NVH-1064 | −/− | 200/10 | 10/90 | 300/200 |
| NVH-1065 | −/− | −/10 | 200/600 | 20/1000 |
| NVH-1066 | −/− | 20/− | 200/30 | 30/20 |
| NVG-1067 | −/− | −/− | 200/10 | 40/100 |
| NVH-1068 | −/− | −/− | −/− | −/− |
| NVH-1069 | −/− | −/− | −/− | −/− |
| NVH-1070 | −/− | −/− | −/− | −/− |
| NVH-1071 | −/− | −/− | −/− | −/− |
| NVH-1072 | −/− | −/− | −/− | −/− |
| NVH-1073 | −/− | −/− | −/− | −/− |
| NVH-1074 | −/− | −/− | −/− | −/− |
| NVH-1075 | −/− | −/− | −/− | 500/20 |
| NVH-1076 | −/− | −/− | −/− | −/− |
| NVH-1077 | 30/100 | −/− | 1200/400 | 30/700 |
| NVH-1078 | 2000/100 | 500/2000 | 10000/8000 | 3000/8000 |
| NVH-1079 | −/− | −/− | −/− | −/− |
| NVH-1080 | −/− | −/− | −/− | −/− |
| NVH-1081 | −/− | −/− | 10/50 | 30/20 |
| NVH-1083 | −/− | −/− | −/− | −/− |
| NVH-1084 | −/− | −/− | 30/30 | −/− |
| NVH-1085 | −/− | −/− | −/− | 30/200 |
| NVH-1086 | −/− | 70/− | 50/300 | 100/20 |
| NVH-1087 | −/− | −/− | −/− | −/− |
| NVH-1088 | −/− | 10000/50000 | 10000/10000 | 8000/9000 |
| NVH-1089 | −/− | −/− | −/− | −/− |
| NVH-1090 | 40/200 | 100/400 | 4000/3000 | 3000/2000 |
| NVH-1091 | −/− | −/− | −/− | −/− |
| NVH-1092 | −/− | −/− | −/− | −/− |
| NVH-1093 | −/− | −/− | 60/40 | 30/80 |
| NVH-1094 | −/− | −/− | −/− | − |
The phage was propagated on three different strains (the original outbreak strain NIPH-11060424, the laboratory E. coli strain C600 and the commensal E. coli strain NVH1090), and two different concentrations of phages (MOI 0.005 and MOI 0.5) were used. The results are presented as the number of lysogens/ml. Two replicates were performed for all conditions.
−, no lysogens detected.
lysogens made under this condition were selected for further examination of phage production (Figure 2).
Semi-quantification of Stx2 levels by VTEC-RPLA kit
A VTEC RPLA-toxin detection kit (Oxoid Limited, Basingstoke, UK) was used to determine the Stx2 production by NIPH-11060424 and C600:ϕ734. The assay was performed according to the manufacturer's instruction. The amount of sample in each test well was reduced 2-fold at each dilution. The Stx2 titer was defined as the reciprocal of the highest dilution causing latex agglutination.
Western blot
Proteins were separated by electrophoresis using the NuPAGE Novex Bis-Tris gel systems (Invitrogen) and SeeBlue Plus2 Pre-Stained Standard (Invitrogen) as molecular weight marker. After electrophoresis, the proteins were transferred to a PVDF membrane (Millipore) according to standard protocols (Harlow and Lane, 1988). Stx2 in culture supernatants was detected using monoclonal antibodies against Stx2 (STX2-11E10, TOXIN TECHNOLOGY, INC., Sarasota, FL) diluted 1:1000. Biotin-conjugated anti-mouse antibodies from goat (Amersham Biosciences) were used as secondary antibodies (1:3000). A complex of streptavidin (Bio-Rad) and biotinylated alkaline phosphatase (Bio-Rad) was used at a dilution of 1:3000 prior to development with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Bio-Rad).
Statistical analysis
Student's t-test was used to determine significant differences between groups. A P ≤ 0.05 was considered significant.
Results
Susceptibility of commensal E. coli strains to lytic infection by the Stx2-converting phage ϕ734
Thirty-eight commensal E. coli strains were tested for susceptibility to lytic infection by ϕ734 or ϕ734 Cm propagated on either in EHEC NIPH-11060424 or on the laboratory strain E. coli C600. None of the commensal E. coli strains were susceptible to lytic infection by any of the two phages propagated on NIPH-11060424 or E. coli C600 at any of the tested concentrations. Previous studies have shown that NIPH-11060424 carries a phi-like phage (TL-2011b) in addition to the Stx2 phage (L' Abée-Lund et al., 2012). This phi-like phage is 53% identical to bacteriophage Φ V10, a temperate phage that specifically infects E. coli of serogroup O157:H7 (Perry et al., 2009). TL-2011b was shown by spot assay and following hybridization using a phi-phage specific probe to infect E. coli of serogroup O103:H25, while none of the commensal strains tested were susceptible for lytic infection by this phage (Supplementary Table 2). This indicates that phage TL-2011b is serotype specific.
Susceptibility of commensal E. coli strains to lysogenic infection by the Stx2-converting phage ϕ734 Cm
A total of 15 out of 38 (39%) commensal E. coli isolates were susceptible to lysogenic infection by ϕ734 Cm (Table 3). The number of lysogenic cells recovered varied considerably, from 10 CFU/ml to 104 CFU/ml, between the different isolates. Two of the tested isolates (E. coli NVH-1078 and E. coli NVH-1088) seemed particularly susceptible to the ϕ734 Cm phage. The bacterial host in which the phage was produced also influenced the lysogenicity, as 8% (3/38) of the commensal isolates were susceptible to lysogenic infection by ϕ734 Cm propagated on NIPH-11060424 while 18% (7/38) was susceptible to ϕ734 Cm propagated on C600 when the multiplicity of infection were the same (MOI of 0.005) (Table 3). Within isolates, the number of lysogens increased with increasing phage concentration. The number of strains susceptible to ϕ734 Cm propagated on C600 increased from 18 to 34% (13/38) when the MOI was increased from 0.005 to 0.5 (Table 3). When the commensal isolates were infected with ϕ734 Cm, propagated on the commensal lysogenic E. coli strain 1090 at an MOI of 0.5, the number of strains susceptible to lysogenic infection increased to 39% (15/38) (Table 3).
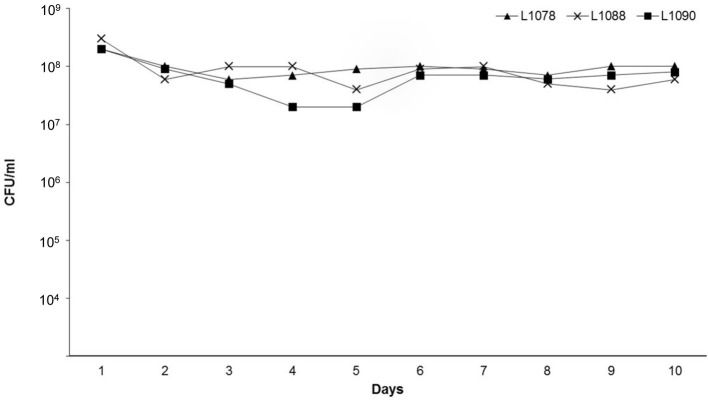
The level of Cm resistant colonies remained constant during all cultural passages of the three lysogens (L1078, L1088, and L1090) that were tested for stability (Figure 1). This shows that ϕ734 Cm was stably maintained in the commensal hosts.
Figure 1.
Stability of the commensal E. coli lysogens L1078, L1088, and L1090 during 10 cultural passages in LB broth without chloramphenicol. After each passage, the bacterial cultures were examined for loss of prophage by determining the number of Cm resistant colonies (CFU/ml).
Phage production by ϕ734 Cm lysogens under non-induced conditions and following treatment with MMC or H2O2
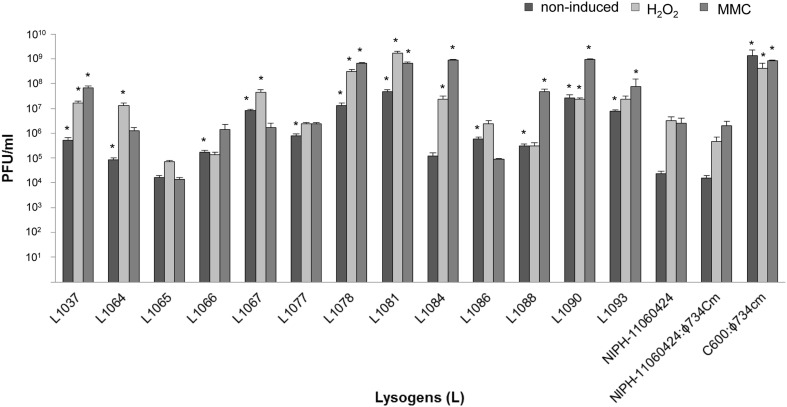
The 13 commensal E. coli isolates that were susceptible to lysogenic infection by ϕ734 Cm propagated on E. coli C600 were selected for further studies (Table 3). These isolates were tested for phage production during spontaneous (non-induced) prophage induction and after induction with mitomycin C (MMC) or H2O2 (Figure 2). There was no difference in phage production between NIPH-11060424 carrying the original ϕ734 phage and NIPH-11060424 carrying ϕ734 Cm. Twelve out of 13 commensal lysogens (L1037, L1064, L1066, L1067, L1077, L1078, L1081, L1084, L1086, L1088, L1090, and L1093) produced significantly more phages than NIPH-11060424 under one or more of the tested conditions. The remaining commensal lysogen (L1065) produced less Stx2-phages compared to NIPH-11060424. The differences between non-induced and induced phage production (either by MMC or H2O2) were less than 2 log for all lysogens except L1084, which showed one of the highest MMC induced phage productions (109 phages/ml). The non-induced culture of NIPH-11060424:ϕ734 Cm produced about 2 log less phages than the MMC or H2O2 induced cultures, which produced approximately equal numbers of phages. All the commensal E. coli lysogens produced more than 104 phages/ml without induction, and L1081 and L1090 produced nearly as much as 108 phages/ml in the non-induced cultures (Figure 2). Three lysogens (L1065, L 1067, and L1086) produced either equal amounts or more phages in the non-induced cultures than in the MMC induced cultures. These lysogens also showed 1–2 log greater phage production after induction with H2O2 than with MMC. Prior to the experiments, all the commensal E. coli strains were tested for the ability to produce phages after MMC induction by testing the culture filtrates in plaque assay (data not shown). Three of the commensal stains (NVH-1064, NVH-1077, and NVH-1086) carried MMC inducible phages naturally, of which none were of Stx type. The level of phage production in these strains was negligible (<103 PFU/ml) compared to the phage production after ϕ734 Cm infection (>105). Furthermore, the naturally carried phages formed larger plaques compared to the characteristic pin-point plaques formed by the Stx2-phages (results not shown) which made them easy to exclude when counting plaques formed by ϕ734.
Figure 2.
Bar chart showing Stx2-phage production by NIPH-11060424, NIPH-11060424:ϕ734 Cm, C600:ϕ734 Cm and 13 commensal E. coli ϕ734 Cm lysogens under non-induced, MMC induced or H2O2 induced conditions. The error bars represent the standard error of the mean (SEM) of three independent experiments. An asterisk indicates statistical significant difference (P < 0.05) in phage titer from lysogen compared to corresponding phage titer from NIPH-11060424.
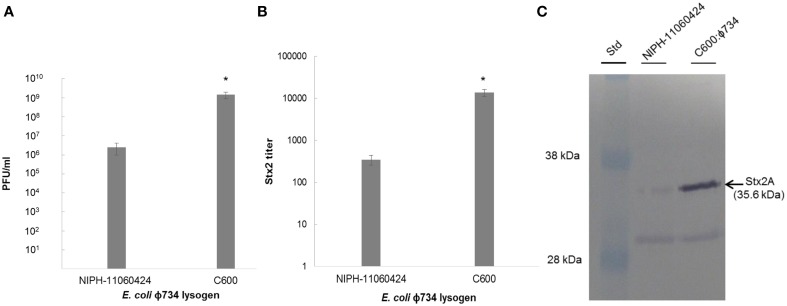
Phage production and Stx2 expression by E. coli C600:ϕ734
While the production of phages was approximately 3 log higher in C600:ϕ734 than in NIPH-11060424 after MMC induction (Figure 3A), the Stx2 titer indicated that Stx2 production was 40 times higher in E. coli C600:ϕ734 than in NIPH-11060424 (Figure 3B). Western blot analysis of the phage filtrates confirmed the high Stx2 production by C600:ϕ734 (Figure 3C).
Figure 3.
Phage production and Stx2 expression by NIPH-11060424 and E. coli C600: ϕ734 after MMC induction. (A) Phage production measured as plaque forming units. (B) Stx2 titer measured by reverse passive latex agglutination. (C) Stx2 production visualized by Western blot. The arrow indicates the Stx2A band. The error bars represent the standard error of the mean (SEM) of three independent experiments. An asterisk indicates statistical significant difference (P < 0.05) in phage production and Stx2 expression between C600:ϕ734 and NIPH-11060424.
Discussion
Children are usually more susceptible to EHEC infections and development of HUS than other groups. While some individuals exposed to the bacteria become ill others carry the bacteria asymptomatically, and the reason for this is still unknown. There is increasing evidence that commensal E. coli strains infected with Stx2-converting phages can contribute to Stx production in the intestine, and thereby increase the pathogenicity during EHEC infection (Gamage et al., 2003, 2004, 2006; Toth et al., 2003; Cornick et al., 2006). In this report, we provide results which suggest that some commensal E. coli have the potential to be significant producers of Stx and could have contributed to the extraordinary pathogenicity of strain NIPH-11060424 during the Norwegian 2006 EHEC outbreak.
We showed that 39% of commensal E. coli isolates from children were susceptible to lysogenic infection by a chloramphenicol resistant derivative of ϕ734. No lytic infection of the commensal E. coli isolates was observed which is consistent with the low rate of lytic infection by Stx2-encoding phages observed in other studies (Schmidt et al., 1999; James et al., 2001; Gamage et al., 2004; Reyes et al., 2012). The lysogenic infection rate observed here is comparable to the rates reported in other studies (Gamage et al., 2004). Gamage et al. (2004) found that 35% of E. coli isolates were susceptible to lysogenic infection by the Stx2-converting phage W933. The E. coli isolates tested in that study were of both clinical and non-clinical origin from animals and humans, and were therefore distinct from our study population. Recently, Tozzoli et al. (2014) showed that E. coli isolates representing the main E. coli pathogroups [enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAggEC) and extraintestinal pathogenic E. coli (ExPEC)] were susceptible to infection by Stx2- phages. However, in contrast to the commensal E. coli isolates studied here, the pathogenic E. coli strains were only able to carry the Stx2-phages transiently (Tozzoli et al., 2014).
In accordance with other studies, we observed that an increased MOI resulted in an increased formation of lysogens (Zeng and Golding, 2011). However, we also observed that the strain used for ϕ734 Cm phage production influenced the susceptibility of the recipient strain to lysogenic infection. When ϕ734 Cm was produced in either E. coli strain C600 or L1090 it seemed to tolerate a broader host range compared to when it was produced in NIPH-11060424 (Table 3).
Phage production by strains NIPH-11060424 and NIPH-11060424:ϕ734 Cm was very similar under all tested conditions (Figure 2), indicating that replacing stx2A with the chloramphenicol resistance gene (cat) did not influence the behavior of the phage. The selective marker was convenient in the phage experiments, as it made retrieval of lysogens more feasible, but, the recombinant phage was of course unsuitable in experiments for Stx production. Unfortunately, due to the relatively low infection rate, we were not able to isolate a commensal E.coli strain lysogenized by the wild-type ϕ734 phage. However, we were able to retrieve the ϕ734 phage in E. coli C600 (C600:ϕ734). This lysogen enabled determination of Stx2 production in another genetic background than the original EHEC outbreak strain. Under the same conditions, E. coli C600:ϕ734 produced about 1000 times more Stx2-converting phage than the original EHEC outbreak strain, and about 40 times more Stx2 (Figure 3). Stx2 measurements could not be done in the commensal E. coli lysogens, however, based on the close linkage between phage production and toxin synthesis (Neely and Friedman, 1998; Unkmeir and Schmidt, 2000; Zhang et al., 2000; Wagner et al., 2002) we assume that the number of phage produced in these lysogens will mirror the amount of Stx2 that would have been produced by the native Stx2-converting phage. A similar discrepancy between increased phage-production compared to increased Stx2 production has been shown earlier by Zhang et al. (2000), where ciprofloxacin induction of an O157:H7 strain resulted a 1000 fold increase in phage production while the Stx2 production only increased 58 fold.
The laboratory strain E. coli C600 lysogenized with ϕ734 Cm produced as much as 109 PFU/ml under non-induced conditions, which was the highest level of phage production observed during this study (Figure 2). Phage-production in the commensal E. coli ϕ734 Cm lysogens ranged from 104 to nearly 108 PFU/ml under both induced and non-induced conditions. This means that some commensal E. coli produced a considerably higher amount of Stx phage than NIPH-11060424, and also higher levels than EHEC O157:H7 EDL933, which produced about 106 PFU/ml under identical non-induced conditions (Imamovic and Muniesa, 2012). The reason why different E. coli strains lysogenized by an identical phage, produce different amounts of phage is not known. However, the amount of phages produced is most probably dependent on the genetic background of the host strain e.g., the regulation of the SOS response and the phage repressor system in each strain will have an impact on phage production.
Since the Stx-prophage induction is closely linked to activation of the bacterial SOS-response and expression of host-encoded RecA protein (Fuchs et al., 1999; Kimmitt et al., 2000), the SOS-response inducing agent MMC is frequently used to activate the phage- and Stx production in EHEC (Fuchs et al., 1999; Schmidt et al., 1999; Muniesa et al., 2004). However, H2O2 may represent a more natural inducing agent, as it is produced in the gut as part of the innate immune response (Wagner et al., 2001). Five of the commensal E. coli ϕ734 Cm lysogens demonstrated higher phage production after H2O2 induction than after MMC induction. The levels of phage production in the non-commensal isolates NIPH-11060424, NIPH-11060424:ϕ734 Cm and C600:ϕ734 Cm were similar after H2O2 and MMC induction. The strong inducing capability by H2O2 seen in the commensal E. coli lysogens may have implications for disease, as H2O2 release occurs during in vivo EHEC infection (Wagner et al., 2001). Surprisingly, we also observed high production of phage in some of the lysogens under non-induced conditions. Five of the commensal E. coli ϕ734 Cm lysogens produced a higher amount of phage non-induced, than NIPH-11060424 did under either H2O2 or MMC induced conditions.
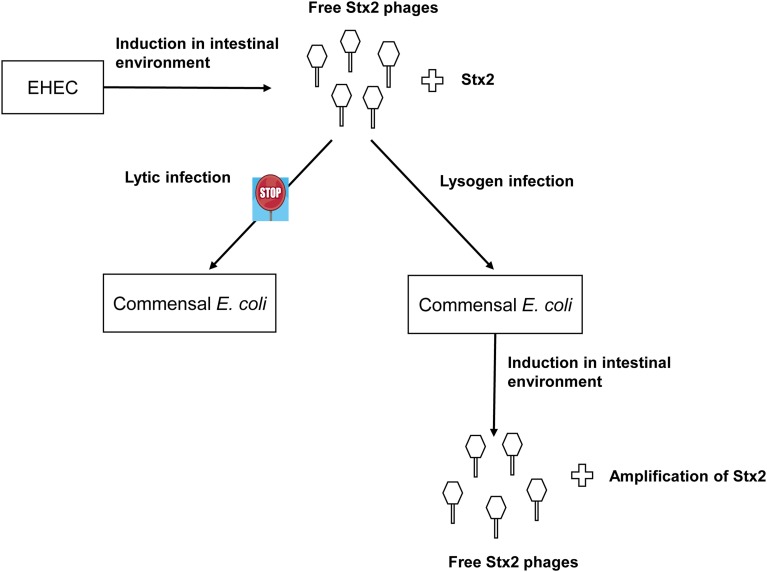
The observed lack of lytic infection by the ϕ734 phage in the commensal E. coli isolates contrasts the high level of non-induced phage production in the corresponding lysogens. However, together these results indicate that commensal E. coli strains might contribute to Stx2 production through first becoming lysogenized and then subsequently enter the lytic cycle at a high frequency (Figure 4). It has previously been reported that spontaneous induction occurs more readily in Stx-phages than in other lambdoid phages (Livny and Friedman, 2004; Aertsen et al., 2005; Shimizu et al., 2009). However, spontaneous induction to the extent observed in this study has, to our knowledge, not previously been reported.
Figure 4.
Suggested model of commensal E. coli contribution to Stx2 production in the intestinal tract. The Stx2-phage ϕ734 are produced by its EHEC host and infect susceptible commensal E. coli strains lysogenically. The commensal E. coli ϕ734 lysogens enter the lytic cycle either spontaneously or after exposure to inducing agents present in the intestinal environment. The commensal lysogens produce phages at a high frequency leading to a concomitant increase in Stx2 production.
Since efficient Stx production only occurs after prophage induction followed by lysis and death of the host cell, one may expect that EHEC carrying these phages will eventually die out. Recently, Loś et al. suggested that prophages are induced at a low frequency in the gut which does not compromise the persistence of the EHEC population (Loś et al., 2012). There are various repressor systems that interfere with phage production in strains carrying several prophages (Burz et al., 1994; Serra-Moreno et al., 2008). The lower production of phages by NIPH-11060424 compared to strain C600:ϕ734 and several of the commensal E. coli lysogens may result from the presence of repressor systems originating from other prophages in the genome of NIPH-11060424. These repressor systems may act to keep a balance between the lysogenic and lytic infection and thereby benefit the survival of the EHEC population.
In conclusion, we observed that a high proportion of commensal E. coli is susceptible to infection by ϕ734 Cm and that some isolates were infected at a higher frequency than others. The ϕ734 Cm phage infected the commensal E. coli isolates only via the lysogenic pathway. Some of the commensal E. coli lysogens produced considerably higher amounts of phage particles than EHEC NIPH-11060424. These lysogens would also likely have produced high levels of Stx2 if they were lysogenized with the original Stx2-converting phage ϕ734 as modeled in Figure 4. This study supports the hypothesis that Stx2-converting phages are able to infect commensal E. coli strains, and thereby enhance Stx2 production during EHEC infection. Together our data strongly endorse that Stx2-converting phages released from EHEC in the gut can lysogenize commensal E. coli and turn them into effective Stx producers and thus enhance the pathogenicity of the EHEC infection. Therefore, it would be interesting to examine commensal E. coli isolates from asymptomatic EHEC carriers and from EHEC triggered HUS patients for Stx phage susceptibility and for the presence of lysogenic Stx-phages.
Author contributions
All authors contributed to the design of the study, and to interpretation and analyses of the data. Hildegunn Iversen did the experiments and drafted the manuscript. Toril Lindbäck assisted in the experiments and in drafting the manuscript. Trine M. L' Abée-Lund, Lotte P. S. Arnesen and Marina Aspholm assisted in drafting the manuscript. All authors have read and approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Kristin O'Sullivan (Norwegian University of Life Sciences) for technical assistance.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fcimb.2015.00005/abstract
References
- Acheson D. W., Reidl J., Zhang X., Keusch G. T., Mekalanos J. J., Waldor M. K. (1998). In vivo transduction with shiga toxin 1-encoding phage. Infect. Immun. 66, 4496–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aertsen A., Faster D., Michiels C. W. (2005). Induction of Shiga toxin-converting prophage in Escherichia coli by high hydrostatic pressure. Appl. Enviro. Microbiol. 71, 1155–1162. 10.1128/AEM.71.3.1155-1162.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison H. E. (2007). Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2, 165–174. 10.2217/17460913.2.2.165 [DOI] [PubMed] [Google Scholar]
- Appleyard R. K. (1954). Segregation of New Lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Martin A. (2012). Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J. Food Prot. 75, 408–418. 10.4315/0362-028X.JFP-11-452 [DOI] [PubMed] [Google Scholar]
- Burz D. S., Beckett D., Benson N., Ackers G. K. (1994). Self-assembly of bacteriophage lambda cI repressor: effects of single-site mutations on the monomer-dimer equilibrium. Biochemistry 33, 8399–8405. 10.1021/bi00194a003 [DOI] [PubMed] [Google Scholar]
- Cornick N. A., Helgerson A. F., Mai V., Ritchie J. M., Acheson D. W. (2006). In vivo transduction of an Stx-encoding phage in ruminants. Appl. Enviro. Microbiol. 72, 5086–5088. 10.1128/AEM.00157-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datz M., Janetzki-Mittmann C., Franke S., Gunzer F., Schmidt H., Karch H. (1996). Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl. Environ. Microbiol. 62, 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erill I., Campoy S., Barbe J. (2007). Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31, 637–656. 10.1111/j.1574-6976.2007.00082.x [DOI] [PubMed] [Google Scholar]
- Friedrich A. W., Bielaszewska M., Zhang W. L., Pulz M., Kuczius T., Ammon A., et al. (2002). Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185, 74–84. 10.1086/338115 [DOI] [PubMed] [Google Scholar]
- Fuchs S., Muhldorfer I., Donohue-Rolfe A., Kerenyi M., Emody L., Alexiev R., et al. (1999). Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 27, 13–23. 10.1006/mpat.1999.0279 [DOI] [PubMed] [Google Scholar]
- Gamage S. D., Patton A. K., Hanson J. F., Weiss A. A. (2004). Diversity and host range of Shiga toxin-encoding phage. Infect. Immun. 72, 7131–7139. 10.1128/IAI.72.12.7131-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage S. D., Patton A. K., Strasser J. E., Chalk C. L., Weiss A. A. (2006). Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect. Immun. 74, 1977–1983. 10.1128/IAI.74.3.1977-1983.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage S. D., Strasser J. E., Chalk C. L., Weiss A. A. (2003). Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 71, 3107–3115. 10.1128/IAI.71.6.3107-3115.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. M., O'Brien C. L. (2006). Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152, 3239–3244. 10.1099/mic.0.28690-0 [DOI] [PubMed] [Google Scholar]
- Gyles C. L. (2007). Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85, E45–E62. 10.2527/jas.2006-508 [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1985). Techniques for transformation of Escherichia coli, In DNA Cloning: a Practical Approach, 1st Edn., ed Glover D. M. (McLean, VA: IRL Press; ), 109–129. [Google Scholar]
- Harlow E., Lane D. (1988). Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Herold S., Karch H., Schmidt H. (2004). Shiga toxin-encoding bacteriophages genomes in motion. Int. J. Med. Microbiol. 294, 115–121. 10.1016/j.ijmm.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Hughes A. K., Stricklett P. K., Schmid D., Kohan D. E. (2000). Cytotoxic effect of Shiga toxin-1 on human glomerular epithelial cells. Kidney Int. 57, 2350–2359. 10.1046/j.1523-1755.2000.00095.x [DOI] [PubMed] [Google Scholar]
- Imamovic L., Muniesa M. (2012). Characterizing RecA-independent induction of Shiga toxin2-encoding phages by EDTA treatment. PLoS ONE 7:e32393. 10.1371/journal.pone.0032393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacewicz M., Clausen H., Nudelman E., Donohue-Rolfe A., Keusch G. T. (1986). Pathogenesis of shigella diarrhea. XI. Isolation of a shigella toxin-binding glycolipid from rabbit jejunum and HeLa cells and its identification as globotriaosylceramide. J. Exp. Med. 163, 1391–1404. 10.1084/jem.163.6.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C. E., Stanley K. N., Allison H. E., Flint H. J., Stewart C. S., Sharp R. J., et al. (2001). Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 67, 4335–4337. 10.1128/AEM.67.9.4335-4337.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmali M. A., Petric M., Lim C., Fleming P. C., Arbus G. S., Lior H. (1985). The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151, 775–782. 10.1093/infdis/151.5.775 [DOI] [PubMed] [Google Scholar]
- Karmali M. A., Petric M., Lim C., Fleming P. C., Steele B. T. (1983). Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet 2, 1299–1300. 10.1016/S0140-6736(83)91167-4 [DOI] [PubMed] [Google Scholar]
- Kimmitt P. T., Harwood C. R., Barer M. R. (2000). Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6, 458–465. 10.3201/eid0605.000503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L' Abée-Lund T. M., Jorgensen H. J., O'Sullivan K., Bohlin J., Ligard G., Granum P. E., et al. (2012). The highly virulent 2006 Norwegian EHEC O103:H25 outbreak strain is related to the 2011 German O104:H4 outbreak strain. PLoS ONE 7:e31413. 10.1371/journal.pone.0031413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J., Friedman D. I. (2004). Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 51, 1691–1704. 10.1111/j.1365-2958.2003.03934.x [DOI] [PubMed] [Google Scholar]
- Loś J. M., Loś M., Wegrzyn A., Wegrzyn G. (2010). Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunol. Med. Microbiol. 58, 322–329. 10.1111/j.1574-695X.2009.00644.x [DOI] [PubMed] [Google Scholar]
- Loś J. M., Loś M., Wegrzyn A., Wegrzyn G. (2012). Altruism of Shiga toxin-producing Escherichia coli: recent hypothesis versus experimental results. Front. Cell. Infect. Microbiol. 2:166. 10.3389/fcimb.2012.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loś J. M., Loś M., Wegrzyn G., Wegrzyn A. (2009). Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb. Pathog. 47, 289–298. 10.1016/j.micpath.2009.09.006 [DOI] [PubMed] [Google Scholar]
- Luna-Gierke R. E., Griffin P. M., Gould L. H., Herman K., Bopp C. A., Strockbine N., et al. (2014). Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 142, 2270–2280. 10.1017/S0950268813003233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M., Blanco J. E., de S. M., Serra-Moreno R., Blanch A. R., Jofre J. (2004). Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 150, 2959–2971. 10.1099/mic.0.27188-0 [DOI] [PubMed] [Google Scholar]
- Muniesa M., Jofre J. (1998). Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl. Environ. Microbiol. 64, 2443–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M., de S. M., Prats G., Ferrer D., Panella H., Jofre J. (2003). Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71, 4554–4562. 10.1128/IAI.71.8.4554-4562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely M. N., Friedman D. I. (1998). Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28, 1255–1267. 10.1046/j.1365-2958.1998.00890.x [DOI] [PubMed] [Google Scholar]
- Obrig T. G. (2010). Escherichia coli Shiga Toxin mechanisms of action in Renal Disease. Toxins (Basel) 2, 2769–2794. 10.3390/toxins2122769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrig T. G., Karpman D. (2012). Shiga toxin pathogenesis: kidney complications and renal failure. Curr. Top. Microbiol. Immunol. 357, 105–136. 10.1007/82_2011_172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T., Tokuda N., Numata S., Ito M., Ohta M., Kawamura K., et al. (2006). Targeted disruption of Gb3/CD77 synthase gene resulted in the complete deletion of globo-series glycosphingolipids and loss of sensitivity to verotoxins. J. Biol. Chem. 281, 10230–10235. 10.1074/jbc.M600057200 [DOI] [PubMed] [Google Scholar]
- Perry L. L., SanMiguel P., Minocha U., Terekhov A. I., Shroyer M. L., Farris L. A., et al. (2009). Sequence analysis of Escherichia coli O157:H7 bacteriophage PhiV10 and identification of a phage-encoded immunity protein that modifies the O157 antigen. FEMS Microbiol. Lett. 292, 182–186. 10.1111/j.1574-6968.2009.01511.x [DOI] [PubMed] [Google Scholar]
- Reyes A., Semenkovich N. P., Whiteson K., Rohwer F., Gordon J. I. (2012). Going viral: next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 10, 607–617. 10.1038/nrmicro2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. W., Remis R. S., Helgerson S. D., McGee H. B., Wells J. G., Davis B. R., et al. (1983). Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308, 681–685. 10.1056/NEJM198303243081203 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor. [Google Scholar]
- Sandvig K., van Deurs B. (2000). Entry of ricin and Shiga toxin into cells: molecular mechanisms and medical perspectives. EMBO J. 19, 5943–5950. 10.1093/emboj/19.22.5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer B., Nygard K., Eriksen H. M., Lassen J., Lindstedt B. A., Brandal L. T., et al. (2008). Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect. Dis. 8:41. 10.1186/1471-2334-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Bielaszewska M., Karch H. (1999). Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage phi3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65, 3855–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotland S. M., Smith H. R., Rowe B. (1985). Two distinct toxins active on Vero cells from Escherichia coli O157. Lancet 2, 885–886. 10.1016/S0140-6736(85)90146-1 [DOI] [PubMed] [Google Scholar]
- Serra-Moreno R., Acosta S., Hernalsteens J. P., Jofre J., Muniesa M. (2006). Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol. Biol. 7:31. 10.1186/1471-2199-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R., Jofre J., Muniesa M. (2008). The CI repressors of Shiga toxin-converting prophages are involved in coinfection of Escherichia coli strains, which causes a down regulation in the production of Shiga toxin 2. J. Bacteriol. 190, 4722–4735. 10.1128/JB.00069-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Ohta Y., Noda M. (2009). Shiga toxin 2 is specifically released from bacterial cells by two different mechanisms. Infect. Immun. 77, 2813–2823. 10.1128/IAI.00060-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Sato T., Kawakami S., Ohta T., Noda M., Hamabata T. (2007). Receptor affinity, stability and binding mode of Shiga toxins are determinants of toxicity. Microb. Pathog. 43, 88–95. 10.1016/j.micpath.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Shin I. S., Ishii S., Shin J. S., Sung K. I., Park B. S., Jang H. Y., et al. (2009). Globotriaosylceramide (Gb3) content in HeLa cells is correlated to Shiga toxin-induced cytotoxicity and Gb3 synthase expression. BMB Rep. 42, 310–314. 10.5483/BMBRep.2009.42.5.310 [DOI] [PubMed] [Google Scholar]
- St-Pierre F., Endy D. (2008). Determination of cell fate selection during phage lambda infection. Proc. Natl. Acad. Sci. U.S.A. 105, 20705–20710. 10.1073/pnas.0808831105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr P. I., Gordon C. A., Chandler W. L. (2005). Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086. 10.1016/S0140-6736(05)71144-2 [DOI] [PubMed] [Google Scholar]
- Tesh V. L., Burris J. A., Owens J. W., Gordon V. M., Wadolkowski E. A., O'Brien A. D., et al. (1993). Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 61, 3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth I., Schmidt H., Dow M., Malik A., Oswald E., Nagy B. (2003). Transduction of porcine enteropathogenic Escherichia coli with a derivative of a shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69, 7242–7247. 10.1128/AEM.69.12.7242-7247.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzoli R., Grande L., Michelacci V., Ranieri P., Maugliani A., Caprioli A., et al. (2014). Shiga toxin-converting phages and the emergence of new pathogenic Escherichia coli: a world in motion. Front. Cell. Infect. Microbiol. 4:80. 10.3389/fcimb.2014.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkmeir A., Schmidt H. (2000). Structural analysis of phage-borne stx genes and their flanking sequences in shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68, 4856–4864. 10.1128/IAI.68.9.4856-4864.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner P. L., Acheson D. W., Waldor M. K. (1999). Isogenic lysogens of diverse shiga toxin 2-encoding bacteriophages produce markedly different amounts of shiga toxin. Infect. Immun. 67, 6710–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner P. L., Acheson D. W., Waldor M. K. (2001). Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli. Infect. Immun. 69, 1934–1937. 10.1128/IAI.69.3.1934-1937.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner P. L., Livny J., Neely M. N., Acheson D. W., Friedman D. I., Waldor M. K. (2002). Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44, 957–970. 10.1046/j.1365-2958.2002.02950.x [DOI] [PubMed] [Google Scholar]
- Waldor M. K., Friedman D. I. (2005). Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8, 459–465. 10.1016/j.mib.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Zeng L., Golding I. (2011). Following cell-fate in E. coli after infection by phage lambda. J. Vis. Exp. 56:e3363. 10.3791/3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., McDaniel A. D., Wolf L. E., Keusch G. T., Waldor M. K., Acheson D. W. (2000). Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin.production, and death in mice. J. Infect. Dis. 181, 664–670. 10.1086/315239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.