Abstract
The relationship between wound healing and cancer has long been recognized. The mechanisms that regulate wound healing have been shown to promote transformation and growth of malignant cells. In addition, chronic inflammation has been associated with malignant transformation in many tissues. Recently, pathways involved in inflammation and wound healing have been reported to enhance cancer stem cell (CSC) populations. These cells, which are highly resistant to current treatments, are capable of repopulating the tumor after treatment, causing local and systemic recurrences. In this review, we highlight proinflammatory cytokines and developmental pathways involved in tissue repair, whose deregulation in the tumor microenvironment may promote growth and survival of CSCs. We propose that the addition of anti-inflammatory agents to current treatment regimens may slow the growth of CSCs and improve therapeutic outcomes.
Keywords: cancer stem cells, inflammation, tumor microenvironment, developmental pathways
Introduction
Tumors have been described as wounds that do not heal.1 Recently, inflammatory processes that occur during normal wound healing have been linked to the pathological state of many tumors. Normal epithelial tissue exists in a state of homeostasis where tissue regeneration is tightly regulated by epithelial stem cells located within highly specialized niches. During tissue injury, replenishment of epithelial cell loss is ensured by the proliferation of these stem cells and their progeny in response to proinflammatory cytokines.2,3 Additionally, numerous potent growth factors released by macrophages and lymphocytes during wound healing promote stem cell proliferation and plasticity. Among these are morphogens that are commonly associated with embryonic development.4
Many of the factors involved in the inflammatory micro-environment have also been identified as key contributors to the cancer stem cell (CSC) niche. CSCs have been identified in a number of solid tumors and are thought to be a subpopulation of tumor cells that resemble normal stem cells and continuously undergo differentiation to populate the heterogeneous tumor. In this review, we focus on breast CSCs and highlight proinflammatory factors, which are involved in regulation of normal adult stem cells during tissue repair and have also been shown to promote survival and proliferation of CSCs in breast cancer. We propose that the presence of inflammation in the tumor microenvironment may contribute to tumor aggressiveness and treatment resistance by shifting the equilibrium between differentiated and undifferentiated tumor cells toward a CSC phenotype.
Wound Healing, Chronic Inflammation, and the Tumor Microenvironment
Wound healing is a dynamic process that consists of an inflammatory phase followed by epithelial cell proliferation and tissue remodeling. In normal tissue, the inflammatory phase is limited, lasting only 3–14 days. Tissue injury induces immediate recruitment of neutrophils, which are later replaced by macrophages and lymphocytes. Infiltrating leukocytes play a major role in secretion of inflammatory cytokines, growth factors, and chemokines, which stimulate proliferation of progenitor cells and recruitment of keratinocytes and endothelial cells during the proliferative phase of wound healing.2 At this stage, granulation tissue forms, angiogenesis is induced, and new extracellular matrix (ECM) is secreted. Epithelial cells undergo epithelial–mesenchymal transition (EMT) and migrate to the edges of the wound to impart re-epithelialization of the damaged tissue. In the final phase of wound healing, the maturation phase, wound contraction, and differentiation of fibroblasts to myofibroblasts result in the formation of scar tissue. Failure to exit the inflammatory stage results in improper tissue remodeling and is associated with impaired wound healing in many disorders including diabetes mellitus, pressure necrosis, and vasculitis.5
Likewise, chronic inflammation has been linked to tumorigenesis, tumor progression, and metastasis in many different cancers. The tumor microenvironment shares many features of a chronic wound. Infiltrating leukocytes present within the tumor and associated stroma stimulate tumor growth, invasion, and angiogenesis. Tumor-associated macrophages (TAM), tumor-associated dendritic cells (TADC), and tumor infiltrating lymphocytes are a source of proinflammatory mediators in the tumor microenvironment. Environmental stimuli, such as hypoxia and DNA damaging agents, elicit secretion of chemokines from tumor cells that recruit pro-tumor inflammatory cells and help to shape the pro-tumor immune response.6 Wound healing responses associated with tissue injury have been shown to promote the growth of breast cancer (Table 1). It has recently been shown that injection of wound fluid close to the tumor site in mice bearing syngeneic breast cancer xenografts resulted in increased tumor growth.7 Additionally, acute tissue injury associated with biopsy in rodent models of breast cancer is reported to promote lung metastasis.8 It has recently been recognized that the abundance of inflammatory cytokines and growth factors that occur during post-therapy wound healing can also contribute to survival and expansion of resistant CSCs.9–11
Table 1.
Comparison of signaling pathways involved in wound healing and cancer.
| PATHWAY | FUNCTION IN WOUND HEALING | FUNCTION IN CANCER |
|---|---|---|
| Hedgehog | *Regulation of stem cell proliferation and plasticity | *Regulation of cancer stem cell proliferation and plasticity |
| *Induction of EMT to promote epithelial cell migration | *Induction of EMT in cancer cells | |
| *Angiogenesis | *Angiogenesis | |
| *Re-epithelialization and wound closure | *Chemoresistance | |
| Notch | *Cell proliferation and migration of endothelial cells, keratinocytes, and fibroblasts at wound site | *Survival and proliferation of cancer stem cells |
| *Recruitment of inflammatory cells to wound site | ||
| Wnt | *Migration of epithelial cells | *Self-renewal of the cancer stem cells |
| *Self-renewal of stem cells | *Cancer stem cell plasticity | |
| *Progenitor cell plasticity | *Contributes to radioresistance of cancer stem cells | |
| *Angiogenesis | *Angiogenesis | |
| TGF-β | *Inflammatory cell infiltration | *Expansion of cancer stem cell population |
| *Fibroblast proliferation | *Fibroblast proliferation | |
| *Re-epithelialization and wound contraction | *Regulator of EMT in cancer cells | |
| *Induction of EMT | ||
| IL-1 β/iL-6/STAT3 | *Recruitment of inflammatory cells to wound site | *Self-renewal of cancer stem cells |
| *Proliferation of progenitor cells | *Cancer stem cell plasticity | |
| *Initiates re-epithelialization and wound closure through activation of developmental pathways | *Tumor growth and metastasis | |
| *Regulation of genes in cancer progression | ||
| IL-8 | *Chemoattractant to recruit inflammatory cells to wound site | *Cancer stem cell proliferation |
| *Chemoattractant to recruit inflammatory cells to tumor microenvironment |
CSCs and Treatment Resistance
CSCs are a subpopulation of cells within a tumor that share features with normal stem cells in which they have the ability to self-renew and differentiate into multiple lineages.12,13 Initially described in leukemia,14,15 CSCs have been identified in a variety of solid cancers including brain,16 breast,17 prostate,18 pancreas,19 melanoma,20 and colon.21–23 Injections of small numbers of CSCs into immunocompromised mice resulted in formation of heterogeneous tumors, suggesting that these cells are the tumor-initiating population.17 According to the CSC theory, tumors, like normal tissue, are organized in a hierarchy with a small number of CSCs at the top. Only the CSC population has the ability to initiate tumors and create differentiated progeny, which lack the ability to do so, and thus contribute to the heterogeneity of tumors. This is believed to be a dynamic process that is regulated by epigenetic events24 and may be induced by inflammatory factors25 or cellular stress.26 Thus, the number of CSCs present within the tumor is highly variable and influenced by the tumor microenvironment.
The presence of CSCs in breast cancer specimens has been associated with poor prognosis. Higher levels of CSCs, identified by the phenotype CD44high/CD24−/low/ESA+,27,28 and high expression of the enzyme aldehyde dehydrogenase (ALDH)29 predict poor clinical outcome for breast cancer patients.30–32 Investigation of patients with surgically resected invasive ductal carcinoma after chemotherapy showed that patients with CD44high/CD24−/low tumor cells had shorter cumulative disease-free and overall survival, suggesting that this population may be an important factor of malignant relapse in these patients.33
Expression of CSC markers in breast cancer is predictive of metastasis.34,35 Breast CSCs have been linked to metastasis through enhanced expression of EMT proteins. Investigation of gene expression patterns of CD44high/CD24−/low cells revealed upregulated expression of genes associated with EMT compared to non-stem like cells.36 Overexpression of the EMT-associated protein Twist, or treatment with TGF-β, a known inducer of EMT, leads to an increased number of cells bearing CSC phenotypes in non-tumorigenic human mammary epithelial cells.37 Likewise, inhibition of EMT has been shown to reduce the number of CD44high/CD24−/low cells as well as limit self-renewal properties in mammosphere forming assays.38 Therefore, a higher amount of EMT-related proteins may promote increases in the breast CSC population and pose a greater risk of breast cancer metastasis.
Growing evidence suggests that breast CSCs are chemo-and radioresistant. CD44high/CD24−/lowESA+ breast cancer cells were found to be more resistant to chemotherapeutic agents, such as paclitaxel and 5-fluorouracil, compared to the bulk of tumor cells.39 Chemotherapy has been found to be less effective on breast CSCs because of multiple mechanisms that include overexpression of ATP-binding cassette transporters (eg, ABCG2), upregulation of multidrug resistance transporters (eg, MDR1), and enhanced ability to survive and repair damaged DNA.32,40–42 The size of the breast CSC population may therefore determine chemotherapeutic sensitivity. Expression of CSC markers has been shown to be predictive of response to neoadjuvant chemotherapy in breast cancer patients, further confirming the resistance of this population to standard chemotherapy.
The breast CSC population has also been shown to be resistant to radiation therapy.40,43 CD44high/CD24−/low breast CSCs have an increased ability to scavenge free-radicals and overcome oxidative stress.44,45 Although radiation preferentially kills non-tumorigenic cells resulting in the enrichment of CSCs,40 the increase in the number of breast CSCs following ionizing radiation is not fully explained by preferential killing of non-tumorigenic cells.46,47 It has been proposed that differentiated cancer cells may acquire stem cell traits under certain microenvironmental stressors.48,49 Radiation-induced re-programming of CSCs has been observed in vitro;47,50 however, in vivo conversion of non-stem cells into CSCs may require a complex network of signals from the microenvironment and signaling changes within the cancer cells.
The initiation and progression of breast cancer rely on changes within the malignant epithelial cells as well as the tumor microenvironment. Normal and tumorigenic non-stem cells have the ability to spontaneously convert to a stem-like state.51 In addition, it has been shown that in vivo, differentiated cells can de-differentiate/re-program spontaneously under the right microenvironment conditions.52 Conversion of non-stem cells to CSCs either occurring spontaneously or therapy induced is tightly regulated by the tumor microenvironment. Thus, changes in the cellular populations or signals within the tumor microenvironment may drive proliferation and survival of the CSC population, promoting tumorigenicity, tumor recurrence following treatment, and metastasis.
Inflammatory Cytokines in Wound Healing and CSCs
Inflammatory cytokines play an important role in wound healing and tissue repair. Cytokines such as interleukin (IL)-1β, IL-6, and IL-8 are strongly upregulated during the inflammatory phase of wound healing. They help to initiate re-epithelialization and wound closure through activation of developmental pathways.3 These cytokines have also been reported to be present in the tumor microenvironment and contribute to tumor growth and increased stemness of tumor cells.
IL-1β is one of the first cytokines to be released during response to tissue injury. It is induced by platelet aggregation and leads to the recruitment of inflammatory cells to the site of tissue injury.3,53 IL-1β signaling stimulates the production of IL-6 and IL-8 in inflammatory cells via an NF-κβ-dependent pathway.54 Activation of IL-6 and IL-8 signaling pathways results in phosphorylation and nuclear activation of signal transducer and activator of transcription 3 (STAT3), which is a key regulator of genes involved in tissue repair and cancer progression.55 Levels of IL-6 and IL-8 at the injury site increase exponentially during the initial phases of wound healing and decrease following re-epithelialization.53 Elevated levels of IL-1β, IL-6, and IL-8 have been associated with chronic inflammation in many disorders including rheumatoid arthritis,56 sepsis,57 cardiovascular disease,58 and diabetes.59,60 IL-8 serves as a potent chemoattractant and is required for the recruitment of inflammatory cells to the site of injury.61 IL-6 also plays a role in macrophage recruitment and is important for wound closure and re-epithelialization.62 In addition, IL-6 knock-out mice demonstrate impaired wound healing. Punch biopsies in these mice took up to three times longer period to heal than those of wild-type mice and exhibited delayed reepi-thelialization.63,64 Likewise, targeted knock-down of STAT3 resulted in impaired proliferation of epidermal progenitor cells and delayed wound healing, resembling that of the IL-6 knockouts.65
In breast cancer, high levels of IL-6 and IL-8 are associated with poor prognosis.66,67 Activation of these signaling pathways within the tumor cells and the tumor microenvironment has been linked to tumor growth and metastasis.68,69 High levels of IL-6 are critical for NF-κβ-mediated transformation, mammosphere formation, and tumorigenesis.70 Treatment of normal mammary tissues with recombinant IL-6 or IL-8 results in increased self-renewal and expression of stem cell markers.71–73 Inhibition of IL-8 receptor binding68 and STAT3 activation74 has been shown to decrease tumorigenicity of breast cancer cell lines. Within the tumor microenvironment, IL-6 and IL-8 are secreted by TAM, TADC, mesenchymal stem cells, and CSCs.75 Activation of IL-6 in tumor cells has been shown to activate a positive feedback loop, increasing both the secretion of IL-6 and the binding of IL-6 to its receptor.72 In addition, STAT3 signaling in inflammatory cells induces transcriptional activation of NF-κβ, leading to increased release of IL-6 and IL-8 within the tumor microenvironment.76 Interestingly, activation of IL-6-STAT3 signaling induces a stem-like phenotype in non-stem cells.25,77 IL-6 was shown to activate Oct-4 signaling and increase xenograft formation in non-CSCs, suggesting an effect on stem cell plasticity. Thus, inflammatory signaling within the tumor microenvironment may promote changes in differentiated cancer cells by increasing expression of stem cell markers and self-renewal properties. This may represent a change in function of existing tumor cells or the emergence of CSCs within inflammatory microenvironments.
Developmental Pathways in Wound Healing and CSCs
In addition to the initiation of inflammatory cascades, wounding results in reactivation of developmental pathways that, in some ways, recapitulate the tissue generation that occurs during embryogenesis. Developmental pathways are activated by inflammatory factors at the beginning of the proliferative phase of wound healing and are crucial to wound closure and tissue regeneration, which occur in the later phases. Similar to wound healing, developmental pathways can be activated in the tumor microenvironment by immune cells,78 inflammatory signaling cascades,79 and in response to cellular stress.80 These pathways are important for the proliferation and survival of breast CSCs, as well as stimulation of angiogenesis and initiation of metastasis. Key developmental pathways that are activated during wound healing are described in the following subsections.
Hedgehog
The hedgehog (HH) pathway is a key morphogenic pathway involved in many aspects of embryonic development, including regulation of embryonic stem cells, tissue patterning, and angiogenesis.81,82 HH pathway activation occurs when a secreted ligand (Sonic, Desert, or Indian) binds to the surface receptor Patched (PTCH). This binding releases the inhibition of the transmembrane receptor SMOOTH-ENED (SMO), setting off a signaling cascade that results in the nuclear translocation of the Gli family of transcription factors. Transcriptional targets for Gli include genes controlling proliferation, angiogenesis, differentiation, EMT, and survival. Recently, a role for HH in wound healing and tissue repair has become evident. Activation of HH signaling has been reported to promote proliferation of epithelial progenitor cells during airway injury83 and contributes to epithelial cell migration by inducing EMT.84 Diabetic mice treated with topical Sonic hedgehog (SHH) experienced improvements in re-epithelialization and wound closure.85 Inhibition of HH signaling has been shown to impair cutaneous wound healing both by inhibiting angiogenesis and by decreasing proliferation of progenitor populations.86 HH signaling is activated in the area surrounding the wound and promotes plasticity of epidermal cells during re-epithelialization, leading to increases in stem cell populations.86
Prolonged activation of HH during cutaneous wound healing results in suppression of differentiation and reprogramming of cells to resemble interfollicular epidermal progenitor cells. These de-differentiated cells behave as tumor initiating CSCs, resulting in the formation of basal cell carcinoma (BCC).87 In addition to BCC, HH signaling has been shown to be important for maintenance of both normal and malignant stem cells in the skin,88,89 prostate,89 pancreas,90 brain,91 and breast.92,93 Similar to its role in wound healing, HH signaling in malignant tissues induces proliferation of progenitor populations and induces EMT within the stem cell compartment.94,95 Although some tumors have constitutive activation of HH signaling, activation of the pathway can also arise from the tumor microenvironment. Release of SHH ligand by TAM has been reported in breast cancer models and promotes expansion of CSC populations and drug resistance through HH-mediated regulation of drug transporters.78
HH signaling plays a role in stem cell expansion, both in wound healing and within the tumor microenvironment. Components of the pathway including Gli-1, Gli-2, and PTCH are highly expressed in normal and malignant mammary stem cells. Activation of the HH signaling is necessary for mammosphere formation and for treatment of mammary stem cells with SHH increasing the size of mammosphere, indicating that the HH signaling is involved in regulation of self-renewal and proliferation of mammary stem cells.92 We have recently shown that release of SHH from tumor cells is induced by chemotherapy treatment in breast cancer cells and serves to expand stem-like populations.9 Breast cancer cell lines treated with docetaxel showed an expansion of cells bearing the CD44highCD24−/low phenotype, as well as increased mammosphere formation and expression of the stemness gene OCT4. This expansion is dependent on the HH signaling as it can be blocked with SHH blocking antibodies or SMO inhibitors. It is unknown whether the expansion of CSCs driven by chemotherapy-induced HH is due to the exit of CSC from G0 and increased proliferation, or enhanced resistance of the CSC populations through SHH-induced upregulation of drug transporters and DNA repair enzymes. Alternatively, it is possible that HH stimulation, induced by inflammation associated with chemotherapeutic stress, may alter the plasticity of non-stem like populations, causing them to acquire a CSC phenotype. Several studies have shown that lineage committed cells may have the capacity to acquire stemness features during alterations in homeostasis such as those that occur during wound healing and the tumor microenvironment.96–100 For example, release of SHH from peri-follicular sensory nerve endings has been shown to result in permanent conversion of hair follicle progenitor cells into intrafollicular epidermal stem cells during wound healing. The progeny of these converted cells is capable of generating long-lasting epidermal clones suggesting the conversion of committed follicular cells into self-renewing epidermal stem cells.98,99 Further research is needed to determine if SHH contributes to plasticity of breast cancer cells in the context of an inflammatory microenvironment.
WNT pathway
Canonical WNT signaling occurs when secreted WNT glycoproteins bind to the membrane protein Frizzled and low-density lipoprotein receptor-related protein 5/6. This binding stimulates a downstream signaling mediator, Dishevelled, which stabilizes the transcriptional co-activator β-catenin, leading to increased nuclear levels of β-catenin. In the absence of stimulation by WNT ligands, β-catenin is targeted for proteasomal degradation by glycogen synthase kinase 3β. WNT signaling is an important mediator of embryonic development and regulates differentiation and EMT. During tissue injury, WNT ligands are highly expressed during the early phases of wound healing and promote migration of epithelial cells.101 WNT signaling is also necessary for formation of new hair follicles during cutaneous wound healing by driving self-renewal of interfollicular epidermal stem cells102 and initiating de-differentiation of epidermal cells to follicular progenitor cells.103
Activation of WNT signaling is predictive of poor prognosis in breast cancer.104 Immunohistochemical expression of nuclear β-catenin has been associated with a basal-like gene signature in breast cancer, and higher levels of cytoplasmic and nuclear β-catenin have been found in cells bearing the CD44highCD-24low/− phenotype.105 Evidence of the role of the WNT pathway in the maintenance of the CSC population comes from transgenic mouse models in which overexpression of WNT1 under control of the Mouse Mammary Tumor Virus promoter leads to an increase in Thy1+CD24+ tumor progenitor cells.106 Additionally, WNT signaling has been shown to promote de-differentiation of normal mammary cells, suggesting that it may also play a role in stem cell plasticity.107 Targeted knockdown of WNT1 in breast cancer cell lines decreases the expression of stem cell markers such as ALDH, inhibits sphere formation, and decreases in vivo tumor forming ability.108 In addition, WNT signaling has been implicated in the radioresistance of CSCs. Overexpression of WNT/β-catenin signaling promotes survival of mammary epithelial progenitor cells after exposure to clinically relevant doses of radiation through upregulation of survivin.43 These findings suggest that the presence of WNT ligands in the tumor microenvironment may promote survival and resistance of CSCs.
TGF-β
The transforming growth factor-beta (TGF-β) superfamily consists of a large number of structurally related proteins that include TGF-β cytokines (TGF-β1, TGF-β2, and TGF-β3), bone morphogenic proteins (BMP), anti-mullerian hormones, and activins among other growth factors. Although originally discovered in malignant tissues, TGF-βs have many physiological functions in normal tissue processes such as embryonic development, immune responses, and wound healing. TGF-βs are secreted from many cell types during tissue injury including platelets, macrophages, endothelial cells, keratinocytes, and fibroblasts.70 They can also be released upon disruption of the ECM.109,110 During the wound healing response, TGF-βs stimulate pleiotropic effects that are dependent on cell type, spatial concentration, and temporal distribution.111 TGF-βs are involved in angiogenesis, inflammatory cell infiltration, fibroblast proliferation, and wound contraction. TGF-β1 knockout mice exhibit defects in re-epithelialization and formation of granulation tissue.112 Activation of TGF-β1 is important for induction of EMT in keratinocytes during re-epithelialization of cutaneous wounds.113
For years, it has been known that TGF-β is a key regulator of EMT in cancer cells.114,115 Because of its role in EMT, TGF-β is involved in the acquisition of CSC-like properties, which is necessary for breast cancer cell metastasis.37,114,116,117 Treatment of immortalized human mammary epithelial cells with TGF-β increases the CD44high/CD24−/low population and the ability of cells to form tumorspheres. Treatment of breast cancer cells with TGF-β not only increases stem cell populations but also induces a mesenchymal phenotype, suggesting that they have entered into EMT.37 This link between EMT and breast CSC properties may be a prerequisite for metastasis and TGF-β is believed to be the driving force behind this.117
Important sources of TGF-β are present in the tumor microenvironment. Stromal cells, cancer cells, and cancer cell-associated platelets are among the cell types in the niche responsible for secreting TGF-β.118,119 Once CSCs metastasize to different sites, they primarily produce TGF-β and induce EMT in order to create their own niche.120 In addition, TGF-β signaling can be induced by chemotherapy and leads to increases in IL-8 expression as well as increases in cells bearing CSC phenotypes. Inhibition of TGF-β signaling is able to block IL-8 induced expansion of CSC and sensitizes breast cancer xenografts to chemotherapy.11 Thus, TGF-β within the tumor microenvironment may regulate the breast CSC population to aid in chemoresistance.
NOTCH
The Notch signaling pathway regulates cell fate decisions during development, including cell fate specification, differentiation, proliferation, and survival.121–124 Studies have shown that Notch signaling is critical for normal embryonic development since the absence of Notch or Notch ligands in mice is embryonically lethal because of angiogenic vascular remodeling defects that affect the embryo, yolk sac, and the placenta.125,126 Notch pathway activation occurs when any of the four transmembrane Notch receptors (Notch1–4) interact with one of the five membrane-bound ligands from the protein families of Delta or Jagged (Jagged-1 and -2, Delta-like (DLL)-1, -3, and -4), which are located on the surface of adjacent cells.127–129 Following binding of the ligand to the Notch receptor, cleavage of the extracellular Notch domain by the metalloprotease TNF-α-converting enzyme (ADAM 17), a member of the ADAM (a disintegrin and metalloprotease domain) family of metalloproteases,130 generates a short-lived intermediate that is then cleaved by the γ-secretase complex.131,132 This final cleavage releases the active intracellular domain of Notch, NICD, which translocates to the nucleus and functions as a transcriptional activator to upregulate the expression of a number of genes that are associated with differentiation and survival including the family of transcription factors, HES and HEY,133,134 cyclin D1,135 and c-Myc.136 Recent studies suggest that Notch signaling is also crucial for tissue homeostasis in adults including angiogenesis and vascular homeostasis,123 lymphocyte expansion and immune function,137 synaptic plasticity,138 and neural cell responses to injury.139
There is also evidence of a role for Notch signaling in wound healing. Notch1 and Jagged-1 are highly expressed in the epidermis after tissue injury140 and inhibition of Notch signaling has been shown to decrease cell proliferation and migration of cultured vascular endothelial cells, keratinocytes, and fibroblasts in a mechanical scratch wound assay. When Notch signaling is reduced or inhibited by a γ-secretase inhibitor in mice, the healing of dermal wounds is delayed, whereas treatment with the Notch ligand, Jagged peptide, enhances wound healing.141 However, in healing corneal epithelium, Notch1 expression is reduced at the leading edge of the wound and inhibition of Notch signaling increases cell migration in vitro and accelerates corneal epithelial wound closure in vivo,142 indicating that the role of Notch signaling in wound healing may be tissue dependent. Notch has also been found to be involved in the inflammatory response of wound healing. Myeloid-specific Notch1 deletion in mice results in decreased macrophage recruitment and TNF-α expression in a wound healing assay.143 In addition, both activation and deletion of Notch1 disrupt the epidermal barrier and can trigger an inflammatory infiltrate.144,145 Although Notch plays a role in epidermal differentiation in development,133,146 the effects of Notch signaling on differentiation of epidermal progenitors during wound healing have not fully been investigated. However, Notch has been shown to crosstalk with HH and WNT pathways,133 both of which are involved in regulation of progenitor proliferation and plasticity during cutaneous wound healing.
In adult skin, loss of Notch1 expression can lead to tumor development through alterations of the stroma and creation of a wound-like environment in the skin,147 indicating that the changes in Notch signaling consistent with increases in inflammation can promote tumor growth. Expression of Notch receptors and its ligands has been found to be upregulated in a number of hematopoietic and solid tumors including colon, lung, pancreas, and breast.148–153 In breast cancer, high levels of Notch1 are associated with poor prognosis.154,155 Likewise, expression of Jagged-1 protein or mRNA has been associated with lower disease-free survival, basal-like phenotype, and poor prognosis in breast cancer.156,157
Inhibiting Notch signaling by γ-secreatase inhibitors, anti-Notch1, or anti-DLL-4 monoclonal antibodies has been shown to result in antitumor activity in a variety of cancers.158–161 Promotion of tumor development through altered Notch signaling may be in part through survival and proliferation of CSCs. Specific inhibitors to Notch receptors or ligands have been reported to reduce breast tumor initiating cell frequency or depletion of stem-like breast cancer cells.162–164 Inhibiting Notch1 in triple-negative breast tumors results in tumor growth inhibition, decreased tumor growth upon reimplantation, and delayed tumor recurrence possibly due to reduction in the CSC population.165 In addition, knockdown of the Notch1 receptor inhibits breast cancer cell growth by induction of apoptosis and can enhance chemosensitivity of breast cancer tumors through reduction of CSC frequency.166 Therefore, including Notch inhibitors in the therapeutic regimen for certain cancers may be useful to target the CSCs to enhance chemo- and/or radiosensitivity and prevent cancer recurrence.
Pharmacological Inhibition of Inflammatory and Developmental Pathways
Recently, new therapies targeting developmental pathways such as WNT, Notch, and HH have entered clinical trials (Table 3). Additionally, many anti-inflammatory agents that are currently under development for treatment of chronic inflammatory conditions may also be beneficial to cancer patients (Table 2). Multiple strategies have been used to target the IL-1β–IL-6–STAT3 pathway for both autoimmune diseases and cancer. Inhibition of the IL-1 signaling cascades via IL-1 receptor antagonists (IL-1Ra; Kineret(R) or anakinra/Amgen, Inc) or blocking antibodies Rilonacept (IL-1 Trap/Arcalyst Regeneron Pharmaceuticals) effectively block IL-6–STAT3 signaling in autoimmune disorders and have some success in the treatment of inherited autoimmune diseases.167 These agents are currently in clinical trials for treatment of multiple myeloma, as well as some solid tumors. Additionally, IL-6 blocking antibodies are in clinical trials for various diseases. Siltuximab, a chimeric antibody to IL-6 developed by Johnson and Johnson, recently gained Food and Drug Administration (FDA) approval for treatment of Castleman’s disease and is currently in clinical trials for cancers such as multiple myeloma and prostate cancer as well as other solid tumors. Recently, results from a double-blinded randomized clinical trial indicated improvement in response rates to standard therapy in patients with multiple myeloma, but no change in overall survival or progression-free survival.168 Phase I/II dose escalation trials in solid tumors have shown that the drug is well tolerated, but so far the advantage of including Siltuximab in standard therapy regimens has not been clinically proven.169 Antibodies to the IL-6 receptor gp-130 are also undergoing clinical trials for treatment of autoimmune diseases. Tocilizumab, an IL-6R blocking antibody developed by Hoffmann-La Roche AG, has recently gained FDA approval for treatment of rheumatoid arthritis. Although it has been shown to have an effect on cachexia associated with non-small cell lung tumors, its role as a treatment for solid tumors has not yet been investigated clinically. Additionally, several inhibitors of STAT3 are currently under preclinical/clinical development for treatment of solid tumors.170–172 To date, Phase I dose escalation studies of STAT3 inhibitors have shown poor pharmacokinetics and severe dose-limiting side effects.173
Table 3.
Current clinical trials of pharmacological inhibitors of developmental pathways.
| TARGET | INHIBITOR | OTHER NAME | CONDITION | PHASE | NCT NUMBER |
|---|---|---|---|---|---|
| Hedgehog | Vismodegib | GDC-0449 | Advanced and/or metastatic solid tumors | I/II | NCT02073838; NCT01088815; NCT01713218; NCT00887159; NCT00957229; NCT00959647; NCT00636610; NCT00739661; NCT00968981; NCT00991718; NCT00833417; NCT01546519; NCT01209143; NCT01174264; NCT01064622; NCT00878163; NCT01195415 |
| Ipilimumab | IPI-926 | Advanced and/or metastatic solid tumors | I/II | NCT01255800; NCT01371617; NCT01130142; NCT00761696; NCT01310816 | |
| Erismodegib | LDE225 | Advanced and/or metastatic solid tumors | I/II | NCT02151864; NCT01911416; NCT01579929; NCT01431794; NCT01757327; NCT01954355; NCT01694589; NCT01125800; NCT01787552; NCT01350115; NCT00880308; NCT01208831; NCT01769768; NCT01576666 | |
| PF-04449913 | – | Advanced and/or metastatic solid tumors | I/II | NCT01842646; NCT01841333; NCT01286467; NCT01546038; NCT04449913; NCT0203877; NCT02226172 | |
| LEQ506 | – | Advanced solid tumor, recurrent or refractory medulloblastoma or locally advanced or metastatic basal cell carcinoma | I | NCT01106508 | |
| TAK-441 | – | Advanced nonhematologic malignancies | I | NCT01204073 | |
| Itraconazole | – | Relpased prostate cancer | II | NCT01787331 | |
| LY2940680 | – | Small cell lung carcinoma | I/II | NCT01722292 | |
| Sonidegib | – | Myeloid malignancies | I | NCT02129101 | |
| BMS-833923 | – | Extensive stage small cell lung cancer (ES-SCLC) | I | NCT00927875 | |
| Notch | Notch inhibitor | – | Advanced and/or metastatic solid tumors | I/II | NCT01158404; NCT02069730 |
| MK0752 | – | Advanced and/or metastatic solid tumors | I/II | NCT00106145; NCT01098344; NCT00645333 | |
| BMS-906024 | – | Advanced and/or metastatic solid tumors | I | NCT01653470; NCT01363817; NCT01292655; NCT01986218 | |
| PF-03084014 | – | Desmoid tumors/aggressive fibromatosis; AIDS-related Kaposi sarcoma; Advanced solid tumor malignancy and t-cell acute lymphoblastic leukemia/lymphoblastic lymphoma | I/II | NCT01981551; NCT02137564; NCT00878189 | |
| RO4929097 | – | Advanced and/or metastatic solid tumors | I/II | NCT01238133; NCT01122901; NCT01196416; NCT01151449; NCT01198184; NCT01120275; NCT01232829; NCT01193881; NCT01218620; NCT01119599; NCT01145456; NCT01131234; NCT01096355; NCT01158274; NCT01141569; NCT01189240; NCT01200810; NCT01198535; NCT01149356; NCT01175343; NCT01217411; NCT01116687 | |
| Notch & Hedgehog | Vismodegib & RO4929097 | GDC-0449 & RO4929097 | Metastatic or unresectable breast cancer; Advanced or metastatic sarcoma | I/II | NCT01071564; NCT01154452 |
| Wnt signaling pathway | Resveratrol | – | Colon cancer | I | NCT02137421; NCT00256334; NCT00578396 |
| Diclofenac & Calcitriol | Diclofenac & topical vitamin D3 | Basal cell carcinoma | III | NCT01358045 | |
| Genistein | Metastatic colon cancer | I/II | NCT01985763 |
Table 2.
Current clinical trials of anti-inflammatory agents in chronic inflammatory diseases and cancer.
| TARGET | INHIBITOR | OTHER NAME | CONDITION | PHASE | NCT NUMBER |
|---|---|---|---|---|---|
| IL-1 | Kineret | Anakinra | Rheumatoid arthritis; Juvenile rheumatoid arthritis; Myocardial infarction and heart failure; Type I and II Diabetes; Advanced cancers including breast, pancreatic, colorectal | I/II/III/IV | NCT02018458; NCT01802970; NCT02021422; NCT01175018; NCT00789724; NCT00645840; NCT01950299; NCT00117091; NCT02236481; NCT02021422; NCT02090101; NCT01300650; NCT00213538; NCT01936844; nCt00037648 |
| Ilaris | Canakinumab; ACZ885 | Autoinflammatory disease; Systemic juvenile idiopathic arthritis; Schnitzler syndrome; Type I and II diabetes; Atrial fibrillation; COPD; Gout | I/II/III | NCT01390350; NCT01676948; NCT00891046; NCT00947427; NCT01080131; NCT00663169; NCT01356602; NCT02204293; NCT00819585; NCT00927810; NCT01068860; NCT00605475; NCT01805960; NCT00581945; NCT00487825; NCT00424346; nCt00504595 | |
| Arcalyst | Rilonacept; IL-1 Trap | Autoinflammatory disease; Gout; Systemic sclerosis; Juvenile Idiopathic arthritis; Cryopyrin-associated periodic syndromes; Type I Diabetes; Chronic kidney disease; Cardiovascular disease | I/II/III/IV | NCT00962026; NCT00288704; NCT00417417; NCT01663103; NCT00534495; NCT00855920; NCT01538719; NCT00094900; NCT00582907 | |
| IL-6 | Tocilizumab | Actemra | Rheumatoid arthritis; Juvenile idiopathic arthritis; schizophrenia; Type I diabetes; HIV infection; systemic sclerosis; Non-ST elevation myocardial infarction; recurrent ovarian cancer | I/II/III/IV | NCT02087696; NCT01194414; NCT02010216; NCT01941095; NCT01347983; NCT01696929; NCT02034474; NCT00868751; NCT01603355; NCT00642460; NCT01904292; NCT02165345; NCT01637532; NCT01532869; NCT01491074; NCT02049437; NCT02293837 |
| Sirukumab | CNTO 136 | Rheumatoid arthritis; Lupus nephritis; Lupus erythematosus | I/II/III | NCT01606761; NCT01273389; NCT01702740 | |
| IL-8 | Reparixin | – | Rheumatoid arthritis; Pancreatic islet transplantation in type 1 Diabetes Mellitus; delayed graft function after kidney or lung transplantation; HER2 negative breast cancer; stage II breast cancer | I/II/III | NCT01220856; NCT02001974; NCT00248040; NCT01861054; NCT01967888; NCT01817959; NCT00224406 |
| Danirixin | GSK1325756 | COPD: Respiratory syncytial virus (RSV) infection | I | NCT02130193; NCT02201303 | |
| AZD5069 | Asthma; COPD; Bronchiectasis | I/II | NCT01890148; NCT01704495; NCT01962935; NCT01233232; NCT01255592 | ||
| Navarixin | SCH 527123; MK-7123 | Asthma; COPD; Psoriasis | I/II | NCT00632502; NCT00688467; NCT00441701; NCT00684593 | |
| SB-656933 | COPD; Cystic fibrosis; Ulcerative colitis | I/II | NCT00551811; NCT00748410; NCT00903201; NCT00605761 | ||
| STAT3 | OPB-31121 | Advanced and/or metastatic solid tumors | I/II | NCT00955812; NCT01406574 | |
| AZD9150 | ISIS-STAT3Rx; ISIS 481464 | Advanced and/or metastatic solid tumors | I/II | NCT01563302; NCT01839604 | |
| Daraprim | Pyrimethamine | Chronic lymphocytic/small lymphocytic lymphoma | I/II | NCT01066663 | |
| OPB-51602 | Advanced and/or metastatic solid tumors and hematologic malignancies | I | NCT02058017; NCT01423903; NCT01184807; NCT01867073; NCT01344876 |
Several agents targeting developmental pathways are also in clinical development. The first HH inhibitor to gain clinical approval for the treatment of BCC was Vismodegib (GDC-0449) developed by Genentech. This agent was found to have a significant effect on metastatic and resistant BCC. Preliminary findings indicate that this agent may also have anti-tumoral activity in medulloblastoma.174 Phase II trials of Vismodegib for treatment of other solid tumors are currently underway. Several other inhibitors to the HH pathway are also under clinical development, including Sonidegib (also known as Erismodegib or LDE225, Novartis) which has shown acceptable toxicity profiles in Phase I trials and is currently progressing into Phase II trials to examine efficacy in solid tumors.175 Likewise, multiple agents are under preclinical/clinical development for inhibition of γ-secretase, a key enzyme involved in Notch signaling. Phase I trials of RO4929097, a γ-sectrease inhibitor developed by Roche, have shown acceptable toxicity profiles and some evidence of anti-tumor activity.176
Significant crosstalk exists between developmental pathways in both wound healing and the tumor microenvironment. For this reason, combinatorial approaches with targeted inhibitors for multiple developmental pathways are currently being explored. A Phase II study is currently underway to determine the effects of concurrent inhibition of the HH and Notch pathways in breast cancer. If successful, the combination of Vismodegib and RO4929097 may offer new treatment options for advanced or metastatic tumors. Additionally, crosstalk between developmental and inflammatory pathways within the tumor microenvironment has been reported. Currently, a Phase I trial of combination therapy using Vismodegib and the immunosuppressive agent Sirolimus (rapamycin; Pfizer) are underway. Advancement of this approach will depend on an acceptable toxicity profile, but may yield promising results for the treatment of solid tumors including pancreatic and breast cancer. It is likely that combination therapies targeting both developmental and inflammatory pathways may lead to better responses. Additionally, studies have shown that treatment with radiation and standard chemotherapy induces a substantial wound healing response, which may further promote resistance and expansion of CSCs.9,10,177,178 Activation of Notch signaling during post-surgical wound healing has been linked to increased metastasis.10 Thus, combinations of anti-inflammatory drugs with standard treatment approaches may prevent activation of developmental pathways responsible for cancer resistance and metastasis and lead to better clinical outcomes.
Conclusion
During the course of malignancy, tumor cells invade neighboring tissues, stimulate angiogenesis, remodel the ECM, undergo EMT, and metastasize. In doing so, they activate a chronic inflammatory response involving numerous cytokines, developmental pathways, and growth factors involved in the normal wound healing process. The presence of these factors within the tumor microenvironment is linked to an increase in the proportion of cells bearing stem-like phenotypes, enhanced tumor-initiating properties, and increased resistance to standard therapies.
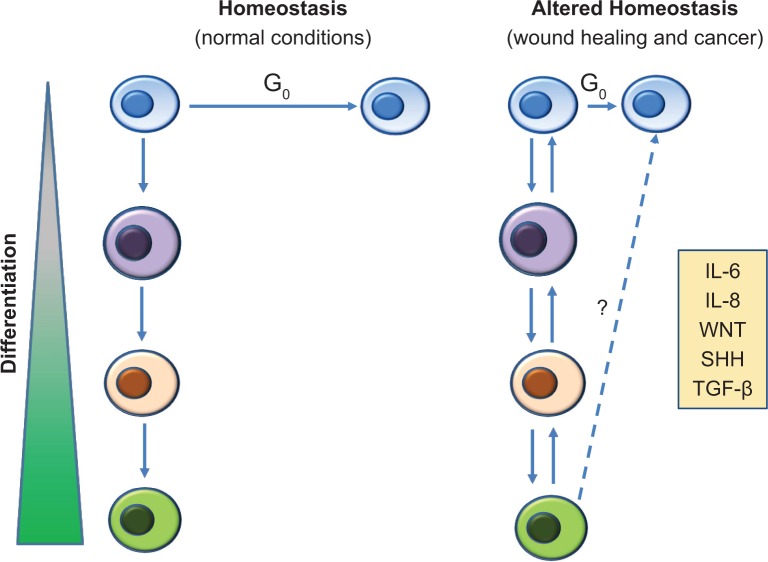
It is currently unclear whether the tumor microenvironment confers a survival advantage through selection of resistant CSC or through upregulation of stem-like properties in non-stem cells. Although evidence for cells bearing stem cell signatures in resistant breast cancer is compelling, the origin of these cells is controversial. The question remains as to whether signals within the tumor microenvironment lead to proliferation of a small population of cells arising from genetically altered stem cells, or do they induce a stem-like phenotype in differentiated cells through upregulation of developmental signaling pathways and stem cell markers. Further, does this change in phenotype confer a functional change in differentiated cells so that they now behave as CSC, exhibiting self-renewal and tumor-initiating properties? As shown in Figure 1, we propose that when normal homeostasis is disrupted, either by tissue injury or by the tumor microenvironment, activation of inflammatory and developmental pathways alters the ratio of stem cells to non-stem cells in two possible ways. The first is by driving existing slow-cycling, quiescent stem cell populations into the proliferative phase of the cell cycle. The second is by altering plasticity of differentiated cells so that they obtain stemness properties. The latter of these processes is supported by recent evidence showing significant cellular plasticity in normal tissue during wound healing processes. Lineage tracing experiments have shown that committed progeny in the lung and intestine can revert to a stem-like phenotype and contribute to tissue regeneration.4,96,179 Likewise, interconversions between stem-like and differentiated states have been reported in glioblastoma26 and breast cancer180 cells after chemotherapy. Although both wound healing and the tumor microenvironment represent states of disrupted homeo-stasis in which stem cell numbers may be increased, they differ significantly in that the level of stemness during wound healing is tightly regulated, with a number of stem-like cells returning to normal during the final phases of the healing process. In the tumor microenvironment, stem cell plasticity may be constant with an overall increase in stem-like cells.
Figure 1.
Cellular plasticity in normal vs altered homeostasis. (Left panel) Under normal conditions, stem cell/non-stem cell ratios are maintained at a consistent level via slow cycling stem cells (blue), which undergo a unidirectional differentiation to lineage restricted cells. (Right panel) In conditions of altered homeostasis, the ratio of stem cell/non-stem cell is increased by proliferation of stem cells and a bidirectional differentiation whereby lineage restricted cells are able to dedifferentiate and acquire stem-like features. In both wound healing and the tumor microenvironment, this cellular plasticity is driven by inflammatory and developmental factors. However, in wound healing, expression of these factors is transient, homeostasis returns, and the ratio of stem/non-stem cells returns to normal levels. In the tumor microenvironment, continuous expression of these factors may lead to a permanent expansion of stem-like cells.
The effect of inflammatory signals within the microenvironment on the cellular plasticity of tumor cells has yet to be fully determined. Thus, it will be important to investigate whether blocking specific inflammatory signals, alone or in combination with other stem cell pathway inhibitors, can decrease CSC populations in a therapeutic setting and promote better responses to standard therapies. The answer to this question would help to uncover a link between inflammation and CSCs and determine whether, together or separately, they impact the response to therapy.
Footnotes
ACADEMIC EDITOR: Marc D. Basson, Editor in Chief
FUNDING: This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number U54-GM104941. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE). Provenance: the authors were invited to submit this paper.
Author Contributions
Contributed to the writing of the manuscript: KMA, LMO, JSM. Jointly developed the structure and arguments for the paper: JSM, DF. Made critical revisions and approved final version: JSM, DF. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 2.Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. 2007;127(3):514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 3.Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83(3):835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 4.Blanpain C, Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344(6189):1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999;341(10):738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 6.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuelten CH, Barbul A, Busch JI, et al. Acute wounds accelerate tumorigenesis by a T cell-dependent mechanism. Cancer Res. 2008;68(18):7278–7282. doi: 10.1158/0008-5472.CAN-08-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobson J, Gummadidala P, Silverstrim B, et al. Acute inflammation induced by the biopsy of mouse mammary tumors promotes the development of metastasis. Breast Cancer Res Treat. 2013;139(2):391–401. doi: 10.1007/s10549-013-2575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sims-Mourtada J, Opdenaker LM, Davis J, Arnold KM, Flynn D. Taxane-induced hedgehog signaling is linked to expansion of breast cancer stem-like populations after chemotherapy. Mol Carcinog. 2014 Sep 27; doi: 10.1002/mc.22225. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Taketo MM. Reflections on the spread of metastasis to cancer prevention. Cancer Prev Res (Phila) 2011;4(3):324–328. doi: 10.1158/1940-6207.CAPR-11-0046. [DOI] [PubMed] [Google Scholar]
- 11.Bhola NE, Balko JM, Dugger TC, et al. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest. 2013;123(3):1348–1358. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 13.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 14.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 15.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 16.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 20.Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature. 2008;451(7176):345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci U S A. 2010;107(8):3722–3727. doi: 10.1073/pnas.0915135107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 24.Lluis F, Cosma MP. Resetting epigenetic signatures to induce somatic cell reprogramming. Cell Mol Life Sci. 2013;70(8):1413–1424. doi: 10.1007/s00018-012-1137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A. 2011;108(4):1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auffinger B, Tobias AL, Han Y, et al. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21(7):1119–1131. doi: 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36(suppl 1):59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 29.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ginestier C, Wicha MS. Mammary stem cell number as a determinate of breast cancer risk. Breast Cancer Res. 2007;9(4):109. doi: 10.1186/bcr1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong Y, Wang J, Huo L, Wei W, Ueno NT, Woodward WA. Aldehyde dehydrogenase 1 expression in inflammatory breast cancer as measured by immunohistochemical staining. Clin Breast Cancer. 2014;14(3):e81–e88. doi: 10.1016/j.clbc.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y, Zhong Y, Guan H, Zhang X, Sun Q. CD44+/CD24− phenotype contributes to malignant relapse following surgical resection and chemotherapy in patients with invasive ductal carcinoma. J Exp Clin Cancer Res. 2012;31:59. doi: 10.1186/1756-9966-31-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcato P, Dean CA, Pan D, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29(1):32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 35.Giordano A, Gao H, Cohen EN, et al. Clinical relevance of cancer stem cells in bone marrow of early breast cancer patients. Ann Oncol. 2013;24(10):2515–2521. doi: 10.1093/annonc/mdt223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhat-Nakshatri P, Appaiah H, Ballas C, et al. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24− phenotype. BMC Cancer. 2010;10:411. doi: 10.1186/1471-2407-10-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollier BG, Tinnirello AA, Werden SJ, et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013;73(6):1981–1992. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98(24):1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 41.Shi W, Harris AL. Notch signaling in breast cancer and tumor angiogenesis: cross-talk and therapeutic potentials. J Mammary Gland Biol Neoplasia. 2006;11(1):41–52. doi: 10.1007/s10911-006-9011-7. [DOI] [PubMed] [Google Scholar]
- 42.Al-Hajj M. Cancer stem cells and oncology therapeutics. Curr Opin Oncol. 2007;19(1):61–64. doi: 10.1097/CCO.0b013e328011a8d6. [DOI] [PubMed] [Google Scholar]
- 43.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104(2):618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh S, Brocker C, Koppaka V, et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2012;56:89–101. doi: 10.1016/j.freeradbiomed.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lagadec C, Vlashi E, Della Donna L, et al. Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Res. 2010;12(1):R13. doi: 10.1186/bcr2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagadec C, Vlashi E, Della Donna L, Dekmezian C, Pajonk F. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012;30(5):833–844. doi: 10.1002/stem.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charles N, Holland EC. The perivascular niche microenvironment in brain tumor progression. Cell Cycle. 2010;9(15):3012–3021. doi: 10.4161/cc.9.15.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8(20):3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghisolfi L, Keates AC, Hu X, Lee DK, Li CJ. Ionizing radiation induces stemness in cancer cells. PLoS One. 2012;7(8):e43628. doi: 10.1371/journal.pone.0043628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12(2):79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 53.Kondo T, Ohshima T. The dynamics of inflammatory cytokines in the healing process of mouse skin wound: a preliminary study for possible wound age determination. Int J Legal Med. 1996;108(5):231–236. doi: 10.1007/BF01369816. [DOI] [PubMed] [Google Scholar]
- 54.Sugawara T, Gallucci RM, Simeonova PP, Luster MI. Regulation and role of interleukin 6 in wounded human epithelial keratinocytes. Cytokine. 2001;15(6):328–336. doi: 10.1006/cyto.2001.0946. [DOI] [PubMed] [Google Scholar]
- 55.Dauer DJ, Ferraro B, Song L, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24(21):3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 56.Gao W, McCormick J, Connolly M, Balogh E, Veale DJ, Fearon U. Hypoxia and STAT3 signalling interactions regulate pro-inflammatory pathways in rheumatoid arthritis. Ann Rheum Dis. 2014 Feb 13; doi: 10.1136/annrheumdis-2013-204105. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 57.Kruttgen A, Rose-John S. Interleukin-6 in sepsis and capillary leakage syndrome. J Interferon Cytokine Res. 2011;32(2):60–65. doi: 10.1089/jir.2011.0062. [DOI] [PubMed] [Google Scholar]
- 58.Ridker PM, Luscher TF. Anti-inflammatory therapies for cardiovascular disease. Eur Heart J. 2014;35(27):1782–1791. doi: 10.1093/eurheartj/ehu203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao R, Zhou H, Su SB. A critical role for interleukin-1beta in the progression of autoimmune diseases. Int Immunopharmacol. 2013;17(3):658–669. doi: 10.1016/j.intimp.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 60.Pradhan Nabzdyk L, Kuchibhotla S, Guthrie P, et al. Expression of neuropeptides and cytokines in a rabbit model of diabetic neuroischemic wound healing. J Vasc Surg. 2013;58(3):766–775. e712. doi: 10.1016/j.jvs.2012.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rennekampff HO, Hansbrough JF, Kiessig V, Dore C, Sticherling M, Schroder JM. Bioactive interleukin-8 is expressed in wounds and enhances wound healing. J Surg Res. 2000;93(1):41–54. doi: 10.1006/jsre.2000.5892. [DOI] [PubMed] [Google Scholar]
- 62.McFarland-Mancini MM, Funk HM, Paluch AM, et al. Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J Immunol. 2010;184(12):7219–7228. doi: 10.4049/jimmunol.0901929. [DOI] [PubMed] [Google Scholar]
- 63.Gallucci RM, Simeonova PP, Matheson JM, et al. Impaired cutaneous wound healing in interleukin-6-deficient and immunosuppressed mice. FASEB J. 2000;14(15):2525–2531. doi: 10.1096/fj.00-0073com. [DOI] [PubMed] [Google Scholar]
- 64.Sato M, Sawamura D, Ina S, Yaguchi T, Hanada K, Hashimoto I. In vivo introduction of the interleukin 6 gene into human keratinocytes: induction of epidermal proliferation by the fully spliced form of interleukin 6, but not by the alternatively spliced form. Arch Dermatol Res. 1999;291(7–8):400–404. doi: 10.1007/s004030050429. [DOI] [PubMed] [Google Scholar]
- 65.Sano S, Itami S, Takeda K, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18(17):4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benoy IH, Salgado R, Van Dam P, et al. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res. 2004;10(21):7157–7162. doi: 10.1158/1078-0432.CCR-04-0812. [DOI] [PubMed] [Google Scholar]
- 67.Cho YA, Sung MK, Yeon JY, Ro J, Kim J. Prognostic role of interleukin-6, interleukin-8, and leptin levels according to breast cancer subtype. Cancer Res Treat. 2013;45(3):210–219. doi: 10.4143/crt.2013.45.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ginestier C, Liu S, Diebel ME, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120(2):485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dethlefsen C, Hojfeldt G, Hojman P. The role of intratumoral and systemic IL-6 in breast cancer. Breast Cancer Res Treat. 2013;138(3):657–664. doi: 10.1007/s10549-013-2488-z. [DOI] [PubMed] [Google Scholar]
- 70.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao C, Lin Y, Chua MS, et al. Interleukin-8 modulates growth and invasiveness of estrogen receptor-negative breast cancer cells. Int J Cancer. 2007;121(9):1949–1957. doi: 10.1002/ijc.22930. [DOI] [PubMed] [Google Scholar]
- 72.Sansone P, Storci G, Giovannini C, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25(3):807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- 73.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin L, Hutzen B, Lee HF, et al. Evaluation of STAT3 signaling in ALDH+ and ALDH+/CD44+/CD24− subpopulations of breast cancer cells. PLoS One. 2014;8(12):e82821. doi: 10.1371/journal.pone.0082821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vendramini-Costa DB, Carvalho JE. Molecular link mechanisms between inflammation and cancer. Curr Pharm Des. 2012;18(26):3831–3852. doi: 10.2174/138161212802083707. [DOI] [PubMed] [Google Scholar]
- 76.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim SY, Kang JW, Song X, et al. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell Signal. 2013;25(4):961–969. doi: 10.1016/j.cellsig.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jinushi M, Chiba S, Yoshiyama H, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci U S A. 2011;108(30):12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su YC, Li SC, Wu YC, Wang LM, Chao KS, Liao HF. Resveratrol downregulates interleukin-6-stimulated sonic hedgehog signaling in human acute myeloid leukemia. Evid Based Complement Alternat Med. 2013;2013:547430. doi: 10.1155/2013/547430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xing F, Okuda H, Watabe M, et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene. 2011;30(39):4075–4086. doi: 10.1038/onc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2(5):361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 82.Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6(1):21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- 83.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422(6929):313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 84.Crosby LM, Waters CM. Epithelial repair mechanisms in the lung. Am J Physiol Lung Cell Mol Physiol. 2010;298(6):L715–L731. doi: 10.1152/ajplung.00361.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asai J, Takenaka H, Kusano KF, et al. Topical sonic hedgehog gene therapy accelerates wound healing in diabetes by enhancing endothelial progenitor cell-mediated microvascular remodeling. Circulation. 2006;113(20):2413–2424. doi: 10.1161/CIRCULATIONAHA.105.603167. [DOI] [PubMed] [Google Scholar]
- 86.Le H, Kleinerman R, Lerman OZ, et al. Hedgehog signaling is essential for normal wound healing. Wound Repair Regen. 2008;16(6):768–773. doi: 10.1111/j.1524-475X.2008.00430.x. [DOI] [PubMed] [Google Scholar]
- 87.Youssef KK, Lapouge G, Bouvrée K, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat Cell Biol. 2012;14(12):1282–1294. doi: 10.1038/ncb2628. [DOI] [PubMed] [Google Scholar]
- 88.Tan DW, Barker N. Stem cell reprogramming as a driver of basal cell carcinoma. Nat Cell Biol. 2012;14(12):1246–1247. doi: 10.1038/ncb2631. [DOI] [PubMed] [Google Scholar]
- 89.Chari NS, Romano RA, Koster MI, et al. Interaction between the TP63 and SHH pathways is an important determinant of epidermal homeostasis. Cell Death Differ. 2013;20(8):1080–1088. doi: 10.1038/cdd.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang FT, Zhuan-Sun YX, Zhuang YY, et al. Inhibition of hedgehog signaling depresses self-renewal of pancreatic cancer stem cells and reverses chemoresistance. Int J Oncol. 2012;41(5):1707–1714. doi: 10.3892/ijo.2012.1597. [DOI] [PubMed] [Google Scholar]
- 91.Manoranjan B, Venugopal C, McFarlane N, et al. Medulloblastoma stem cells: modeling tumor heterogeneity. Cancer Lett. 2012;338(1):23–31. doi: 10.1016/j.canlet.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 92.Liu S, Dontu G, Mantle ID, et al. Hedgehog signaling and Bmi-1 regulate self-renewalof normal and malignant human mammary stem cells. Cancer Res. 2006;66(12):6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 94.Xu X, Su B, Xie C, et al. Sonic hedgehog-Gli1 signaling pathway regulates the epithelial mesenchymal transition (EMT) by mediating a new target gene, S100A4, in pancreatic cancer cells. PLoS One. 2014;9(7):e96441. doi: 10.1371/journal.pone.0096441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med. 2009;9(7):873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 96.van Es JH, Sato T, van de Wetering M, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14(10):1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rawlins EL, Okubo T, Xue Y, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4(6):525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brownell I, Guevara E, Bai CB, Loomis CA, Joyner AL. Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell. 2011;8(5):552–565. doi: 10.1016/j.stem.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Levy V, Lindon C, Zheng Y, Harfe BD, Morgan BA. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 2007;21(7):1358–1366. doi: 10.1096/fj.06-6926com. [DOI] [PubMed] [Google Scholar]
- 100.Kumar SM, Liu S, Lu H, et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31(47):4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Okuse T, Chiba T, Katsuumi I, Imai K. Differential expression and localization of WNTs in an animal model of skin wound healing. Wound Repair Regen. 2005;13(5):491–497. doi: 10.1111/j.1067-1927.2005.00069.x. [DOI] [PubMed] [Google Scholar]
- 102.Lim X, Tan SH, Koh WL, et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342(6163):1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ito M, Yang Z, Andl T, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447(7142):316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 104.Geyer FC, Lacroix-Triki M, Savage K, et al. Beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2010;24(2):209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 105.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176(6):2911–2920. doi: 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cho RW, Wang X, Diehn M, et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26(2):364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 107.Constantinou T, Baumann F, Lacher MD, Saurer S, Friis R, Dharmarajan A. SFRP-4 abrogates Wnt-3a-induced beta-catenin and Akt/PKB signalling and reverses a Wnt-3a-imposed inhibition of in vitro mammary differentiation. J Mol Signal. 2008;3:10. doi: 10.1186/1750-2187-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Choi AR, Park JR, Kim RJ, et al. Inhibition of Wnt1 expression reduces the enrichment of cancer stem cells in a mouse model of breast cancer. Biochem Biophys Res Commun. 2012;425(2):436–442. doi: 10.1016/j.bbrc.2012.07.120. [DOI] [PubMed] [Google Scholar]
- 109.Margadant C, Sonnenberg A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010;11(2):97–105. doi: 10.1038/embor.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schultz GS, Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 2009;17(2):153–162. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 111.Finnson KW, Arany PR, Philip A. Transforming growth factor beta signaling in cutaneous wound healing: lessons learned from animal studies. Adv Wound Care (New Rochelle) 2014;2(5):225–237. doi: 10.1089/wound.2012.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brown RL, Ormsby I, Doetschman TC, Greenhalgh DG. Wound healing in the transforming growth factor-beta-deficient mouse. Wound Repair Regen. 1995;3(1):25–36. doi: 10.1046/j.1524-475X.1995.30108.x. [DOI] [PubMed] [Google Scholar]
- 113.Weber CE, Li NY, Wai PY, Kuo PC. Epithelial-mesenchymal transition, TGF-beta, and osteopontin in wound healing and tissue remodeling after injury. J Burn Care Res. 2012;33(3):311–318. doi: 10.1097/BCR.0b013e318240541e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 115.Wharton K, Derynck R. TGFbeta family signaling: novel insights in development and disease. Development. 2009;136(22):3691–3697. doi: 10.1242/dev.040584. [DOI] [PubMed] [Google Scholar]
- 116.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98(10):1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20(5):576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ikushima H, Miyazono K. Cellular context-dependent “colors” of transforming growth factor-beta signaling. Cancer Sci. 2010;101(2):306–312. doi: 10.1111/j.1349-7006.2009.01441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fazilaty H, Gardaneh M, Bahrami T, Salmaninejad A, Behnam B. Crosstalk between breast cancer stem cells and metastatic niche: emerging molecular metastasis pathway? Tumour Biol. 2013;34(4):2019–2030. doi: 10.1007/s13277-013-0831-y. [DOI] [PubMed] [Google Scholar]
- 121.Simpson P. Developmental genetics. The Notch connection. Nature. 1995;375(6534):736–737. doi: 10.1038/375736a0. [DOI] [PubMed] [Google Scholar]
- 122.Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9(6):583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- 123.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterio-scler Thromb Vasc Biol. 2003;23(4):543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 124.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66(10):1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee JB, Werbowetski-Ogilvie TE, Lee JH, et al. Notch-HES1 signaling axis controls hemato-endothelial fate decisions of human embryonic and induced pluripotent stem cells. Blood. 2013;122(7):1162–1173. doi: 10.1182/blood-2012-12-471649. [DOI] [PubMed] [Google Scholar]
- 126.Krebs LT, Iwai N, Nonaka S, et al. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 2003;17(10):1207–1212. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80(6):909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 128.Shawber C, Boulter J, Lindsell CE, Weinmaster G. Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol. 1996;180(1):370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- 129.Dunwoodie SL, Henrique D, Harrison SM, Beddington RS. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124(16):3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- 130.Brou C, Logeat F, Gupta N, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5(2):207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 131.De Strooper B, Annaert W, Cupers P, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 132.Saxena MT, Schroeter EH, Mumm JS, Kopan R. Murine notch homologs (N1–4) undergo presenilin-dependent proteolysis. J Biol Chem. 2001;276(43):40268–40273. doi: 10.1074/jbc.M107234200. [DOI] [PubMed] [Google Scholar]
- 133.Okuyama R, Tagami H, Aiba S. Notch signaling: its role in epidermal homeo-stasis and in the pathogenesis of skin diseases. J Dermatol Sci. 2008;49(3):187–194. doi: 10.1016/j.jdermsci.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 134.Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr Opin Cell Biol. 2008;20(2):171–179. doi: 10.1016/j.ceb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21(17):5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kared H, Adle-Biassette H, Foïs E, et al. Jagged2-expressing hematopoietic progenitors promote regulatory T cell expansion in the periphery through notch signaling. Immunity. 2006;25(5):823–834. doi: 10.1016/j.immuni.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 138.Wang Y, Chan SL, Miele L, et al. Involvement of Notch signaling in hippocampal synaptic plasticity. Proc Natl Acad Sci U S A. 2004;101(25):9458–9462. doi: 10.1073/pnas.0308126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arumugam TV, Chan SL, Jo DG, et al. Gamma secretase-mediated Notch signaling worsens brain damage and functional outcome in ischemic stroke. Nat Med. 2006;12(6):621–623. doi: 10.1038/nm1403. [DOI] [PubMed] [Google Scholar]
- 140.Thelu J, Rossio P, Favier B. Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2002;2:7. doi: 10.1186/1471-5945-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chigurupati S, Arumugam TV, Son TG, et al. Involvement of notch signaling in wound healing. PLoS One. 2007;2(11):e1167. doi: 10.1371/journal.pone.0001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Movahedan A, Majdi M, Afsharkhamseh N, et al. Notch inhibition during corneal epithelial wound healing promotes migration. Invest Ophthalmol Vis Sci. 2012;53(12):7476–7483. doi: 10.1167/iovs.12-10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Outtz HH, Wu JK, Wang X, Kitajewski J. Notch1 deficiency results in decreased inflammation during wound healing and regulates vascular endothelial growth factor receptor-1 and inflammatory cytokine expression in macrophages. J Immunol. 2010;185(7):4363–4373. doi: 10.4049/jimmunol.1000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Demehri S, Liu Z, Lee J, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6(5):e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ambler CA, Watt FM. Adult epidermal Notch activity induces dermal accumulation of T cells and neural crest derivatives through upregulation of jagged 1. Development. 2010;137(21):3569–3579. doi: 10.1242/dev.050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10(3):207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16(1):55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pece S, Serresi M, Santolini E, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167(2):215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Miele L, Golde T, Osborne B. Notch signaling in cancer. Curr Mol Med. 2006;6(8):905–918. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- 150.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66(3):1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 151.Galluzzo P, Bocchetta M. Notch signaling in lung cancer. Expert Rev Anticancer Ther. 2011;11(4):533–540. doi: 10.1586/era.10.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bertrand FE, Angus CW, Partis WJ, Sigounas G. Developmental pathways in colon cancer: crosstalk between WNT, BMP, Hedgehog and Notch. Cell Cycle. 2012;11(23):4344–4351. doi: 10.4161/cc.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tang SC, Chen YC. Novel therapeutic targets for pancreatic cancer. World J Gastroenterol. 2014;20(31):10825–10844. doi: 10.3748/wjg.v20.i31.10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Reedijk M. Notch signaling and breast cancer. Adv Exp Med Biol. 2012;727:241–257. doi: 10.1007/978-1-4614-0899-4_18. [DOI] [PubMed] [Google Scholar]
- 155.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14(5):779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 156.Dickson BC, Mulligan AM, Zhang H, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20(6):685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 157.Reedijk M, Pinnaduwage D, Dickson BC, et al. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008;111(3):439–448. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 158.Wei P, Walls M, Qiu M, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9(6):1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 159.Konishi J, Kawaguchi KS, Vo H, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67(17):8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 160.Hoey T, Yen WC, Axelrod F, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5(2):168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 161.Lewis HD, Leveridge M, Strack PR, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14(2):209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 162.Harrison H, Farnie G, Howell SJ, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70(2):709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kondratyev M, Kreso A, Hallett RM, et al. Gamma-secretase inhibitors target tumor-initiating cells in a mouse model of ERBB2 breast cancer. Oncogene. 2012;31(1):93–103. doi: 10.1038/onc.2011.212. [DOI] [PubMed] [Google Scholar]
- 164.Sharma A, Paranjape AN, Rangarajan A, Dighe RR. A monoclonal antibody against human Notch1 ligand-binding domain depletes subpopulation of putative breast cancer stem-like cells. Mol Cancer Ther. 2012;11(1):77–86. doi: 10.1158/1535-7163.MCT-11-0508. [DOI] [PubMed] [Google Scholar]
- 165.Qiu M, Peng Q, Jiang I, et al. Specific inhibition of Notch1 signaling enhances the antitumor efficacy of chemotherapy in triple negative breast cancer through reduction of cancer stem cells. Cancer Lett. 2013;328(2):261–270. doi: 10.1016/j.canlet.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 166.Zang S, Chen F, Dai J, et al. RNAi-mediated knockdown of Notch-1 leads to cell growth inhibition and enhanced chemosensitivity in human breast cancer. Oncol Rep. 2010;23(4):893–899. doi: 10.3892/or_00000712. [DOI] [PubMed] [Google Scholar]
- 167.Jesus AA, Goldbach-Mansky R. IL-1 blockade in autoinflammatory syndromes. Annu Rev Med. 2014;65:223–244. doi: 10.1146/annurev-med-061512-150641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Orlowski RZ, Gercheva L, Williams C, et al. A phase II, randomized, double-blind, placebo-controlled study of Siltuximab (Anti-IL-6 mAb) and bortezomib versus bortezomib alone in patients with relapsed or refractory multiple myeloma. Am J Hematol. 2014 doi: 10.1002/ajh.23868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Angevin E, Tabernero J, Elez E, et al. A phase I/II, multiple-dose, dose-escalation study of siltuximab, an anti-interleukin-6 monoclonal antibody, in patients with advanced solid tumors. Clin Cancer Res. 2014;20(8):2192–2204. doi: 10.1158/1078-0432.CCR-13-2200. [DOI] [PubMed] [Google Scholar]
- 170.Rath KS, Naidu SK, Lata P, et al. HO-3867, a safe STAT3 inhibitor, is selectively cytotoxic to ovarian cancer. Cancer Res. 2014;74(8):2316–2327. doi: 10.1158/0008-5472.CAN-13-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tierney BJ, McCann GA, Cohn DE, et al. HO-3867, a STAT3 inhibitor induces apoptosis by inactivation of STAT3 activity in BRCA1-mutated ovarian cancer cells. Cancer Biol Ther. 2012;13(9):766–775. doi: 10.4161/cbt.20559. [DOI] [PubMed] [Google Scholar]
- 172.Assi HH, Paran C, VanderVeen N, et al. Preclinical characterization of signal transducer and activator of transcription 3 small molecule inhibitors for primary and metastatic brain cancer therapy. J Pharmacol Exp Ther. 2014;349(3):458–469. doi: 10.1124/jpet.114.214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bendell JC, Hong DS, Burris HA, III, et al. Phase 1, open-label, dose-escalation, and pharmacokinetic study of STAT3 inhibitor OPB-31121 in subjects with advanced solid tumors. Cancer Chemother Pharmacol. 2014;74(1):125–130. doi: 10.1007/s00280-014-2480-2. [DOI] [PubMed] [Google Scholar]
- 174.LoRusso PM, Rudin CM, Reddy JC, et al. Phase I trial of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin Cancer Res. 2011;17(8):2502–2511. doi: 10.1158/1078-0432.CCR-10-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rodon J, Tawbi HA, Thomas AL, et al. A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin Cancer Res. 2014;20(7):1900–1909. doi: 10.1158/1078-0432.CCR-13-1710. [DOI] [PubMed] [Google Scholar]
- 176.Tolcher AW, Messersmith WA, Mikulski SM, et al. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J Clin Oncol. 2012;30(19):2348–2353. doi: 10.1200/JCO.2011.36.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sims-Mourtada J, Izzo JG, Apisarnthanarax S, et al. Hedgehog: an attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin Cancer Res. 2006;12(21):6565–6572. doi: 10.1158/1078-0432.CCR-06-0176. [DOI] [PubMed] [Google Scholar]
- 178.Pasi F, Facoetti A, Nano R. IL-8 and IL-6 bystander signalling in human glioblastoma cells exposed to gamma radiation. Anticancer Res. 2010;30(7):2769–2772. [PubMed] [Google Scholar]
- 179.Tata PR, Mou H, Pardo-Saganta A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503(7475):218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gupta PB, Fillmore CM, Jiang G, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]