High-density fibrillar collagen matrix induces invadopodia formation in both fibroblasts and carcinoma cell lines through a kindlin2-dependent mechanism that drives local ECM remodeling.
Abstract
Cell interactions with the extracellular matrix (ECM) can regulate multiple cellular activities and the matrix itself in dynamic, bidirectional processes. One such process is local proteolytic modification of the ECM. Invadopodia of tumor cells are actin-rich proteolytic protrusions that locally degrade matrix molecules and mediate invasion. We report that a novel high-density fibrillar collagen (HDFC) matrix is a potent inducer of invadopodia, both in carcinoma cell lines and in primary human fibroblasts. In carcinoma cells, HDFC matrix induced formation of invadopodia via a specific integrin signaling pathway that did not require growth factors or even altered gene and protein expression. In contrast, phosphoproteomics identified major changes in a complex phosphosignaling network with kindlin2 serine phosphorylation as a key regulatory element. This kindlin2-dependent signal transduction network was required for efficient induction of invadopodia on dense fibrillar collagen and for local degradation of collagen. This novel phosphosignaling mechanism regulates cell surface invadopodia via kindlin2 for local proteolytic remodeling of the ECM.
Introduction
Dynamic bidirectional interactions between cells and their surrounding ECM can regulate cell migration, invasion, proliferation or death, and differentiation (Bissell et al., 1982; Hay, 1991; Hynes, 2009; Schultz et al., 2011). Distinct physical properties of an ECM, such as its biochemical composition, stiffness, elasticity, density, or extent of cross-linking can alter cell behavior; conversely, cells can extensively remodel ECM locally using proteases (e.g., see Hotary et al., 2006; Tang et al., 2013). The detailed molecular signaling mechanisms that regulate these processes are still relatively poorly understood.
Extracellular matrix organization and homeostasis are often dramatically dysregulated in advanced malignancy. Fibrillar collagen, particularly collagen type I, is deposited densely in stroma adjacent to tumors (Zhu et al., 1995; Kauppila et al., 1998, 1999; van Kempen et al., 2008; Huijbers et al., 2010; Shields et al., 2012). This dense desmoplastic microenvironment promotes tumor progression and metastasis, and it correlates with poor prognosis in cancer patients (Provenzano et al., 2008; Conklin and Keely, 2012). Tumor cells can locally remodel the ECM; for example, intravital imaging of fibrillar collagen at the carcinoma tumor–stroma interface reveals that during tumorigenesis, collagen fibrils in a dense ECM network become radially aligned to facilitate tumor cell migration away from the tumor (Provenzano et al., 2006). Increased collagen density renders fibrillar collagen matrix stiffer (Roeder et al., 2002). Besides changes in collagen density, enzymatic cross-linking of collagen during tumor progression can also lead to matrix stiffening, and stiffened cross-linked fibrillar collagen promotes invasion by oncogene-initiated epithelium (Levental et al., 2009). However, mechanisms by which the density of collagen fibrils per se might promote an invasive or matrix-remodeling phenotype remain to be explored.
To locally degrade and sometimes to invade ECM barriers, cells use protrusions termed invadosomes, which consist of invadopodia or podosomes (Chen, 1989; Linder et al., 2011). Invadopodia are dynamic microscopic protrusions of plasma membrane rich in proteases with a diameter of ∼1 µm and ≤5 µm length. Invadopodial internal structure is complex and includes an actin-rich core with actin-nucleating machinery including the Arp2/3–neuronal WASP (Wiskott–Aldrich syndrome protein)–WASP-interacting protein complex; regulators of actin bundling and turnover such as cortactin, cofilin, fascin, and RhoGTPases; and a variety of adaptor proteins mediating protein complexes within the actin core such as AFAP-110 and the Tks family (Bharti et al., 2007; Diaz et al., 2009; Li et al., 2010; Oser et al., 2010; Schoumacher et al., 2010; Hu et al., 2011; Monteiro et al., 2013; Sharma et al., 2013; Razidlo et al., 2014; Williams et al., 2014). Invadopodia are now considered to be hubs of coordinated cell adhesion, signaling, actin polymerization and remodeling, directional endo/exocytosis, and ECM proteolysis. ECM rigidity alone can influence the matrix-degrading activity of invadopodia via a myosin II–FAK–Cas pathway (Alexander et al., 2008). The composition of the ECM can also affect invadosomes. For example, collagen fibrils can promote the formation of linear arrays of invadopodia along stress fibers (Juin et al., 2012), and the blunt invadosomes termed podosomes can be induced in megakaryocytes by interaction with a collagen substrate (Schachtner et al., 2013).
In general, invadopodial and invadosome mechanosensing, structure, function, and regulation have been studied using model systems based on gelatin, globular fibronectin, low-concentration fibrillar collagen and polyacrylamide matrices, or intact basement membranes (Artym et al., 2009; Weaver et al., 2013). We describe new assay systems based on high-density fibrillar collagen (HDFC) or decellularized tumor tissue containing high concentrations of collagen to mimic the dense stromal collagenous cancer environment to search for novel mechanisms regulating invadopodia formation. Unexpectedly, even in the absence of significantly altered gene or specific protein expression, dense fibrillar collagen can itself activate a complex, posttranscriptional integrin regulatory network to induce robust induction of invadopodia and local matrix degradation. This induction by a specific type of ECM stimulus occurs in both human cancer cells and in nonmalignant primary human fibroblasts. The large magnitude of this response without any requirement for growth factor stimulation provided an opportunity to characterize new regulators and resulted in identification of a key role for kindlin2 phosphorylation.
Results
HDFC is a potent inducer of invadopodia
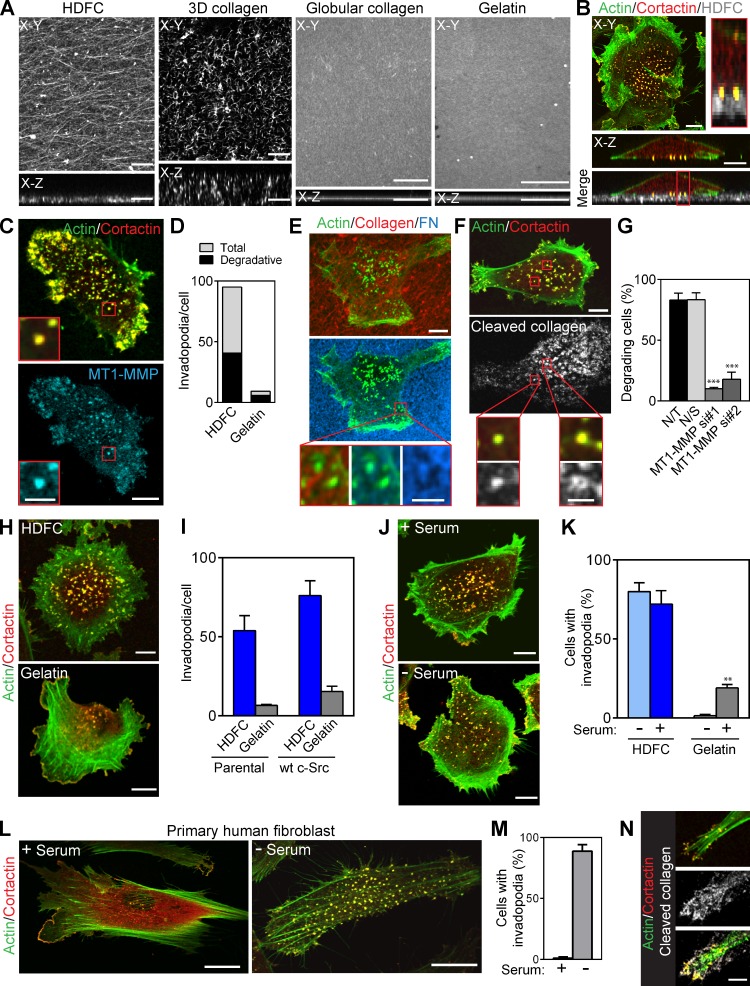
Formation of invadopodia by invading cancer cells has been studied extensively using artificial gelatin or globular fibronectin substrates in vitro. To explore the role of the density of native fibrillar collagen on invadopodia formation and function, we developed an in vitro HDFC matrix system (Fig. 1 A). HDFC matrix consists of a 4–5-µm-thick layer of densely packed fibrillar collagen type I compressed by centrifugation. HDFC is thin enough for high-resolution microscopy, while providing sufficient depth for 3D invadopodial penetration and proteolytic degradation of fibrillar collagen. Direct comparisons with a noncompressed collagen gel, a layer of globular collagen, and a conventional thin gelatin layer confirms that HDFC is a fine layer of a dense meshwork with well-preserved fibrillar topography (Fig. 1 A).
Figure 1.
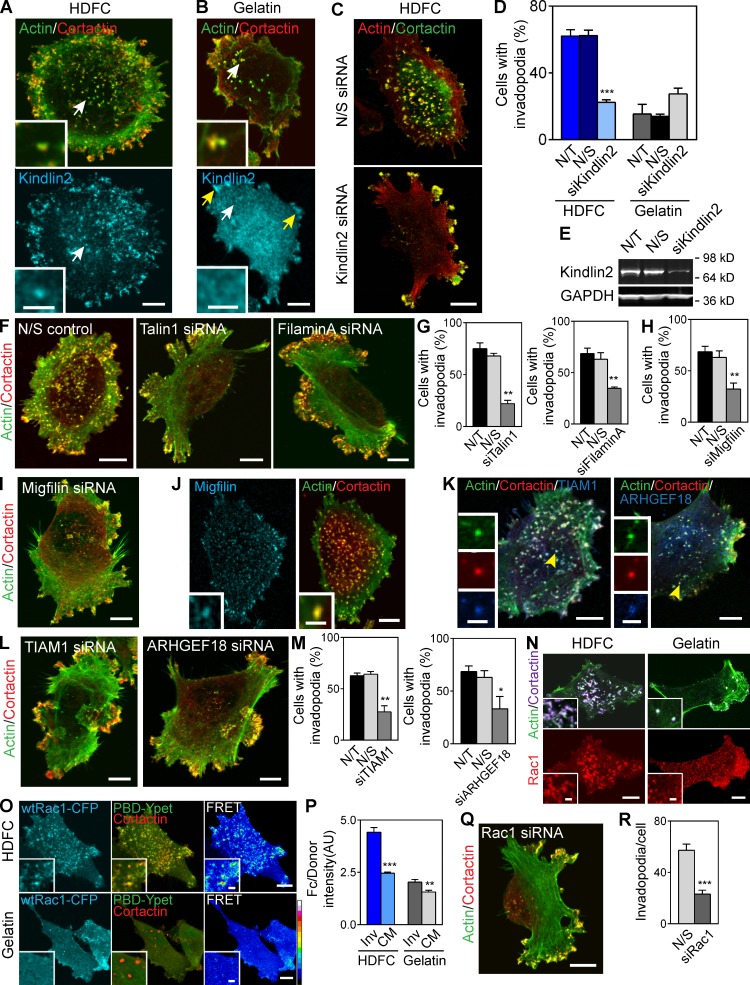
Potent induction of invadopodia by HDFC. (A) Topography and orthogonal confocal views of HDFC, thin 3D collagen, globular collagen, and gelatin matrices labeled with Alexa Fluor 568F. Confocal z stacks of the matrices were deconvolved using blind deconvolution algorithm set to 10 iterations (AutoDeblur software; Media Cybernetics). (B) Confocal and orthogonal views of MDA-MB-231 carcinoma cell invading HDFC matrix. Invadopodia appear as yellow dots with colocalized actin and cortactin that penetrate into fluorescently labeled HDFC matrix. Inset (red rectangle) shows a magnified orthogonal view of the invadopodia. (C) Endogenous MT1-MMP accumulation at invadopodia of MDA-MB-231 cells on HDFC. Insets (red outlines) show magnified views of the invadopodia. (D) Quantification of total and degradative invadopodia in MDA-MB-231 cells invading HDFC and gelatin matrices. Total invadopodia were identified as aggregates of colocalized actin and cortactin at the cell membrane adherent to the matrix, and degradative invadopodia were actin/cortactin aggregates with colocalized MT1-MMP. Mean number of invadopodia per cell with 19–20 cells analyzed per condition. (E) Proteolytic degradation as indicated by black holes in a layer of fibronectin (FN) bound from serum to HDFC by MDA-MB-231 cells invading the matrix. Degradation is present at invadopodia (actin aggregate). Magnified views of the invadopodia and corresponding area of matrix degradation are shown in the red-framed insets. (F) Immunofluorescence labeling of 1/4 collagen type I fragment. Proteolytic cleavage of fibrillar collagen by MDA-MB-231 cells invading HDFC was readily detected in serum-free medium (gray–white localization). Insets (red boxes) show enlarged views of the invadopodia and localized collagen degradation. (G) Effect of MT1-MMP silencing in MDA-MB-231 cells on HDFC degradation identified by immunofluorescence labeling of cleaved 1/4 collagen type I fragment under cells. Cells were transfected with two different single duplex siRNAs specific to MT1-MMP (MT1-MMP si#1 and si#2). Controls were mock-transfected (not transfected [N/T]) or transfected with nonspecific control siRNA (N/S). Mean ± SEM from each of 100–106 cells from each of three independent experiments. ***, P < 0.0001. (H) Confocal images of MDA-MB-231 cells invading HDFC and 2D gelatin matrices. (I) Quantification of invadopodia in parental and c-Src–expressing MDA-MB-231 cells invading HDFC or gelatin matrices. Means ± SEM from each of 12–15 cells. wt, wild type. (J) Immunostaining of MDA-MB-231 carcinoma cells invading HDFC matrix in medium with or without serum. (K) Quantification of invadopodia formation in MDA-MB-231 cells invading HDFC or gelatin in media with or without serum. Means ± SEM of 80–130 cells from each of three independent experiments. **, P < 0.001. (L) Confocal images of primary HFFs invading HDFC matrix in the presence or absence of serum. (M) Quantification of results from L showing the number of invadopodia-forming HFFs plated on HDFC matrix in the presence or absence of serum in culture medium. Means ± SEM from each of 90–100 cells from each of three independent experiments. (N) Confocal images of the HFF invadopodia and associated HDFC degradation as detected by immunofluorescence of 1/4 collagen type I fragments. Bars: (A–C [main images], E and F [main images], H, I, and N) 10 µm; (C and F, insets) 3 µm; (E, insets) 2 µm; (L) 20 µm.
HDFC matrix induced multiple invadopodia in a variety of tumor cells, including breast carcinoma MDA-MB-231 (Fig. 1 B), fibrosarcoma HT-1080, and prostate carcinoma PC3 (Fig. S1, A and B). Invadopodia were easily identified as actin–cortactin-rich aggregates associated with cell membrane adherent to HDFC and penetrating deep into the matrix (Fig. 1 B, X-Z inset). Degradative invadopodia displaying colocalization of the protease MT1–matrix metalloproteinase (MMP) with the classical actin–cortactin invadopodial markers comprised ∼45% of the invadopodia (Fig. 1, C and D; and Fig. S1 C). As expected, HDFC-induced invadopodia were capable of matrix degradation, e.g., detected as foci of degraded fibronectin that had bound to HDFC from serum (Fig. 1 E). Conversely, an antibody specific for cleaved collagen I stained the regions of HDFC cleaved by invadopodia (Fig. 1 F). Knockdown of MT1-MMP inhibited matrix degradation by these tumor cells, indicating an MT1-MMP requirement for local proteolysis (Fig. 1 G and Fig. S1 D). A close examination of HDFC-induced invadopodia revealed varying morphologies: they ranged from actin–cortactin-rich aggregates similar to invadopodia found on 2D gelatin to prominent ruffles and slender protrusions emanating from actin–cortactin cores penetrating the HDFC matrix (Fig. S1 E).
In comparisons with a standard gelatin invadopodia assay, HDFC displayed substantially more robust induction of invadopodia in a variety of cancer cells (Fig. 1, H–K; and Fig. S1, A and B). Growth of these cancer cells on HDFC for even short periods of time induced a strong invadopodial response, with substantially higher percentages of invadopodia-positive cells (Fig. 1 K) and six- to sevenfold more invadopodia per cell than on the conventional 2D gelatin matrix commonly used to study invadopodia (Fig. 1, H and I). Surprisingly, the formation of invadopodia on HDFC remained equally efficient even in the absence of serum, unlike the formation of invadopodia on classical 2D gelatin substrates, which is known to require serum (Fig. 1, J and K; and Fig. S1, A and B). Moreover, many more invadopodia were induced by HDFC with or without serum than by the commonly used approach of artificially overexpressing Src to boost invadopodia on gelatin substrates (Fig. 1 I).
When primary human fibroblasts were cultured on HDFC, they unexpectedly responded with a massive production of invadopodia that was at least as strong as in tumor cells (Fig. 1 L, right). The invadopodial response was suppressed by fetal bovine serum, which instead enhanced focal adhesions (FAs) and stress fiber formation on HDFC (Fig. 1, L and M). The human fibroblasts on HDFC displayed prominent local foci of HDFC collagen degradation, e.g., at individual invadopodia (Fig. 1 N).
Dense collagen polymerized in vitro or isolated from cancerous tissues induces abundant invadopodia
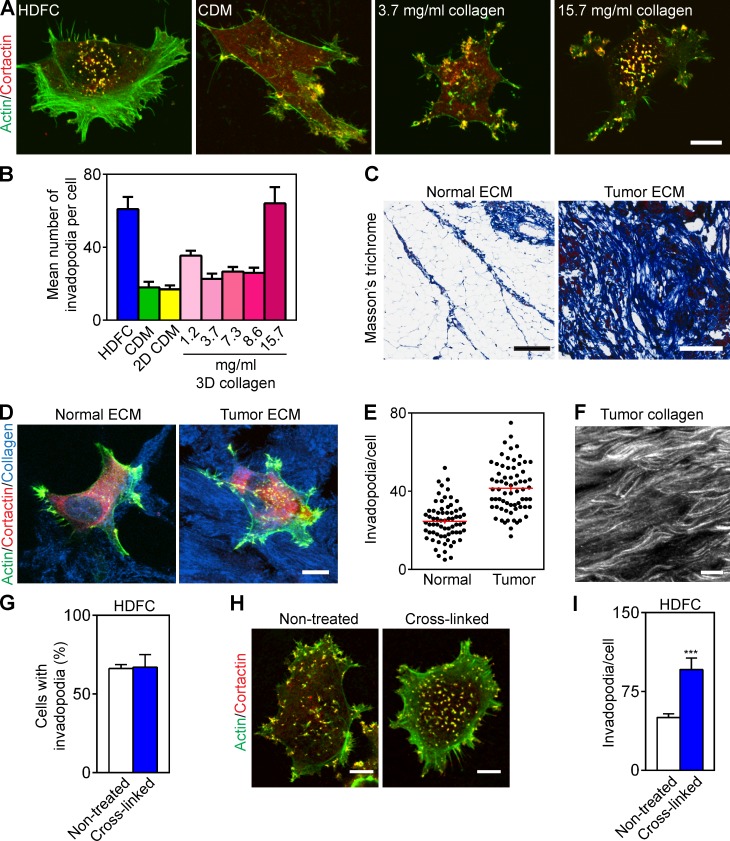
We tested the hypothesis that invadopodia are induced by the density of fibrillar collagen using thin gels of 3D fibrillar collagen type I matrix at varying concentrations compared with a cell-derived matrix (CDM). Only dense 3D collagen (15–16 mg/ml) could induce high numbers of invadopodia per cell, whereas fibrillar collagen polymerized at concentrations of 1.2–8.6 mg/ml and dense CDM containing fibrillar fibronectin could not (Fig. 2, A and B).
Figure 2.
HDFC mimics tumor desmoplastic collagen. (A) Maximum-intensity projections of immunofluorescence confocal images of MDA-MB-231 cells invading HDFC, cell-derived fibrillar fibronectin matrix (CDM), or thin 3D layers of fibrillar collagen type I polymerized at low versus high concentrations. Invadopodia are yellow dots of overlaid cortactin and actin staining. (B) Quantification of results from A showing invadopodia per MDA-MB-231 cell invading HDFC, CDM, flattened 2D CDM, or thin 3D fibrillar collagen matrices at the specified concentrations. Means ± SEM of each of 10–30 cells/condition from three independent experiments. (C) Masson’s trichrome staining of acellular ECM from normal and tumor breast sections visualizing collagen in blue. (D) Immunostaining of MDA-MB-231 cells invading acellular normal and tumor breast ECM immunolabeled for collagen type I. Invadopodia are yellow dots with colocalized actin and cortactin. (E) Invadopodial response of MDA-MB-231 cells plated on acellular normal or malignant breast ECM. The values (red bars) are mean ± SEM of total invadopodia in each of 68 cells from three independent repeat experiments for each condition. (F) Topography of desmoplastic collagen immunostained for collagen type I. (G) Quantification of invadopodia in cells invading nontreated or chemically cross-linked (4% paraformaldehyde) HDFC matrix. Mean ± SEM of each of 100–111 cells/condition from three independent experiments. (H) Immunostaining of invadopodia in MDA-MB-231 cells invading nontreated or cross-linked HDFC. Invadopodia are yellow dots of colocalized invadopodial markers, actin, and cortactin. (I) Quantification of invadopodia in MDA-MB-231 cells invading nontreated and cross-linked HDFC as in H. Means ± SEM of 19–20 cells/condition. ***, P < 0.0001. Bars: (A, D, F, and H) 10 µm; (C) 300 µm.
The ECMs of advanced metastatic tumors are often characterized by increased deposition of fibrillar collagen (Fig. 2 C and Fig. S1, F and G). We compared the effect of decellularized ECMs isolated from normal and malignant human breast tissue, characterized by low and high concentrations of fibrillar collagen, respectively, on induction of invadopodia. We found that dense collagen matrix isolated from human breast tumors stimulates a doubling of the numbers of invadopodia on breast carcinoma MDA-MB-231 cells compared with normal breast ECM (Fig. 2, D and E); the total numbers were similar to those induced by HDFC. Consequently, high-concentration, dense fibrillar collagen from human patients induces a similar invadopodial response as HDFC. Moreover, immunostaining of decellularized human tumor ECMs for collage type I revealed similarities between HDFC and human desmoplastic ECM in fibrillar matrix topography and density of fibrillar collagen organization (Fig. 2 F and Fig. 1 A).
Dense fibrillar collagen matrix becomes cross-linked in the course of tumorigenesis. We therefore compared the effect of HDFC cross-linking on invadopodia formation (Fig. 2, G–I). Both nontreated HDFC and HDFC chemically cross-linked with paraformaldehyde induced abundant invadopodia in the same high numbers of tumor cells (∼65%; Fig. 2, G and H), though cross-linking further increased total numbers of invadopodia per cell (Fig. 2 I).
We used atomic force microscopy (AFM) to compare the stiffness of nontreated and cross-linked HDFC matrices with 2D gelatin and globular collagen matrices as well as thin 3D collagen matrices polymerized at 1.3–15 mg/ml (Table 1). Chemical cross-linking of HDFC matrix increased its elastic modulus ∼10-fold. Although cross-linked HDFC matrices were much more pliable than 2D gelatin and globular collagen matrices adsorbed onto glass surfaces, they induced many more invadopodia. Moreover, 5% gelatin cushions, ∼1-µm-thick gelatin films cross-linked with glutaraldehyde (Alexander et al., 2008), had elastic moduli close to cross-linked HDFC matrix as measured with AFM. Yet, they elicited invadopodial responses closer to 2D gelatin matrices (Fig. S1 H), which were substantially less than the strong invadopodial induction by nontreated or cross-linked HDFC matrices (Fig. 2 H).
Table 1.
Stiffness of matrices as determined by AFM
| Matrix | Elastic modulusc | SD | Na | nb |
| kPa | kPa | |||
| HDFC, nontreated | 4 | 1 | 4 | 685 |
| HDFC, cross-linked | 56 | 29 | 8 | 1,474 |
| Gelatin | 236 | 121 | 8 | 803 |
| 5% Gelatin cushions | 30 | 4 | 2 | 283 |
| Globular collagen | 216 | 24 | 2 | 345 |
| 3D collagen, 1.3 mg/ml | 1.7 | 2.6 | 4 | 893 |
| 3D collagen, 8.6 mg/ml | 6.3 | 2.3 | 7 | 1,785 |
| 3D collagen, 15 mg/ml | 17.7 | 9.8 | 10 | 2,402 |
The elastic modulus is given as means ± SD. The raw AFM data used to derive mean and SD values of matrix stiffness are provided in dataset 1.
Number of independent sites measured in two to four independent experiments.
Number of independent points.
The differences between treatment groups (except for gelatin vs. globular collagen) were statistically significant according to Kruskal–Wallis test (P < 0.001) and Dunn’s post-test (P < 0.05).
Integrins required for invadopodial induction
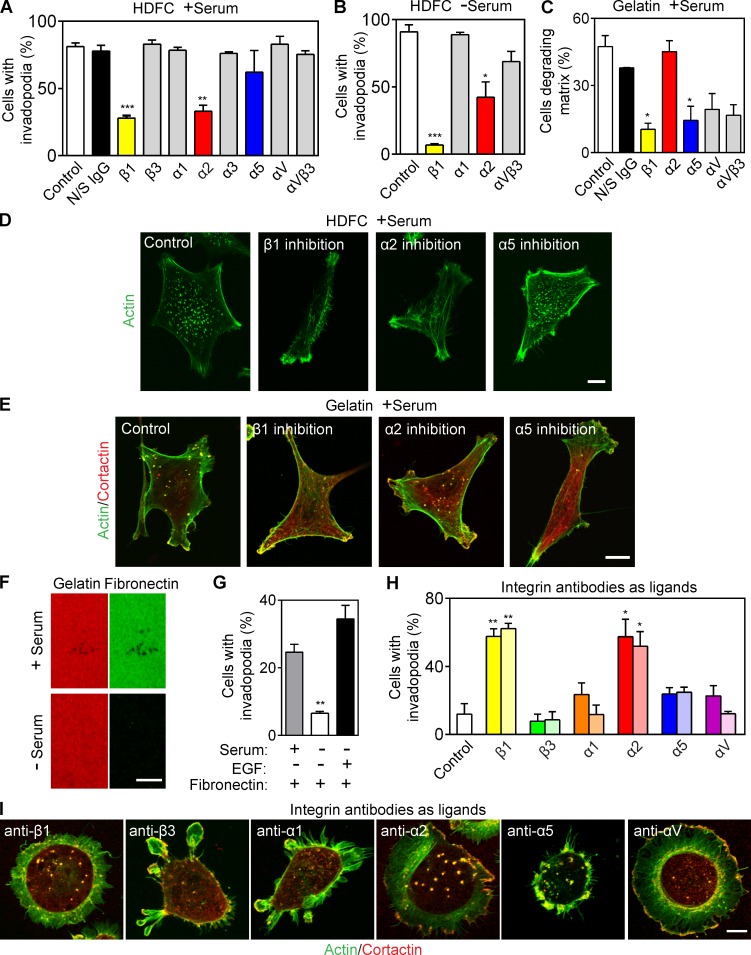
Human breast carcinoma MDA-MB-231 cells express multiple cell surface integrin subunits (Fig. S2 A). We tested an array of inhibitory antibodies against specific integrins for their ability to block invadopodia formation; only inhibition of β1 or α2 integrin subunits inhibited HDFC induction of invadopodia (Fig. 3, A, B, and D), regardless of the presence of serum. Importantly, the concentration of antibody used did not produce any inhibition of cell migration (Fig. S2, B and C). Inhibition of α1, α5, or αV integrins did not prevent induction of invadopodia by HDFC. Immunostaining for α2 and activated β1 integrins colocalized both of these subunits to the invadopodia of carcinoma cells invading HDFC matrix (Fig. S2 F).
Figure 3.
α2β1 integrin is the receptor for HDFC and is a potent inducer of invadopodia. (A) Effect of specific integrin inhibition on invadopodia formation in MDA-MB-231 cells invading HDFC in serum-containing medium. Means ± SEM from 100–150 cells for each condition with three independent repeats. N/S, nonspecific. (B) Effect of integrin inhibition by mAbs on invadopodia formation in MDA-MB-231 cells invading HDFC in serum-free medium. Means ± SEM of 100 cells/condition with three repeats. (C) Effect of specific integrin inhibition on invadopodia formation in MDA-MB-231 cells invading 2D gelatin in the presence of serum. The values are mean number of cells with ECM-degrading invadopodia ± SEM of 100–150 cells/condition, with three independent repeats. (D) Representative images for integrin inhibitory experiments of MDA-MB-231 cells invading HDFC in the presence of serum in A. (E) Representative images for integrin inhibitory studies in C. (F) Immunostaining of fibronectin (green) bound to the gelatin matrix with or without serum in the medium. (G) Effect of fibronectin-coated gelatin matrix on invadopodia induction in c-Src–expressing MDA-MB-231 cells. Serum-starved cells were plated on the fibronectin-coated gelatin in the presence or absence of serum or with 5 nM EGF. Means ± SEM of 90–100 cells per condition with three independent experiments. (H) Percentage of MDA-MB-231 cells with invadopodia after adhesion to antibody-coated substrates targeting individual integrin subunits. Means ± SEM of ∼100 cells per condition with three experiments each, comparing antibodies at 10 µg (dark bar) or 200 µg (light bar) per 14-mm-diameter coverslip. (I) Representative micrographs of invadopodia formation by MDA-MB-231 carcinoma cells induced by adhesion to antibody-coated substrates targeting individual integrin subunits as in H. Invadopodia are yellow dots of colocalized actin and cortactin immunostaining. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. Bars, 10 µm.
On the contrary, integrin-inhibitory experiments identified that cells degrading 2D gelatin matrices required functional α5 and β1 subunits (the fibronectin receptor) to induce invadopodia (Fig. 3, C and E; and Fig. S2, D and E) but not α2 integrins. Fibronectin from serum binds to gelatin (Fig. 3 F) to serve as a ligand for this α5β1 integrin–mediated induction of invadopodia on gelatin matrix, and formation of invadopodia on gelatin was inhibited by omission of serum (Fig. 1 K) or by removal of fibronectin from serum (Fig. S2 G). As expected, activated β1 and α5 integrin localized to invadopodia on gelatin matrices (Fig. S2, H and I). However, α5β1 integrin binding to fibronectin required a costimulatory signal from a growth factor receptor to induce invadopodia (Fig. 3 G). In addition, HDFC induced higher overall levels of β1 integrin activation than gelatin matrix as shown by staining for activated β1 integrin (Fig. S2 J).
As a direct test of integrin specificity for invadopodia induction, MDA-MB-231 carcinoma cells were plated on antibodies against different integrin subunits immobilized on substrates to serve as integrin-specific adhesive ligands (Fig. 3, H and I). Subunit-specific integrin ligation induced invadopodia formation in ∼60% of cells attaching via cell surface β1 or α2 versus only ∼20% in cells with α5 integrin ligation. Thus, the α2β1 integrin collagen receptor is both necessary and sufficient for potent induction of invadopodia.
Potent induction of invadopodia does not require altered gene or protein expression
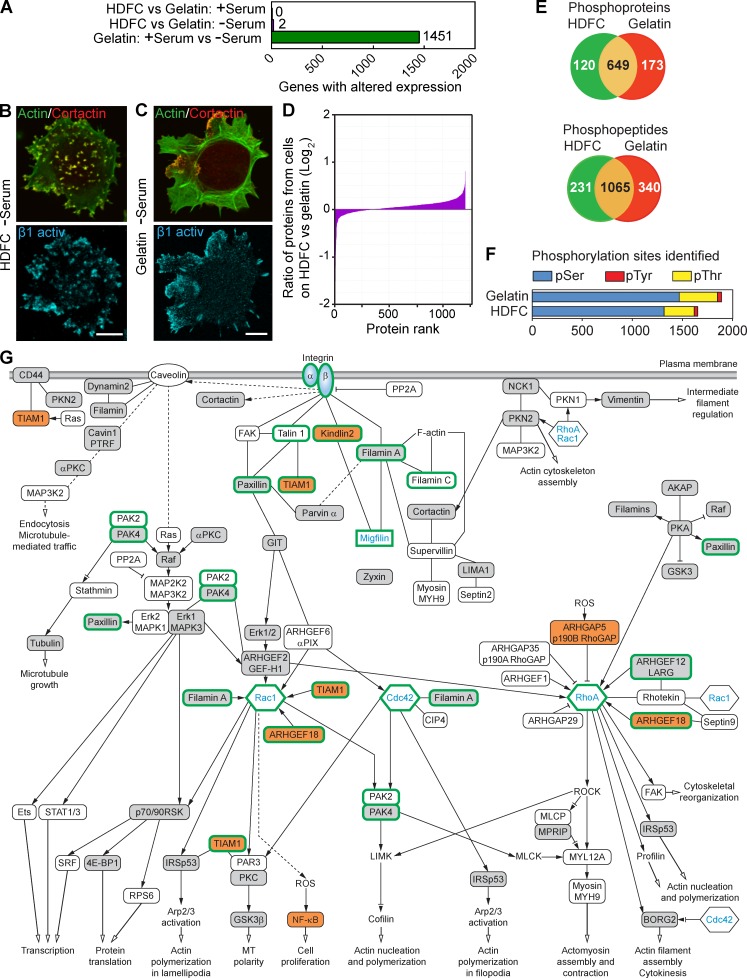
We searched for substrate-specific differential gene expression mediating the substantially enhanced induction of invadopodia by HDFC versus gelatin matrices. Remarkably, whole human genome microarray analysis revealed that the induction of abundant invadopodia by HDFC after overnight stimulation was not associated with any significant change in the pattern of gene expression (Fig. 4 A and Fig. S2 K). As a positive control, serum induced large shifts in gene expression.
Figure 4.
Phosphoproteomics analysis of cells invading HDFC matrix reveals complex downstream signaling. (A) Comparison of whole-genome microarray expression profiles for MDA-MB-231 cells with or without serum, or invading gelatin versus HDFC matrices, based on data pooled from five independent experiments for each condition. (B) Localization of activated β1 integrin to invadopodia of the MDA-MB-231 adherent to HDFC at the absence of serum. Invadopodia are yellow dots with colocalized actin and cortactin. (C) Localization of activated β1 integrin to FAs of MDA-MB-231 cells adherent to gelatin matrix in the absence of serum, showing poor invadopodia formation. (D) Comparative analysis of protein expression levels in MDA-MB-231 cells on HDFC versus cells on gelatin at the absence of serum. Cell lysates from each sample were labeled with specific iTRAQ labels to compare amounts of each protein (protein rank) identified by MS between HDFC and gelatin samples. (E) Numbers of unique and shared phosphoproteins and phosphopeptides in MDA-MB-231 carcinoma cells invading HDFC (green) compared with 2D gelatin (red) matrices in serum-free medium as identified by phosphoproteomics. (F) Quantification of types of phosphorylation sites identified by phosphoproteomics analysis. (G) Proposed signaling network associated with integrin-dependent induction of abundant invadopodia in carcinoma cells invading HDFC matrix. All identified phosphoproteins and their phosphosites are listed in Table S1. Open boxes indicate phosphoproteins identified in both HDFC and gelatin samples with the same phosphorylation sites by comparison of phosphopeptides. Gray boxes denote phosphoproteins identified in HDFC versus gelatin samples with different phosphorylation sites. Orange boxes indicate phosphoproteins unique to the HDFC matrix. Proteins without phosphorylation changes, such as Rac1, Cdc42, RhoA, and αβ integrin, are added to clarify the signaling context of proteins identified by phosphoproteomics. Although not regulated by phosphorylation, migfilin is depicted in the network because it is known to bind directly to both kindlin2 and filamin A. Migfilin and the other proteins highlighted by thick green outlines were verified experimentally in this study to play a role in invadopodia regulation by siRNA knockdown. Solid lines indicate known direct physical binding between proteins. Dashed lines indicate indirect interactions involving intermediate partners. Black arrows at the ends of lines indicate proteins known to stimulate the downstream signaling partner, lines with inhibition symbol indicate down-regulation of activity of the downstream signaling partner, lines with an inhibition symbol plus a black arrow indicate both potential activation and inhibition, and open arrows denote stimulation of the cellular process. Abbreviations used in this figure: ROCK, Rho-associated protein kinase; MLCP, myosin light-chain phosphatase; MLCK, myosin light-chain kinase; MPRIP, myosin phosphatase Rho-interacting protein; ROS, reactive oxygen species; SRF, serum response factor; AKAP, A kinase anchor protein. Bars, 10 µm.
Because gene expression analysis could not explain the prolific induction of invadopodia by dense fibrillar collagen, we applied proteomics approaches to compare cells with abundant invadopodia on HDFC versus extensive FAs on gelatin (Fig. 4, B and C). Consistent with the absence of altered gene expression, we found no differences in protein expression patterns, as determined by mass spectrometry (MS). Specifically, cell lysates of breast carcinoma cells cultured on HDFC versus gelatin substrates were analyzed after differential labeling with isobaric tags for relative and absolute quantitation (iTRAQ) to compare relative amounts of each protein. A total of 1,205 proteins analyzed by MS revealed identical protein profiles on HDFC versus gelatin within a factor of two, and no contamination of the lysates by matrix components from the substrates was detected (Fig. 4 D).
Phosphoproteomics analyses reveal a complex integrin signaling network underlying invadopodia formation
In striking contrast, phosphoproteomics analysis revealed characteristic, major changes in phosphorylation in signaling pathways associated with the potent induction of invadopodia in cells invading HDFC versus 2D gelatin. Phosphoproteomics analysis based on pooled data from two experiments, each with two technical replicates, identified 1,651 phosphorylation sites in 769 phosphoproteins associated with abundant invadopodia formation on HDFC compared with 1,889 phosphorylation sites in 822 phosphoproteins during FA formation with few invadopodia on gelatin (Fig. 4 E). The overall ratios of phosphoserine (∼80%) to phosphothreonine (∼18%) and phosphotyrosine (∼2%) were consistent with other cells (Fig. 4 F; Olsen et al., 2006).
Using Ingenuity Pathway Analysis, we focused on 82 phosphoproteins selected by three criteria: relevance to integrin signaling, known direct physical interactions to form molecular complexes, and potential relevance to invadopodia formation and function in ECM degradation (Table S1). These phosphoproteins could be organized into two signaling networks: a provisional integrin signaling network associated with the formation of invadopodia on dense fibrillar collagen by carcinoma cells (Fig. 4 G) versus a converse signaling network associated with FA formation on gelatin matrix (Fig. S3 A).
Integrin binding to ECM induces the assembly of macromolecular complexes comprised of numerous intracellular signaling and adaptor proteins regulating integrin activation, signaling, and connections to the actin cytoskeleton (CSK; Kim et al., 2011). Out of these many proteins, our phosphoproteomics data for integrin-mediated invadopodia formation by HDFC identified phosphorylation of kindlin2 unique to HDFC and not to gelatin (Fig. 4 G and Fig. S3 B). The analysis of phosphorylated proteins from gelatin samples identified multiple proteins known to function in FA and stress fiber regulation (Fig. S3 A; Vicente-Manzanares and Horwitz, 2011).
Kindlin2 is required for invadopodia formation selectively on dense fibrillar collagen
Kindlin2 plays an important role in activating integrins, but its potential regulation by phosphorylation was unexpected. Kindlin2 binds migfilin, which in turn binds filamin A, both associated with regulation of integrins and actin CSK. To test for a regulatory role for kindlin2 in invadopodia formation, we first examined localization of endogenous kindlin2 in cells invading HDFC versus gelatin (Fig. 5, A and B). Kindlin2 was found to be prominent in the invadopodia induced by HDFC but not in the invadopodia formed on gelatin. Next, we depleted kindlin2 using siRNAs and compared effects on invadopodia formation in breast carcinoma cells plated on HDFC or gelatin in serum-containing media permitting invadopodia assembly on both matrices. We established that kindlin2 was required for efficient invadopodia formation in cells invading HDFC but not gelatin (Fig. 5, C–E; and Fig. S4 D). Invadopodia formation on HDFC in the absence of serum also required kindlin2 (Fig. S4 E). Kindlin2 was also required for efficient HDFC induction of invadopodia in HT-1080 and PC-3 cells (see Fig. 7, C and D).
Figure 5.
Complex signaling network of multiple proteins regulating invadopodia formation on HDFC. (A) Immunostaining of endogenous kindlin2 in MDA-MB-231 cells invading HDFC reveals kindlin2 localization to invadopodia (white arrow and inset). (B) Immunostaining of endogenous kindlin2 in carcinoma cells adherent to gelatin demonstrates kindlin2 accumulation in FAs (yellow arrows) but not in invadopodia (white arrow and insert). (C) Representative images of carcinoma cells invading HDFC after transfection with nonspecific control (N/S) or kindlin2-specific siRNA pools. (D) Effect of knockdown with kindlin2-specific siRNA pool (siKindlin2) on invadopodia formation in MDA-MB-231 cells invading HDFC or gelatin matrices. Control conditions are mock transfection (not transfected [N/T]) or transfection with nonspecific siRNA control pool (N/S). Mean ± SEM of 100 cells/condition with three independent experiments. (E) Representative Western blot of kindlin2 knockdown in MDA-MB-231 cells in D. (F) Representative images of MDA-MB-231 cells transfected with nonspecific control or talin1-, filamin A–specific siRNA pools invading HDFC. (G) Effect of siRNA depletion of talin 1 or filamin A on invadopodia formation in carcinoma cells invading HDFC. Means ± SEM of 100 cells/condition with three independent experiments. (H) Effect of siRNA depletion of migfilin on invadopodia formation in carcinoma cells invading HDFC. Means ± SEM of 100 cells/condition with three independent experiments. (I) Representative images of HDFC-invading MDA-MB-231 cells transfected with migfilin-specific siRNA. (J) Immunostaining of endogenous migfilin in MDA-MB-231 cells localizes migfilin to actin/cortactin-rich invadopodia in cells invading HDFC. Insets show magnified views of invadopodia. (K) Immunofluorescence of endogenous TIAM1 and ARHGEF18 demonstrates localization of both GEFs to invadopodia of carcinoma cells induced by HDFC. Insets show enlarged views of invadopodia. (L) Representative images of TIAM1 and ARHGEF18 knockdown in MDA-MB-231 cells incubated on HDFC matrix. (M) Quantitation of effects of TIAM1 and ARHGEF18 depletion on invadopodia formation in carcinoma cells on HDFC. Means ± SEM of 100 cells/condition with three independent experiments. (N) Immunofluorescence of endogenous Rac1 in MDA-MB-231 carcinoma cells invading HDFC or gelatin. Insets zoom in on individual invadopodia. (O) Rac1 activation status at invadopodia induced by HDFC or gelatin visualized with Rac1 biosensor. Color palette depicts FRET efficiency from low (black) to high (white). Insets show high FRET signal on individual invadopodia induced by HDFC. wt, wild type. (P) Efficiencies of Rac1-CFP/PBD-YPet FRET at invadopodia (Inv) and adjacent cell membrane (CM) in carcinoma cells on HDFC or gelatin. Means ± SEM of 30–36 cells analyzed per condition. AU, arbitrary unit. (Q) Representative micrograph of Rac1-depleted MDA-MB-231 cells on HDFC. (R) Quantitation of Rac1 knockdown on invadopodia formation from Q. The values indicate the mean number of invadopodia per cell ± SEM based on 10 cells/condition. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. Bars: (A and B [main images], C, F, and J and K [main images], L, N and O [main images], and Q) 10 µm; (A and B, insets) 3 µm; (J, K, N, and O, insets) 2 µm.
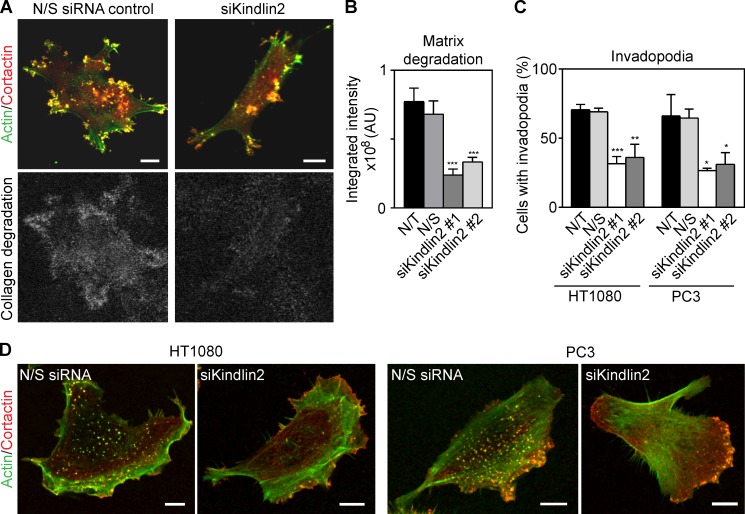
Figure 7.
Kindlin2 is required for invadopodia-mediated degradation of dense collagen. (A) Depletion of kindlin2 using kindlin2-specific single duplex siRNA inhibits degradation of HDFC collagen visualized by immunofluorescence of 1/4 collagen type I fragments. (B) Quantification of effect of kindlin2 depletion by single duplex siRNA on matrix degradation in A. Values are means ± SEM of ∼20 cells. AU, arbitrary unit. (C) Effect of kindlin2 knockdown on invadopodia formation in HT-1080 breast adenocarcinoma and PC3 prostate carcinoma cells. Values are means ± SEM of ∼100 cells/condition with three repeats. (D) Representative images after kindlin2 depletion in HT-1080 and PC3 cells. N/S, nonspecific; N/T, not transfected. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. Bars, 10 µm.
Multiple additional members of a complex invadopodial signaling network are required for invadopodia formation on dense fibrillar collagen
Although dozens of structural and signaling proteins are known to regulate or comprise invadopodia, our phosphoproteomics analyses identified several additional predicted key downstream molecular mediators of invadopodia formation. RhoGTPases are known to regulate invadopodia formation (Bravo-Cordero et al., 2011). Our phosphoproteomics data also revealed substantial changes in regulators of Rac1, Cdc42, and RhoA. TIAM1 and ARHGEF18 showed unique phosphorylation on HDFC matrix and were candidate activators of Rac1 activity downstream of HDFC-ligated integrin. Moreover, phosphorylation of the RhoA regulator ARHGAP5/p190B RhoGAP was enhanced on HDFC matrix. The latter regulates MT1-MMP expression and cell surface presentation (Guegan et al., 2008). Other regulators were phosphorylated on both HDFC and gelatin matrices, including the Rac regulators ARHGEF2/guanine nucleotide exchange factor (GEF)–H1 and ARHGEF6 and multiple RhoA regulators. These results identify selective regulation of the phosphorylation of a subset of protein regulators on HDFC compared with gelatin matrices, consistent with the regulation of invadopodia by signaling without any need for altered gene and protein expression.
We focused on putative signaling systems downstream of integrin and phospho-kindlin2, including migfilin, filamin A, and its HDFC-phosphospecific GEFs TIAM1 and ARHGEF18 (Fig. 4 G). Consistent with critical roles for each of the phosphoproteins implicated by phosphoproteomics, knockdown by siRNA (single siRNAs and pools) of talin1, filamin A, paxillin, TIAM1, or ARHGEF18 inhibited invadopodia formation on HDFC (Fig. 5, F, G, L, and M; and Fig. S4, A–F). Reducing talin1 and kindlin2 inhibited invadopodia but not cell spreading, indicating stronger requirements for invadopodia formation and possible redundancy with other talin and kindlin isoforms for spreading (Debrand et al., 2009; Lai-Cheong et al., 2010). Knockdown of gelatin-specific phosphoproteins filamin B and TIAM2 had no effect on invadopodia (Fig. S4, A–C and F). Localization of TIAM1 and ARHGEF18 to HDFC-induced invadopodia was confirmed by immunolabeling of endogenous proteins (Fig. 5 K). Although migfilin was not phosphorylated, it can interact with kindlin2 and filamin A (Tu et al., 2003). Migfilin depletion by siRNA confirmed its essential role in invadopodia formation on HDFC (Fig. 5, H and I; and Fig. S4, C and F). Confocal microscopy experiments revealed migfilin localization to invadopodia induced by HDFC (Fig. 5 J). Consequently, we introduced migfilin into the integrin signaling network underlying invadopodia formation on HDFC (Fig. 4 G).
Rac1 appeared to be particularly significant because of strong accumulation of endogenous Rac1 protein at invadopodia induced by HDFC but not by gelatin (Fig. 5 N). Cdc42 and RhoA colocalized to invadopodia on HDFC less intensely than Rac1 (Fig. S4 G). Rac1 activity was assayed by a fluorescence resonance energy transfer (FRET) biosensor, which demonstrated accumulation of activated Rac1 at invadopodia (Fig. 5, O and P; and Fig. S4 H). Overexpression of dominant-negative Rac1 or depletion of Rac1 with siRNA inhibited invadopodia formation on HDFC (Fig. S4 I and Fig. 5, Q and R).
The downstream Rac effector PAK4 is phosphorylated uniquely on HDFC, whereas PAK2 is phosphorylated similarly on HDFC and gelatin. Only depletion of PAK4, and not of PAK2, blocked invadopodia formation on HDFC (Fig. S4, A–C and F). These findings confirm that the additional molecules we identified by phosphoproteomics are essential and specific for induction of abundant invadopodia by collagen.
Phosphorylation of kindlin2 regulates invadopodia formation
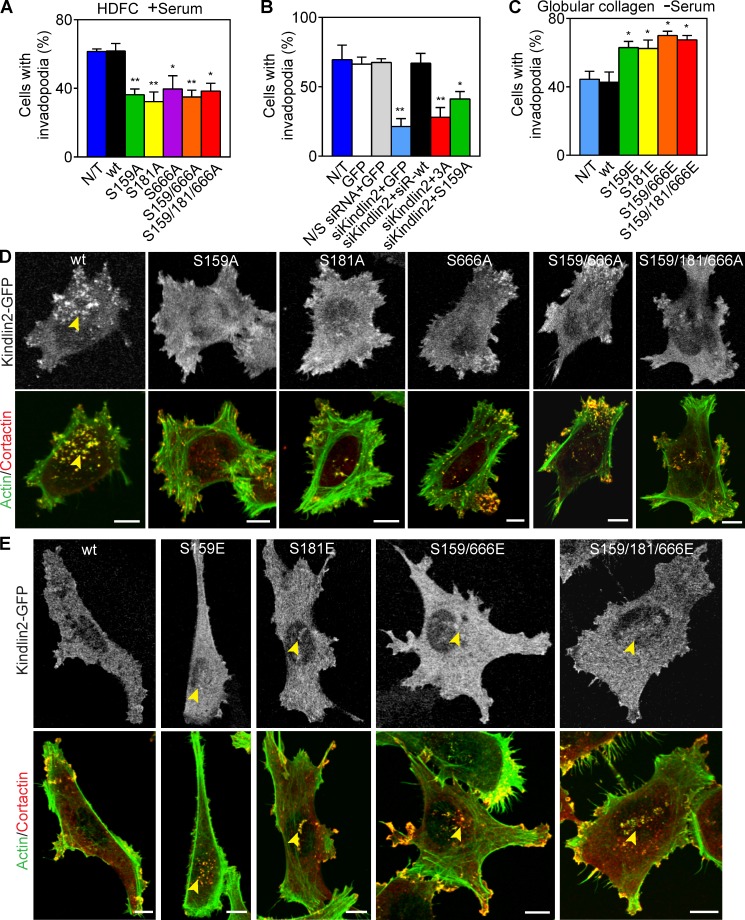
In silico analysis of kindlin2 protein structure predicts 7 threonine, 3 tyrosine, and 11 serine potential phosphorylation sites (Huang et al., 2005). Our proteomics analysis identified kindlin2 phosphoserine at position 159 (Fig. S3 B and Table S2). The phosphorylation of serine 159 in kindlin2 has also been found in cell migration on fibronectin (Schiller et al., 2013), T cell receptor signaling (Mayya et al., 2009), and stem cell differentiation (Rigbolt et al., 2011). Besides this directly determined site, the protein knowledge base UniProt lists serines at positions 181 and 666 as potential phosphosites. We mutated serines 159, 181, and 666 to generate potential dominant-negative mutants (S to A mutations) or phosphomimetics (S to E mutations) to test experimentally the role of kindlin2 serine phosphorylation on invadopodia formation in MDA-MB-231 cells.
All three potential dominant-negative mutants defective for serine phosphorylation significantly inhibited the formation of invadopodia in cells invading HDFC (Fig. 6, A and D). Moreover, in carcinoma cells depleted of endogenous kindlin2 by siRNA, formation of abundant invadopodia was rescued by expression of exogenous wild-type siRNA-resistant kindlin2 but not by expression of the corresponding phospho-null kindlin2 mutants, including S159A and S159/181/666A or 3A mutants (Fig. 6 B). Immunofluorescence detected colocalization of endogenous kindlin2 and migfilin at the tips of the subset of invadopodia (Fig. S5, A and B). However, immunoprecipitation of either kindlin2 or migfilin failed to identify a kindlin2–migfilin molecular complex in MDA-MB-231 cells on HDFC (Fig. S5 C). Nevertheless, overexpression of kindlin2-GFP S159A or S181A mutants resulted in a decrease in kindlin2-migfilin colocalization by immunofluorescence (Fig. S5, D and E), suggesting possible indirect kindlin2-to-migfilin interactions.
Figure 6.
Kindlin2 function in invadopodia formation is regulated by phosphorylation. (A) Effect of expressing dominant-negative phosphomutants of kindlin2 on invadopodia formation in MDA-MB-231 cells invading HDFC in the presence of serum. Means ± SEM of 100 cells/condition with 3–4 independent repeats. (B) Quantification of the effects of kindlin2 knockdown with single duplex siRNA and rescue with siRNA-resistant wild-type kindlin2 (siR-wt) or its mutants S159/181/666A (3A) and S159A on invadopodia formation in MDA-MB-231 cells invading HDFC. Values are means ± SEM of ∼100 cells/condition with three repeats. (C) Effect of expression of kindlin2 phosphomimetic mutants on invadopodia formation in carcinoma cells on globular collagen with no serum. Means ± SEM of 100 cells/condition with 3–4 repeats. (D) Effect of kindlin2-GFP dominant-negative mutants on invadopodia formation in MDA-MB-231 cells invading the HDFC matrix. Cells were cultured on HDFC for 3 h, fixed, and immunolabeled for actin and cortactin to identify invadopodia as actin–cortactin rich aggregates. The yellow arrowhead points to high numbers of invadopodia associated with the cell body. (E) Effect of kindlin2-GFP phosphomimetic mutants on invadopodia formation in MDA-MB-231 cells invading globular collagen matrix at the absence of serum. Cells were cultured on collagen for 3 h, fixed, and immunolabeled for actin and cortactin to identify invadopodia as actin–cortactin-rich aggregates. Yellow arrowheads indicate invadopodia on the cell body. N/T, not transfected; N/S, nonspecific; wt, wild type. *, P < 0.05; **, P < 0.001. Bars, 10 µm.
Conversely, the three gain-of-function kindlin2 phosphomimetic mutants artificially stimulated formation of invadopodia in carcinoma cells cultured on collagen matrices of low ligand density, i.e., a globular collagen layer (Fig. 6, C and E) or a thin 3D layer of 1.5 mg/ml collagen I (Fig. S5 F). The low number of invadopodia formed on globular collagen was independent of serum but required kindlin2 for invadopodia formation (Fig. S5, G–I).
Depletion of kindlin2 by siRNA down-regulated invadopodia formation and suppressed the accompanying local pericellular proteolysis of ECM as determined by the accumulation of collagen proteolytic cleavage fragments generated by MDA-MB-231, HT-1080, and PC3 cancer cells (Fig. 7, A–D). We conclude that dense fibrillar collagen regulates cell surface invadopodia via kindlin2 for local proteolytic remodeling of the ECM.
Discussion
Although the ECM is known to regulate and modulate a wide range of biological functions, less is known about the effects and mechanisms of ECM organization itself on processes that remodel matrix and promote cell invasion. Here, we demonstrate that dense fibrillar collagen is a potent inducer of cell surface invadopodia. We identify activation of a novel phospho-kindlin2–based integrin signaling network that stimulates massive invadopodia formation in tumor cells for local collagen degradation. Our findings provide mechanistic insight into the role of the ECM microenvironment in the regulation of invadopodia by describing a novel in vitro model to study invadopodia under physiologically relevant conditions as well as identifying a new phosphosignaling pathway that regulates invadopodia formation directly without altered gene and protein expression.
Matrix regulation of invadopodia in normal and malignant cells
The ability of cancer cells to locally invade and metastasize correlates with their propensity to form invadopodia in in vitro gelatin and fibronectin invasion assays (Coopman et al., 1998; Clark et al., 2007). The importance of invadopodia in ECM invasion in vivo is supported by live-cell imaging visualizing the formation and extension of invadopodial processes in cancer cells breaching the basement membrane in both in vivo and in in vitro intravasation assays (Gligorijevic et al., 2012). In addition, basement membrane degradation and interstitial ECM invasion depend on the activity of membrane-bound MMPs (Hotary et al., 2006) that must localize to invadopodia to carry out local ECM degradation (Artym et al., 2006). Our finding that dense fibrillar collagen can also induce abundant invadopodia in primary, nontransformed human cells that then degrade matrix locally reveals that invadopodia can be induced readily in “normal” cells. We suggest that this rapid local regulation of invadopodia by dense collagen may be valuable for physiological matrix remodeling but that it may also contribute to deleterious functions of cancer-associated fibroblasts (Gaggioli et al., 2007; Yamaguchi et al., 2014).
The invadopodia-stimulating effect of desmoplastic collagen is provided by its density and is recapitulated by using thin 3D layers of very dense collagen or by a new model of HDFC matrix comprised of fibrillar collagen compacted by centrifugation. Although collagen and fibronectin are the two most abundant ECM proteins deposited during tumor development (Cox and Erler, 2011), we established that it is the dense fibrillar collagen rather than dense fibrillar fibronectin that has this unique stimulatory effect on invadopodia formation. The very high numbers of invadopodia that were observed on HDFC matrix could not be explained simply by the stiffness of the matrix. The effect of HDFC on invadopodia up-regulation exceeded the previously published stimulatory effects of stiff artificial matrices on invadopodia in cancer cells (Alexander et al., 2008; Parekh et al., 2011) or globular collagen absorbed to stiff glass surfaces.
Integrins in invadopodia formation
Integrins were intimately involved in the matrix-induced induction of invadopodia, consistent with their roles as key sensors of ECM physicochemical properties (Hynes, 2002). Integrin-initiated downstream signaling regulates actin CSK, trafficking, and proteolysis as well as invadopodia formation and function. β1, α3, and α6 integrin subunits had previously been localized to invadopodia in vitro, and antibody- or ECM ligand-induced activation of β1 integrin increases degradation (Nakahara et al., 1996; Mueller et al., 1999). Moreover, tumor metastasis can be inhibited by genetically ablating β1 integrin expression, and mesenchymal cell proteolytic invasion is halted by β1 integrin inhibition (Sameni et al., 2008; Friedl and Wolf, 2010). Integrins function through the formation of intracellular macromolecular complexes that link specific integrins via distinct signaling pathways to numerous important cell biological processes (Schwartz and Ginsberg, 2002; Cabodi et al., 2010; Kim et al., 2011).
Interestingly, experimental ligation of the collagen-receptor integrin α2β1 was both necessary and sufficient to induce abundant invadopodia, though lesser contributions from other integrins are possible. This requirement differs from the apparent need for fibronectin and the α5β1 integrin from the formation of the lower numbers of invadopodia in the standard gelatin assays; fibronectin is well-known to bind strongly to gelatin (Engvall and Ruoslahti, 1977). Thus, specific ligand–receptor interactions appear to be crucial: dense fibrillar collagen and the α2β1 integrin are important and potent regulators of invadopodia formation in collagen, but an alternative integrin is important in a fibronectin–gelatin-rich environment.
Parenthetically, the role of α2β1 integrin in tumor metastasis is somewhat controversial. Although expression of α2β1 integrin correlates with metastasis to different organs in vivo (Chan et al., 1991; Ho et al., 1997), malignant transformation of mammary epithelial cells requires α5 but not α2 integrin in vitro (Damiano et al., 2014). Studies on tumorigenesis in α2 integrin–null mice yielded opposite results depending on the tumor model (Ramirez et al., 2011; Tran et al., 2011). However, α2β1 integrin promotes prostate cancer metastasis (Sottnik et al., 2013) and is a prostate cancer stem cell marker (Ricci et al., 2013). α2β1 integrin expression is down-regulated in primary pancreatic tumors but up-regulated in metastases (Goel et al., 2008). Therefore, the ability of the α2β1 integrin to confer or suppress cancer cell metastasis seems to be cell type and ECM dependent.
We found that potent induction of invadopodia after engagement of the α2β1 integrin by HDFC in multiple cancer cell types is notable for its complete independence from added growth factors or other molecules in serum including fibronectin. In contrast, fibronectin and growth factors are needed to form even the lower numbers of invadopodia on gelatin matrix (Beaty et al., 2013). Remarkably, the induction of multiple invadopodia by HDFC occurs without changes in gene expression or cellular protein profiles.
Kindlin2 phosphorylation in invadopodia formation
We discovered that major changes occur in phosphorylation-based signaling involving different levels of signal transduction pathways. Thus, breast carcinoma cell interaction with HDFC altered the phosphorylation state of kindlin2. Conversely, phosphorylation of the FAK-p130CAS–BCAR1-CRK (CT10 regulator of kinase) complex occurred only in cells on gelatin. This latter finding agrees with the previously established role of a myosin II–FAK–p130CAS pathway in invadopodia mechanosensing on rigid gelatin (Alexander et al., 2008).
Kindlin3, another member of the kindlin family, is known to be required for osteoclast-mediated bone resorption by mediating signaling from multiple integrin classes and regulating podosome formation (Schmidt et al., 2011). Here, we demonstrate that kindlin2 is required for strong induction of invadopodia in tumor cells. We found that kindlin2 selectively regulates invadopodia formation via its serine phosphorylation on dense collagen. Kindlin2 is an essential element of bidirectional integrin signaling (Larjava et al., 2008; Meves et al., 2009). It directly binds to β1 integrin cytoplasmic tails and cooperates with talin for integrin activation. Activation of β1 integrins by an activating TS2/16 antibody in cells invading gelatin can stimulate invadopodia formation and matrix degradation (Beaty et al., 2013). Consistent with this role for integrin activation, we found that interaction of carcinoma cells with HDFC is accompanied by a doubling of the amount of activated β1 integrin at the cell surface adherent to the HDFC versus cells on gelatin. In addition, kindlin2 is required for proper assembly of cell–ECM adhesions by bridging integrins and actin CSK-interacting proteins (Larjava et al., 2008).
Kindlin2 binds to migfilin, recruiting it to integrin–ECM adhesions (Tu et al., 2003). Migfilin interacts strongly with filamin A, an inhibitory integrin regulator, displacing filamin from integrins to promote talin–integrin binding and integrin activation. Our knockdown experiments demonstrate that migfilin, talin 1, and filamin A are all required for invadopodia formation on HDFC. We find that kindlin2 and migfilin often colocalize in invadopodia. Moreover, altering kindlin2 phosphorylation alters migfilin localization in invadopodia. Interestingly, kindlin2, filamin A, and talin 1 are associated with increased metastasis in different types of cancers (Lai et al., 2011; Shen et al., 2012; Mahawithitwong et al., 2013; Savoy and Ghosh, 2013). Thus, we suggest that phospho-kindlin2, talin1, filamin A, and migfilin play essential roles in downstream α2β1 integrin signaling for a strong invadopodial response. Our findings represent the first study that kindlin2 function can be regulated by phosphorylation to provide a novel mechanism of invadopodia regulation specific to dense fibrillar collagen. These data underscore the complex nature of regulatory mechanisms driving invadopodia formation, which we demonstrate requires signaling dictated by the ECM environment.
Materials and methods
Cell lines
Parental MDA-MB-231 cells and a MDA-MB-231 stable cell line expressing wild-type chicken c-Src in the pEVX expression vector that uses Moloney murine leukemia virus long terminal repeats to express pp60c-Src (Myoui et al., 2003) were a gift from T. Yoneda (Osaka University, Osaka, Japan). HT-1080 and PC3 cell lines were obtained from the Lombardi Cancer Center Cell Culture Core. Human foreskin fibroblast (HFF) cells were a gift from S. Yamada and were derived from human foreskin tissue samples provided by the Cooperative Human Tissue Network (funded by the National Cancer Institute). All cells were maintained in high glucose-DMEM (HyClone) supplemented with 10% fetal bovine serum (HyClone), 2 mM l-glutamine, 10 U/ml penicillin, and 10 µg/ml streptomycin (Life Technologies). All cells were tested with the mycoplasma detection kit (MycoAlert; Lonza) and found to be mycoplasma free. MDA-MB-231 cells were transfected with cDNA vectors using Lipofectamine 2000 (Invitrogen) and by siRNA using DharmaFECT 4 (Thermo Fisher Scientific) or Cell Line Nucleofector kit V (Amaxa) following the manufacturer’s instructions. HT-1080 cells were transfected with siRNA using DharmaFECT 4, and PC3 cells were transfected with siRNA using Lipofectamine 2000.
Antibodies and reagents
mAbs were obtained from the following sources: BD (β1 integrin rat mAb [mAb13], α5 integrin mouse mAb [IIA1], activated β1 integrin rat mAb [9EG7], paxillin mouse mAb [165], and Cdc42 mouse mAb [610929]), Chemicon (EMD Millipore; mouse IgG [MABC002], β3 integrin mouse mAb [MAB1957Z], β2 integrin mouse mAb [MAB1962Z], α1 integrin mouse mAb [MAB1973Z], α2 integrin mouse mAb [MAB1950Z], α3 integrin mouse mAb [MAB1952Z], and αVβ3 integrin mouse mAb [LM609]), ATCC (αV integrin mouse mAb [L230]), Epitomics (cortactin rabbit mAb [2067–1], PAK2 rabbit mAb [2247–1], and MT1-MMP rabbit mAb [1264Y]), EMD Millipore (cortactin mouse mAb [4F11], PAK4 rabbit pAb [04–583], kindlin2 mouse mAb [MAB2617], Rac1 mouse mAb [23A8], migfilin mouse mAb [clone 43.9], filamin A mouse mAb [MAB1678], and α2 integrin mouse mAb [MAB1988]), Sigma-Aldrich (collagen type I mouse mAb [C2456]), Abcam (RhoA mouse mAb [AB54835]), and generated in house (activated β1 integrin mouse mAb [12G10] and α5 integrin rat mAb [mAb11]).
pAbs were obtained from Abcam (talin 1 rabbit pAb [ab71333], kindlin2 rabbit pAb [ab74030], and filamin A goat pAb [ab11074]), Jackson ImmunoResearch Laboratories, Inc. (goat anti–mouse and goat anti–rat IgG, fragment crystallizable [Fc] fragment-specific), EMD Millipore (TIAM1 rabbit pAb [ST1070]), Sigma-Aldrich (TIAM2 rabbit pAb [HPA013903], ARHGEF12 rabbit pAb [SAB4500976], and ARHGEF18 rabbit pAb [HPA042689]), GeneTex (filamin B rabbit pAb [GTX101206]); EMD Millipore (α2 integrin rabbit pAb [AB1936]), and generated in house (rabbit anti–human plasma fibronectin). Secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. (donkey anti–mouse, anti–rabbit, or anti–rat, each conjugated to Cy2, Cy3, or Cy5) and LI-COR Biosciences (antibodies conjugated to IRDye 800CW or IRDye 680LT). Mouse polyclonal serum was raised in house against the collagen type I 1/4 cleavage site by immunizing mice homozygous for a collagenase-resistant Col1a-1 gene using the peptide IAGQRGV corresponding to the N terminus of the 1/4-length collagen cleavage fragment. This short peptide was conjugated to the T cell epitope of pmp8 protein from Chlamydophila pneumonia. Polyclonal serum was analyzed by Biacore analysis for binding specificity to the IAGQRGV peptide conjugated to BSA or to cleaved collagen. The epitope specificity of the polyclonal serum was also assayed by immunofluorescence confocal microscopy for collagen degradation sites in non–cross-linked HDFC matrices prepared from mouse tail collagen type I isolated from wild-type mice compared with collagen I from mice homozygous for the collagenase-resistant Col1a-1 gene. For experimental analysis, HDFC degradation was performed by MDA-MB-231 tumor cells plated on non–cross-linked HDFC matrices in the absence of serum for incubation at 37°C for 4 h, overnight, or 2 d. Reagents included Alexa Fluor dyes, Alexa Fluor 488– and rhodamine-phalloidin (Invitrogen); BSA (MP Biomedicals); rat tail collagen type I (BD); α/β integrin–mediated cell adhesion arrays (EMD Millipore); Dynabeads protein G immunoprecipitation kit (Invitrogen); and GFP-Trap GFP-binding protein (ChromoTek).
Human tissue biospecimens
Paired human normal and metastatic cancerous breast tissues were obtained from the National Disease Research Interchange, Cooperative Human Tissue Network, and Asterand. Tissue blocks embedded in Optimal Cutting Temperature compound (O.C.T.) were sliced at 12-µm thickness with a cryostat (CM3050 S; Leica), and tissue slices were captured onto positively charged Superfrost Plus/Colorfrost Plus 75 × 25–mm precleaned microslides (Daigger). Extraction of human tissue sections to generate cell-free 3D matrices as well as hematoxylin and eosin (H&E) and Masson’s trichrome staining were performed exactly as previously described (Campbell et al., 2014). In brief, microslides with attached tissue slices were submerged into a dish containing freshly made cell extraction buffer at 37°C consisting of 0.5% (vol/vol) Triton X-100 and 20 mM NH4OH in PBS. The microslide was incubated in extraction buffer on an orbital rocker (DW-150 Waver; VWR) set to 50 cycles/min and a 15° tilt angle to provide slow orbital motion. Tissue extraction was performed with multiple changes of the extraction buffer: two 15-min incubations followed by nine 5-min incubations. After each incubation, used extraction buffer was aspirated, and a fresh aliquot of extraction buffer was added to the dish containing a microslide. Extraction was followed by three 10-min washes with PBS on the orbital shaker. Then, PBS was aspirated together with the gel-like layer of DNA that was found to cover the tissue slices, and microslides were washed with PBS for 30 min on the orbital rocker. Extracted tissues were treated with 0.1% 10-U/ml recombinant DNase I (Roche) in PBS for 30 min on the orbital rocker at room temperature and then washed in sterile PBS overnight. Cells were washed and plated on extracted 3D matrix in serum-free medium and incubated at 37°C for 4 h.
H&E staining was used for quality control of the tissue extraction to visualize collagen fibrils and any residual cellular debris, such as cell nuclei. Staining was performed with microslides placed in a glass staining rack and sequentially dipped into glass staining dishes (Wheaton Science Products) filled with staining solutions or water. The order of staining solutions for extracted tissue slices on slides was two washes in PBS, 3 min each; incubation in hematoxylin solution (Gill no. 3; Sigma-Aldrich) for 2 min; dip in distilled water; 5-min wash in tap water; 12 quick dips into acid ethanol (70% ethanol in 0.25% [vol/vol] HCl); two 1-min washes in tap water; 2-min wash in distilled water; 2-min incubation in acidified Eosin Y solution (aqueous Eosin Y solution containing 0.5% [vol/vol] glacial acetic acid; Sigma-Aldrich); 5-min incubation in 90% ethanol; 5-min incubation in 100% ethanol; and 10-min incubation in xylene. The microslides were covered with glass coverslips using Permount histological mounting medium (Thermo Fisher Scientific).
Trichrome staining was used to visualize fibrillar collagen fibrils and was performed with Masson’s Trichrome Stain kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Glass coverslips were mounted on microslides with Permount histological mounting medium.
siRNA and cDNA vectors
ON-TARGET plus SMARTpool siRNAs for kindlin2, talin1, filamin A, filamin B, migfilin, TIAM1, TIAM2, ARHGEF12, ARHGEF18, paxillin, Rac1, PAK2, PAK4, nonspecific control and single duplex siRNAs for nonspecific control, and all the listed proteins except Rac1 were obtained from Thermo Fisher Scientific (sequence information is provided in Table S3). Single duplex siRNA for Rac1 was Silencer Select 5′-GGAACUAAACUUGAUCUUATT-3′ (Ambion).
Human kindlin2 cDNA (Variant 1; OriGene; RefSeq accession no. NM_006832) was inserted into modified pEGFP-N1 (Takara Bio Inc.) at NheI–BamHI sites to generate pEGFP-kindlin2. Kindlin2 mutant plasmids were generated using the site-directed mutagenesis kit (QuikChange II XL; Agilent Technologies) using the primers listed in Table S4. An siRNA rescue mutant was constructed using this kit by introducing five silent mutations into the kindlin2 cDNA corresponding to On-Target plus siRNA sequence 5′-AAUGAAAUCUGGCUUCGUU-3′ (Thermo Fisher Scientific). All mutations were verified by full-length kindlin2 sequencing.
The pECFP-Rac1 constructs (including wild type, Q61L, and Y40C) were generated by inserting the human Rac1 sequence into the EcoRI and BamHI sites of the pECFP-C1 expression vector (Takara Bio Inc.), with one nucleotide added immediately downstream of the EcoR1 site to maintain the reading frame of CFP (Picard et al., 2009). The YPet–PAK-binding domain (PBD) plasmid encoding the FRET acceptor was constructed by inserting the cDNA encoding amino acids 65–150 of human Pak1 into the EcoR1 and BamH1 sites of YPet-C1 encoding mutant of YFP (YPet; Petrie et al., 2012). YPet-C1 was generated by inserting the cDNA encoding the fluorescent protein YPet into the AgeI and XhoI sites of pEGFP-C1 expression vector. The YPet sequence was generated from pECEP4YPet-MAMM (plasmid 14032; Addgene; Nguyen and Daugherty, 2005). To create CFP-Cdc42, -Cdc42N17, -Cdc42V12, -RhoA, -RhoAN19, and -RhoAL63 vectors, the full-length human cDNAs of wild type or mutants were subcloned from the eukaryotic expression vector pRK5 myc, which contains the myc epitope (Lamarche et al., 1996) into the pECFP-C1 expression vector at EcoRI and BamHI sites (Picard et al., 2009). Chimeric proteins were expressed from pECFP-C1 expression vectors under the cytomegalovirus promoter.
Preparation of matrices for invadopodia assays
2D gelatin matrix.
18-mm-diameter glass coverslips or 35-mm MatTek dishes with 20-mm glass bottoms were coated with a thin layer of gelatin fluorescently labeled with Alexa Fluor dye exactly as previously described (Artym et al., 2009). In brief, glass coverslips or glass-bottom MatTek dishes were coated with a 50-µg/ml solution of poly-l-lysine and incubated for 20 min at room temperature; washed three times with PBS, and incubated in 0.5% glutaraldehyde in PBS solution for 15 min. A mixture of 0.2% gelatin and Alexa Fluor 568–gelatin at an 8:1 ratio was preheated in a 37°C water bath. An 80-µl drop of the fluorescent gelatin mixture was placed on Parafilm, and the 18-mm glass coverslip was inverted onto the drop; alternatively, the cover glass area of a MatTek dish was coated with 200 µl of the gelatin mixture. Either glass slides or MatTek dishes were incubated with gelatin for 10 min at room temperature, washed with PBS three times, treated with 5 mg/ml sodium borohydride for 15 min, washed extensively with PBS to remove any bubbles, and incubated in tissue culture medium without serum for 30 min before cell plating. To generate fibronectin-coated gelatin matrix, gelatin matrix was coated with 10 µg/ml human plasma fibronectin for 1 h and blocked with heat-denatured BSA for 1 h.
HDFC matrix.
Glass bottoms of MatTek dishes (35-mm dish diameter and 20-mm-diameter glass bottom) were prechilled on ice and then coated with 10 µl ice-cold neutralized rat tail collagen (Artym and Matsumoto, 2010) at 8 mg/ml. To create a thin collagen layer, a disposable plastic pipette tip was used to spread the collagen evenly on the glass surface. To neutralize the acidic collagen stock, an aliquot of stock collagen was mixed by pipetting on ice with ice-cold 10× DMEM at 1/8 of the volume of collagen. Then, ice-cold 10× reconstitution buffer (a solution of 2.2 g sodium bicarbonate and 4.8 g Hepes in 100 ml of distilled water) at 1/8 of the collagen volume was mixed in by pipetting. The pH of the neutralized collagen was measured with pH paper. When required, the pH was adjusted with an ice-cold solution of 2 N NaOH or 2 N HCl to 7.1–7.4. The pH of the collagen solution was equilibrated by incubating the neutralized collagen on ice for 5 min, and air bubbles were removed by centrifugation at 9,100 g at 4°C for 3 min. The thin layer of collagen was allowed to polymerize into a fibrillar meshwork at 37°C for 30 min. The dishes were centrifuged using a microplate centrifuge adapter at 3,500 g for 20 min to flatten the collagen meshwork into a 2D layer. To obtain cross-linked HDFC matrices, centrifuged collagen matrices were fixed with 4% paraformaldehyde/5% sucrose in PBS for 20 min, washed with PBS, and blocked with DMEM for 20 min. For nontreated HDFC matrices, the centrifuged collagen meshwork was blocked with DMEM for 20 min
Thin 3D collagen matrix.
Glass bottoms of MatTek dishes were coated with a thin layer of neutralized rat tail collagen (as for HDFC matrix preparation) at final concentrations of 1–16 mg/ml and polymerized into fibrillar meshworks at 37°C for 30 min. The collagen matrix was fixed with 4% paraformaldehyde/5% sucrose in PBS for 20 min, washed extensively with PBS, and blocked with HyQ-DMEM for 20 min. To obtain high concentrations, 9.4 mg/ml rat tail collagen was concentrated using centrifugal filter units (Microcon-10 kD) with Ultracel-10 membrane (EMD Millipore).
CDM.
CDM was prepared exactly as previously described in Cukierman et al. (2001). In brief, glass bottoms of MatTek dishes were coated with 0.2% gelatin in PBS at 37°C for 1 h, washed with PBS, fixed with 1% glutaraldehyde in PBS for 30 min, and blocked with DMEM for 30 min. After PBS washes, HFFs at passages 7–10 were plated to obtain a confluent cell layer overnight. Culture medium supplemented with 50 µg/ml ascorbic acid was changed every day for 10 d. HFF cells were extracted with 20 mM NH4OH and 0.5% Triton X-100 in PBS for 5–10 min. After PBS washes, cell-free matrices were treated with 10 U/ml DNase (Roche) in PBS for 30 min at 37°C, washed with PBS, and stored in PBS supplemented with penicillin/streptomycin at 4°C. To produce 2D CDM, MatTek dishes with CDM were centrifuged at 3,500 g for 20 min, fixed with 4% paraformaldehyde/5% sucrose in PBS for 20 min, washed with PBS, and blocked with HyQ-DMEM for 20 min.
Globular collagen matrix.
Glass coverslips or glass-bottom MatTek dishes were coated with 50 µg/ml poly-l-lysine solution in PBS for 20 min and treated with 0.5% glutaraldehyde for 20 min at room temperature. Then, a solution of collagen type I in PBS at 1.3 or 1.5 mg/ml was applied and incubated 10 min at room temperature followed by blocking with HyQ-DMEM for 20 min.
5% gelatin cushions.
These 1-µm-thick 2D gelatin films were prepared exactly as previously described (Alexander et al., 2008). In brief, MatTek dishes were coated with 5% gelatin/2.5% sucrose in PBS for 1 min. The gelatin solution was then aspirated, and the dishes were inverted and allowed to dry for 30 min at room temperature. Next, gelatin films were cross-linked with ice-cold 0.5% glutaraldehyde/PBS on ice for 15 min and then at room temperature for 30 min, treated with 1 mg/ml sodium borohydride for 3 min, extensively washed with PBS, and coated with 50 µg/ml human plasma fibronectin in PBS for 1 h at room temperature.
Integrin functional ligation using antibodies
Glass bottoms of MatTek dishes were coated with poly-l-lysine at 50 µg/ml for 20 min, washed with PBS, and treated with 0.5% glutaraldehyde for 15 min. After PBS washes, dishes were incubated with anti-Fc species-specific antibodies at 200 µg/ml at room temperature for 1 h. After extensive PBS washes, dishes were blocked with DMEM for 30 min, coated with mAb targeting each indicated integrin subunit at 10 or 200 µg/ml for 1 h, and washed again with PBS. Cells were plated on dishes at 105 cells/ml and incubated for 3 h at 37°C in serum-free medium to induce cell adhesion by targeting specific integrins.
Immunostaining and confocal microscopy
Untransfected or siRNA-transfected cells were fixed with 4% paraformaldehyde/5% sucrose in PBS for 20 min, whereas cells expressing EGFP-protein chimeras were fixed with 2% formaldehyde/5% sucrose for 20 min. The cells were permeabilized for 10 min with 0.5% Triton X-100 in PBS, washed with PBS, and labeled with 10 µg/ml primary antibodies for 1 h followed by labeling with secondary fluorochrome-conjugated antibodies for 30 min. Confocal images were collected using a laser-scanning confocal microscope (LSM 510; Carl Zeiss) with a Plan Apochromat 63×/1.4 NA oil objective (Carl Zeiss) using Zen software (Carl Zeiss). Cells incubated in MatTek dishes were imaged in PBS, whereas coverslips with attached cells were mounted onto slides using aqueous mounting medium with antifading agents (Gel Mount; Biomeda). All imaging was performed at room temperature. Cy2, Cy3, Cy5, rhodamine, Alexa Fluor 488, Alexa Fluor 568, Alexa Fluor 647, DyLight 488, or DyLight 633 was used as the fluorochromes conjugated to the antibodies or ECM proteins for fluorescence labeling of the samples. Cells adherent to matrices were selected for invadopodia analysis from multiple microscopy fields by randomly focusing on multiple areas of the sample.
Image processing and analysis
Images were processed and analyzed using MetaMorph software (Molecular Devices). Image processing for image presentation included low pass or median filtering for smoothing. Image processing for quantification included low pass filtering for smoothing, background correction by subtracting the mean intensity value of the background from the image intensity, sharpen low filtering, and median 3 × 3–pixel filtering. Processed images were thresholded using the standard MetaMorph threshold journal. Numbers of invadopodia per cell were quantified by MetaMorph Integrated Morphometry Analysis (IMA). IMA filters were set to filter circular objects (shape factor ≥ 0.7) with area >0.2 µm2 and <8 µm2. Numbers of invadopodia were quantified by IMA from single z plane or maximum intensity projection images obtained for cortactin or actin. For siRNA experiments, cells on the HDFC matrix were identified as invadopodia negative when only five or fewer invadopodia per cell were identified. When required, confocal z stacks of HDFC matrix were deconvolved with the blind deconvolution algorithm set to 10 iterations and medium filtering.
AFM
The effective elastic modulus of each matrix coated or polymerized in glass-bottom MatTek dishes was measured by performing indentations using a commercial atomic force microscope (AFM) instrument (AFM Catalyst; Bruker). Polystyrene microspheres 5 µm in diameter were attached to tipless cantilevers (MLCT; Bruker) using a UV-curable epoxy adhesive. The spring constants of all cantilevers were estimated using the thermal tune utility of the instrument. For each matrix, indentations were performed over 16 × 16 point grids covering areas of 30 × 30 µm2 that were equal to or larger than a typical cell. A few locations (typically 3–5) lying several millimeters apart from each other were probed. The collected force curves were analyzed using the Hertz model with a custom-written Matlab (MathWorks) software code (supplemental material). The resulting estimates of effective Young’s modulus for each matrix are presented in Table 1.
FRET microscopy
The activation-dependent binding of YPet-PBD to CFP-Rac1 was visualized by sensitized emission FRET microscopy (Kraynov et al., 2000). Cells were imaged using a confocal microscope (LSM 710; Carl Zeiss) outfitted with a 63× 1.0 NA water Plan Apochromat objective (Carl Zeiss). A 514-nm and 457-nm argon laser was used to excite the FRET pair. Ideal acquisition parameters were determined individually for the YPet, CFP, and FRET channels and were maintained in all subsequent FRET experiments. The ratio of bleed through to fluorescence intensity was determined by imaging cells expressing the acceptor or donor alone, with the predetermined acquisition settings. The corrected FRET image (Fc) was derived from the raw FRET image using the LSM FRET tool macro (Carl Zeiss; Gordon et al., 1998). FRET controls for cells expressing the nonfunctional versions of the biosensors indicated that dimerization of the fluorescent proteins did not contribute significantly to the magnitude of the FRET signal generated by wild-type biosensors (Petrie et al., 2012).
Immunoprecipitation and immunoblotting
We followed immunoprecipitation pull-down protocols from the laboratories of C. Wu (University of Pittsburgh, Pittsburgh, PA; Tu et al., 2003), M. Humphries (University of Manchester, Manchester, England, UK; Humphries et al., 2009), D. Bouvard (Institut Albert Bonniot, Grenoble, France; Millon-Frémillon et al., 2013), and our own laboratory (Pankov et al., 2003) to test for binding and coimmunoprecipitation of kindlin2 and migfilin from MDA-MB-231 cells cultured on HDFC matrix overnight. MDA-MB-231 cells were plated on HDFC matrices in 35-mm MatTek dishes with 20-mm-diameter glass coverslip bottoms at a density of 1.5 × 105 cells/dish. Cell lysates were collected from only the glass coverslip section of the dishes coated with HDFC. To prevent contamination, cells on the plastic edges of the bottoms of MatTek dishes were removed using cotton-tipped applicator swabs before cell lysis. The cells were lysed with 200 µl of lysis buffer, taking care to scrape only the HDFC-coated glass area with an 18-mm disposable cell scraper (Costar; Corning). For each experimental condition, cell lysates were collected from five MatTek dishes. All cell lysates were maintained on ice and clarified by centrifugation at 20,000 g for 10 min at 4°C.
We tested the following panel of lysis buffers, plus some variants: (a) radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 10 mM NaF and a mixture of protease and phosphatase inhibitors [cOmplete EDTA-free protease inhibitor cocktail tablets plus PhosSTOP phosphatase inhibitor cocktail tablets, both obtained from Roche]); (b) Nonidet P-40 lysis buffer (137 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1% Nonidet P-40, 10% glycerol, 20 mM Tris-HCl, pH 8.0, 1 mM sodium orthovanadate, 50 mM NaF, and the mixture of protease and phosphatase inhibitors); (c) PBS, pH 7.1, 1% Triton X-100, 1 mM sodium orthovanadate, 25 mM NaCl, and the mixture of protease and phosphatase inhibitors; (d) 50 mM Hepes, pH 7.1, 1% Triton X-100, 150 mM NaCl, 10 mM Na4P207, 10 mM EDTA, 2 mM sodium orthovanadate, 100 mM NaF, and the mixture of protease, and phosphatase inhibitors; (e) CSK buffer (10 mM Pipes, pH 6.8, 50 mM NaCl, 300 mM sucrose, 0.5% Triton X-100, 3 mM MgCl2, 1 mM sodium orthovanadate, 50 mM NaF, and the mixture of protease and phosphatase inhibitors); and (f) modified CSK lysis buffer (10 mM Pipes, pH 6.8, 50 mM NaCl, 150 mM sucrose, 0.5% Triton X-100, 3 mM MgCl2, 1 mM MnCl2, 2 mM sodium orthovanadate, and the mixture of protease and phosphatase inhibitors).
For coimmunoprecipitation of endogenous migfilin with exogenous kindlin2-GFP, cells were lysed with modified CSK lysis buffer, sonicated for 1 s with one pulse of a 100-W sonicator set at power level 3 (Miscroson model XL2000; Misonix) and then maintained on ice for 30 min. After clarification by centrifugation, migfilin was immunoprecipitated using a Dynabeads protein G immunoprecipitation kit (Invitrogen) following the manufacturer’s instructions using beads conjugated to anti-migfilin MAB2618 antibody (EMD Millipore). For coimmunoprecipitation of exogenous kindlin2-GFP with endogenous migfilin, MDA-MB-231 cells transfected with kindlin2-GFP were plated on HDFC at 24 h after transfection and incubated on HDFC overnight. Cells were lysed using one of each of the six lysis buffers listed in the previous paragraph, using the conditions for cell lysis, incubation times for cell lysis, and sonication as listed in the previous paragraph. Lysates were clarified by centrifugation and equalized for protein concentration and volume using Micro BCA Protein Assay kits obtained from Thermo Fisher Scientific. 500 µg of total protein cell lysate was incubated with 40 µl of GFP-Trap conjugated to magnetic beads for 1 h at 4°C with end-over-end rotation. All blots were immunostained with primary anti-kindlin2 MAB2617 antibody and anti-migfilin MAB2618 antibody followed by secondary antibodies conjugated to IRDye 800CW or IRDye 680LT. Blots were imaged using an Odyssey imaging system (LI-COR Biosciences).
Microarray analysis
For microarray analysis, MDA-MB-231 cells were cultured overnight on HDFC or gelatin matrices in MatTek dishes. Five independent repeats were performed for each condition. After mRNA isolated using an RNAqueous-4PCR kits (Ambion), samples were labeled with an Amino Alyl MessageAmp II aRNA Amplification kit (Ambion). Gene expression profiles for MDA-MB-231 cells invading gelatin versus HDFC matrices in the presence or absence of serum in cell culture medium were determined using 4 × 44K Whole Human Genome microarray chips (Agilent Technologies). The readouts of multiple chips were normalized using the quantile normalization method. All probe sets corresponding to one gene were summarized into a single value using Robust Multi-array Average normalization. Unpaired t test and Benjamini–Hochberg false discovery rate–corrected P ≤ 0.05 and fold changes ≥ 2 were used as a cutoff for array comparison for different conditions. Gene expression data were analyzed with GeneSpring Software (Agilent Technologies).
Proteomics and phosphoproteomics
Protein extraction.
MatTek dishes were cleaned with 20% nitric acid, thoroughly rinsed in deionized water, air dried in a tissue-culture hood, and coated with HDFC or gelatin. MDA-MB-231 cells were plated on these matrices in serum-free medium and incubated for 4 h. Cells were lysed with RIPA buffer, and protein concentration was determined by Micro BCA analysis (Thermo Fisher Scientific).
Phosphopeptide analysis.
For phosphoproteomic analyses, ∼500 µg of protein was precipitated from whole cell lysates by addition of ice cold acetone. The proteins were resuspended in 6 M urea, reduced with DTT at 37°C for 1 h, alkylated with iodoacetamide for 1 h at room temperature, and digested with trypsin overnight at 37°C at a trypsin/protein ratio of 1:20. Phosphopeptides were enriched using Fe-NTA phosphopeptide enrichment spin columns (Thermo Fisher Scientific). Enrichment was performed using the protocol provided by the manufacturer. The phosphopeptide-enriched samples were then loaded onto a Zorbax C18 trap column (Agilent Technologies) to desalt the peptide mixture using an online nano–liquid chromatography (LC) ultra HPLC system (NanoLC; Eksigent). The peptides were then separated on a 10-cm PicoFrit BioBasic C18 analytical column (New Objective). Peptides were eluted over a 120-min linear gradient of 5–35% acetonitrile/water containing 0.1% formic acid at a flow rate of 250 nl/min, ionized by electrospray ionization in positive mode, and analyzed on a mass spectrometer (LTQ Orbitrap Velos; Thermo Fisher Scientific). All LC MS analyses were performed in data-dependent mode in which the top 6 most intense precursor ions detected in the MS1 precursor scan (mass/charge [m/z] of 300–2,000) were selected for fragmentation via collision-induced dissociation (CID) or high energy CID (HCD). Precursor ions were measured in the Orbitrap at a resolution of 30,000 (m/z of 400). Fragment ions were measured in the ion trap for CID experiments and in the Orbitrap for all HCD experiments.
LC MS/MS data analysis was performed with the MASCOT algorithm (Matrix Science). For peptide and protein identifications, the data were searched against the mammalian Swiss-Prot protein database. The digestion enzyme was specified as trypsin and set to allow for a maximum of two missed cleavage sites. Cysteine carbamidomethylation was specified as a static modification. Oxidation of methionine and phosphorylation of serine, threonine, and tyrosine residues were chosen as variable modifications. The mass tolerance for precursor ions was set to 20 ppm for all experiments, and fragment ion tolerances for CID and HCD analyses were set at ±0.8 D and ±0.1 D, respectively.
Peptide and protein identifications were further validated using Scaffold 3 software (Proteome Software, Inc.). Probabilities of 80.0% (0.7% false discovery rate) and 20.0% were selected for peptides and proteins, respectively. Only the peptide and protein identifications that exceeded these probability thresholds based on the PeptideProphet (Keller et al., 2002) and ProteinProphet algorithms (Nesvizhskii et al., 2003) were accepted.
Protein assignments containing similar peptides that we could not differentiate by MS/MS analysis were grouped together to satisfy principles of parsimony. Phosphoproteins identified from two independent experiments, each with two technical replicates, were pooled into a single database and analyzed using Ingenuity Pathway Analysis.
iTRAQ analysis.
For iTRAQ experiments, 100 µg of each protein sample was reduced, alkylated, and trypsin digested according to the iTRAQ protocol provided by AB SCIEX. After digestion, tryptic peptide samples were labeled using iTRAQ-8plex reagents at room temperature for 2 h. The labeled peptide samples were combined and separated using offline strong cation exchange chromatography, and fractions were collected and desalted using 1-cc vacuum cartridges (Oasis HLB; Waters Corp.). For the analysis of iTRAQ-labeled peptides by reverse-phase nano-LC MS/MS, each fraction was loaded onto a Zorbax C18 trap column to desalt the peptide mixture using an online nano-LC ultra HPLC system. The peptides were then separated on a 10-cm Picofrit Biobasic C18 analytical column. Peptides were separated over a 90-min linear gradient of 5–40% acetonitrile/water containing 0.1% formic acid at a flow rate of 250 nl/min, ionized by electrospray ionization in positive mode, and analyzed on an LTQ Orbitrap Velos mass spectrometer in data-dependent mode. The top 6 most intense precursor ions detected in the MS1 precursor scan (m/z of 300–2,000) were selected for fragmentation by HCD with a normalized collision energy of 45%. Precursor ions were measured in the Orbitrap at a resolution of 30,000 (m/z of 400), and fragment ions were measured in the Orbitrap at a resolution of 7,500.
LC MS/MS data were analyzed using the MASCOT algorithm. All data were searched against the Human Swiss-Prot protein database for peptide and protein identifications. Trypsin was specified as the digestion enzyme, allowing for up to two missed cleavage sites. Methylthiol (C, cysteine) was set as a static modification, and oxidation (M, methionine) and iTRAQ-8plex (K, lysine; Y, tyrosine; n, N terminus) were selected as variable modifications. Precursor mass tolerances were set to 50 ppm, and fragment ion tolerances were set to 0.05 D.
Statistics
The data are presented as the mean value from at least three pooled independent trials with the corresponding SD or SEM. Statistical tests were two-tailed t test (pair of conditions) or one-way analysis of variance followed by the appropriate post-test (Kruskal–Wallis, Dunn’s, Holm–Sidak, or Duncan’s) performed using SigmaPlot or Prism software (GraphPad Software). The differences were statistically significant at P < 0.0001 (indicated on graphs as triple asterisks), P < 0.001 (indicated as double asterisks), or P < 0.05 (indicated as a single asterisk).
Accession numbers
Primary microarray data are available via Gene Expression Omnibus under accession no. GSE53104, and proteomics data are available via Peptide Atlas under accession no. PASS00371.
Online supplemental material
Fig. S1 shows that HDFC is a potent inducer of invadopodia. Fig. S2 shows integrins and invadopodia formation. Fig. S3 shows phosphoproteomics analysis of cells adherent to gelatin matrix reveals complex downstream signaling network. Fig. S4 shows complex signaling network of multiple cellular proteins is required for invadopodia formation on HDFC. Fig. S5 shows that kindlin2 phosphorylation regulates invadopodia formation in MDA-MB-231 cells on collagen matrices. Table S1 provides integrin signaling network phosphoproteins and their phosphosites. Table S2 is a list of +1 and +2 predicted fragment ions for the kindlin2 phosphopeptide KLDDQSpEDEALELEGPLITPGSGSIYSSPGLYSK. Table S3 is a list of siRNA sequences used in this paper. Table S4 provides a list of primers used for constructing human kindlin2 cDNA. A ZIP file is provided that includes a Matlab code for extracting elastic modulus from indentation data. A dataset is also provided as an Excel (Microsoft) file with AFM data corresponding to Table 1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201405099/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201405099.dv.
Supplementary Material
Acknowledgments
We thank Dr. L. Engelholm for Biacore analysis of immune serum.
This work was supported by Pathway to Independence Award K99CA129205 from National Cancer Institute with American Recovery and Reinvestment Act of 2009 award to V.V. Artym and National Institutes of Health/National Institute of Dental and Craniofacial Research Intramural project ZIA DE 000719 to K.M. Yamada.
The authors declare no competing financial interests.
Author contributions: Conception and design were performed by V.V. Artym and K.M. Yamada. Development of methodology was performed by V.V. Artym, S. Swatkoski, C.B. Campbell, T.H. Bugge, R.J. Petrie, M. Gucek, and K.M. Yamada. Acquisition of data (including providing animals) was conducted by V.V. Artym, S. Swatkoski, C.B. Campbell, E. Dimitriadis, and T.H. Bugge. Analysis and interpretation of data (including statistical analysis) was performed by V.V. Artym, S. Swatkoski, X. Li, and E. Dimitriadis. Writing was performed by V.V. Artym and K.M. Yamada. Technical support was provided by K. Matsumoto, C.B. Campbell, R.J. Petrie, and S. Mueller.
Footnotes
Abbreviations used in this paper:
- AFM
- atomic force microscopy
- CDM
- cell-derived matrix
- CID
- collision-induced dissociation
- CSK
- cytoskeleton
- FA
- focal adhesion
- Fc
- fragment crystallizable
- FRET
- fluorescence resonance energy transfer
- GEF
- guanine nucleotide exchange factor
- HCD
- high energy CID
- HDFC
- high-density fibrillar collagen
- H&E
- hematoxylin and eosin
- HFF
- human foreskin fibroblast
- iTRAQ
- isobaric tags for relative and absolute quantitation
- LC
- liquid chromatography
- MMP
- matrix metalloproteinase
- MS
- mass spectrometry
- m/z
- mass/charge
- PBD
- PAK-binding domain
- RIPA
- radioimmunoprecipitation assay
References
- Alexander N.R., Branch K.M., Parekh A., Clark E.S., Iwueke I.C., Guelcher S.A., and Weaver A.M.. 2008. Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 18:1295–1299 10.1016/j.cub.2008.07.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V.V., and Matsumoto K.. 2010. Imaging cells in three-dimensional collagen matrix. Curr. Protoc. Cell Biol. Chapter 10:Unit 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., and Mueller S.C.. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Artym V.V., Yamada K.M., and Mueller S.C.. 2009. ECM degradation assays for analyzing local cell invasion. Methods Mol. Biol. 522:211–219 10.1007/978-1-59745-413-1_15 [DOI] [PubMed] [Google Scholar]
- Beaty B.T., Sharma V.P., Bravo-Cordero J.J., Simpson M.A., Eddy R.J., Koleske A.J., and Condeelis J.. 2013. β1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol. Biol. Cell. 24:1661–1675: S1–S11 10.1091/mbc.E12-12-0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti S., Inoue H., Bharti K., Hirsch D.S., Nie Z., Yoon H.Y., Artym V., Yamada K.M., Mueller S.C., Barr V.A., and Randazzo P.A.. 2007. Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol. Cell. Biol. 27:8271–8283 10.1128/MCB.01781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell M.J., Hall H.G., and Parry G.. 1982. How does the extracellular matrix direct gene expression? J. Theor. Biol. 99:31–68 10.1016/0022-5193(82)90388-5 [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero J.J., Oser M., Chen X., Eddy R., Hodgson L., and Condeelis J.. 2011. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr. Biol. 21:635–644 10.1016/j.cub.2011.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabodi S., del Pilar Camacho-Leal M., Di Stefano P., and Defilippi P.. 2010. Integrin signalling adaptors: not only figurants in the cancer story. Nat. Rev. Cancer. 10:858–870 10.1038/nrc2967 [DOI] [PubMed] [Google Scholar]
- Campbell C.B., Cukierman E., and Artym V.V.. 2014. 3-D extracellular matrix from sectioned human tissues. Curr. Protoc. Cell Biol. 62:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B.M., Matsuura N., Takada Y., Zetter B.R., and Hemler M.E.. 1991. In vitro and in vivo consequences of VLA-2 expression on rhabdomyosarcoma cells. Science. 251:1600–1602 10.1126/science.2011740 [DOI] [PubMed] [Google Scholar]
- Chen W.T.1989. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J. Exp. Zool. 251:167–185 10.1002/jez.1402510206 [DOI] [PubMed] [Google Scholar]
- Clark E.S., Whigham A.S., Yarbrough W.G., and Weaver A.M.. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227–4235 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Conklin M.W., and Keely P.J.. 2012. Why the stroma matters in breast cancer: insights into breast cancer patient outcomes through the examination of stromal biomarkers. Cell Adhes. Migr. 6:249–260 10.4161/cam.20567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coopman P.J., Do M.T., Thompson E.W., and Mueller S.C.. 1998. Phagocytosis of cross-linked gelatin matrix by human breast carcinoma cells correlates with their invasive capacity. Clin. Cancer Res. 4:507–515. [PubMed] [Google Scholar]
- Cox T.R., and Erler J.T.. 2011. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis. Model. Mech. 4:165–178 10.1242/dmm.004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D.R., and Yamada K.M.. 2001. Taking cell-matrix adhesions to the third dimension. Science. 294:1708–1712 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Damiano L., Stewart K.M., Cohet N., Mouw J.K., Lakins J.N., Debnath J., Reisman D., Nickerson J.A., Imbalzano A.N., and Weaver V.M.. 2014. Oncogenic targeting of BRM drives malignancy through C/EBPβ-dependent induction of α5 integrin. Oncogene. 33:2441–2453 10.1038/onc.2013.220 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Debrand E., El Jai Y., Spence L., Bate N., Praekelt U., Pritchard C.A., Monkley S.J., and Critchley D.R.. 2009. Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms. FEBS J. 276:1610–1628 10.1111/j.1742-4658.2009.06893.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz B., Shani G., Pass I., Anderson D., Quintavalle M., and Courtneidge S.A.. 2009. Tks5-dependent, nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci. Signal. 2:ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engvall E., and Ruoslahti E.. 1977. Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int. J. Cancer. 20:1–5 10.1002/ijc.2910200102 [DOI] [PubMed] [Google Scholar]
- Friedl P., and Wolf K.. 2010. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188:11–19 10.1083/jcb.200909003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., and Sahai E.. 2007. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9:1392–1400 10.1038/ncb1658 [DOI] [PubMed] [Google Scholar]
- Gligorijevic B., Wyckoff J., Yamaguchi H., Wang Y., Roussos E.T., and Condeelis J.. 2012. N-WASP-mediated invadopodium formation is involved in intravasation and lung metastasis of mammary tumors. J. Cell Sci. 125:724–734 10.1242/jcs.092726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel H.L., Li J., Kogan S., and Languino L.R.. 2008. Integrins in prostate cancer progression. Endocr. Relat. Cancer. 15:657–664 10.1677/ERC-08-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G.W., Berry G., Liang X.H., Levine B., and Herman B.. 1998. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74:2702–2713 10.1016/S0006-3495(98)77976-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guegan F., Tatin F., Leste-Lasserre T., Drutel G., Genot E., and Moreau V.. 2008. p190B RhoGAP regulates endothelial-cell-associated proteolysis through MT1-MMP and MMP2. J. Cell Sci. 121:2054–2061 10.1242/jcs.025817 [DOI] [PubMed] [Google Scholar]
- Hay E.D., editor 1991. Cell biology of extracellular matrix. Second edition Plenum Press, New York: 468 pp 10.1007/978-1-4615-3770-0 [DOI] [Google Scholar]
- Ho W.C., Heinemann C., Hangan D., Uniyal S., Morris V.L., and Chan B.M.. 1997. Modulation of in vivo migratory function of α 2 β 1 integrin in mouse liver. Mol. Biol. Cell. 8:1863–1875 10.1091/mbc.8.10.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary K., Li X.Y., Allen E., Stevens S.L., and Weiss S.J.. 2006. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 20:2673–2686 10.1101/gad.1451806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Mukhopadhyay A., Truesdell P., Chander H., Mukhopadhyay U.K., Mak A.S., and Craig A.W.. 2011. Cdc42-interacting protein 4 is a Src substrate that regulates invadopodia and invasiveness of breast tumors by promoting MT1-MMP endocytosis. J. Cell Sci. 124:1739–1751 10.1242/jcs.078014 [DOI] [PubMed] [Google Scholar]
- Huang H.D., Lee T.Y., Tzeng S.W., and Horng J.T.. 2005. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 33(Suppl. 2):W226–W229 10.1093/nar/gki471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers I.J., Iravani M., Popov S., Robertson D., Al-Sarraj S., Jones C., and Isacke C.M.. 2010. A role for fibrillar collagen deposition and the collagen internalization receptor endo180 in glioma invasion. PLoS ONE. 5:e9808 10.1371/journal.pone.0009808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J.D., Byron A., Bass M.D., Craig S.E., Pinney J.W., Knight D., and Humphries M.J.. 2009. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci. Signal. 2:ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R.O.2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Hynes R.O.2009. The extracellular matrix: not just pretty fibrils. Science. 326:1216–1219 10.1126/science.1176009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juin A., Billottet C., Moreau V., Destaing O., Albiges-Rizo C., Rosenbaum J., Génot E., and Saltel F.. 2012. Physiological type I collagen organization induces the formation of a novel class of linear invadosomes. Mol. Biol. Cell. 23:297–309 10.1091/mbc.E11-07-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila S., Stenbäck F., Risteli J., Jukkola A., and Risteli L.. 1998. Aberrant type I and type III collagen gene expression in human breast cancer in vivo. J. Pathol. 186:262–268 [DOI] [PubMed] [Google Scholar]
- Kauppila S., Stenbäck F., Kacinski B.M., Carcangiu M.-L., Risteli J., and Risteli L.. 1999. Characterization of type I collagen synthesis and maturation in uterine carcinosarcomas. Cancer. 86:1299–1306 [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., and Aebersold R.. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Kim C., Ye F., and Ginsberg M.H.. 2011. Regulation of integrin activation. Annu. Rev. Cell Dev. Biol. 27:321–345 10.1146/annurev-cellbio-100109-104104 [DOI] [PubMed] [Google Scholar]
- Kraynov V.S., Chamberlain C., Bokoch G.M., Schwartz M.A., Slabaugh S., and Hahn K.M.. 2000. Localized Rac activation dynamics visualized in living cells. Science. 290:333–337 10.1126/science.290.5490.333 [DOI] [PubMed] [Google Scholar]
- Lai M.T., Hua C.H., Tsai M.H., Wan L., Lin Y.J., Chen C.M., Chiu I.W., Chan C., Tsai F.J., and Jinn-Chyuan Sheu J.. 2011. Talin-1 overexpression defines high risk for aggressive oral squamous cell carcinoma and promotes cancer metastasis. J. Pathol. 224:367–376 10.1002/path.2867 [DOI] [PubMed] [Google Scholar]
- Lai-Cheong J.E., Parsons M., and McGrath J.A.. 2010. The role of kindlins in cell biology and relevance to human disease. Int. J. Biochem. Cell Biol. 42:595–603 10.1016/j.biocel.2009.10.015 [DOI] [PubMed] [Google Scholar]
- Lamarche N., Tapon N., Stowers L., Burbelo P.D., Aspenström P., Bridges T., Chant J., and Hall A.. 1996. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 87:519–529 10.1016/S0092-8674(00)81371-9 [DOI] [PubMed] [Google Scholar]
- Larjava H., Plow E.F., and Wu C.. 2008. Kindlins: essential regulators of integrin signalling and cell-matrix adhesion. EMBO Rep. 9:1203–1208 10.1038/embor.2008.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W., et al. 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 139:891–906 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Dawson J.C., Forero-Vargas M., Spence H.J., Yu X., König I., Anderson K., and Machesky L.M.. 2010. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 20:339–345 10.1016/j.cub.2009.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., Wiesner C., and Himmel M.. 2011. Degrading devices: invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 27:185–211 10.1146/annurev-cellbio-092910-154216 [DOI] [PubMed] [Google Scholar]
- Mahawithitwong P., Ohuchida K., Ikenaga N., Fujita H., Zhao M., Kozono S., Shindo K., Ohtsuka T., Mizumoto K., and Tanaka M.. 2013. Kindlin-2 expression in peritumoral stroma is associated with poor prognosis in pancreatic ductal adenocarcinoma. Pancreas. 42:663–669 10.1097/MPA.0b013e318279bd66 [DOI] [PubMed] [Google Scholar]
- Mayya V., Lundgren D.H., Hwang S.I., Rezaul K., Wu L., Eng J.K., Rodionov V., and Han D.K.. 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2:ra46. [DOI] [PubMed] [Google Scholar]
- Meves A., Stremmel C., Gottschalk K., and Fässler R.. 2009. The Kindlin protein family: new members to the club of focal adhesion proteins. Trends Cell Biol. 19:504–513 10.1016/j.tcb.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Millon-Frémillon A., Brunner M., Abed N., Collomb E., Ribba A.S., Block M.R., Albigès-Rizo C., and Bouvard D.. 2013. Calcium and calmodulin-dependent serine/threonine protein kinase type II (CaMKII)-mediated intramolecular opening of integrin cytoplasmic domain-associated protein-1 (ICAP-1α) negatively regulates β1 integrins. J. Biol. Chem. 288:20248–20260 10.1074/jbc.M113.455956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D., et al. 2013. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203:1063–1079 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.C., Ghersi G., Akiyama S.K., Sang Q.X., Howard L., Pineiro-Sanchez M., Nakahara H., Yeh Y., and Chen W.T.. 1999. A novel protease-docking function of integrin at invadopodia. J. Biol. Chem. 274:24947–24952 10.1074/jbc.274.35.24947 [DOI] [PubMed] [Google Scholar]
- Myoui A., Nishimura R., Williams P.J., Hiraga T., Tamura D., Michigami T., Mundy G.R., and Yoneda T.. 2003. C-SRC tyrosine kinase activity is associated with tumor colonization in bone and lung in an animal model of human breast cancer metastasis. Cancer Res. 63:5028–5033. [PubMed] [Google Scholar]
- Nakahara H., Nomizu M., Akiyama S.K., Yamada Y., Yeh Y., and Chen W.T.. 1996. A mechanism for regulation of melanoma invasion. Ligation of α6β1 integrin by laminin G peptides. J. Biol. Chem. 271:27221–27224 10.1074/jbc.271.44.27221 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., and Aebersold R.. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Nguyen A.W., and Daugherty P.S.. 2005. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23:355–360 10.1038/nbt1066 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M.. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 127:635–648 10.1016/j.cell.2006.09.026 [DOI] [PubMed] [Google Scholar]
- Oser M., Mader C.C., Gil-Henn H., Magalhaes M., Bravo-Cordero J.J., Koleske A.J., and Condeelis J.. 2010. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J. Cell Sci. 123:3662–3673 10.1242/jcs.068163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R., Cukierman E., Clark K., Matsumoto K., Hahn C., Poulin B., and Yamada K.M.. 2003. Specific β1 integrin site selectively regulates Akt/protein kinase B signaling via local activation of protein phosphatase 2A. J. Biol. Chem. 278:18671–18681 10.1074/jbc.M300879200 [DOI] [PubMed] [Google Scholar]
- Parekh A., Ruppender N.S., Branch K.M., Sewell-Loftin M.K., Lin J., Boyer P.D., Candiello J.E., Merryman W.D., Guelcher S.A., and Weaver A.M.. 2011. Sensing and modulation of invadopodia across a wide range of rigidities. Biophys. J. 100:573–582 10.1016/j.bpj.2010.12.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., Gavara N., Chadwick R.S., and Yamada K.M.. 2012. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197:439–455 10.1083/jcb.201201124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M., Petrie R.J., Antoine-Bertrand J., Saint-Cyr-Proulx E., Villemure J.F., and Lamarche-Vane N.. 2009. Spatial and temporal activation of the small GTPases RhoA and Rac1 by the netrin-1 receptor UNC5a during neurite outgrowth. Cell. Signal. 21:1961–1973 10.1016/j.cellsig.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Provenzano P.P., Eliceiri K.W., Campbell J.M., Inman D.R., White J.G., and Keely P.J.. 2006. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4:38 10.1186/1741-7015-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano P.P., Inman D.R., Eliceiri K.W., Knittel J.G., Yan L., Rueden C.T., White J.G., and Keely P.J.. 2008. Collagen density promotes mammary tumor initiation and progression. BMC Med. 6:11 10.1186/1741-7015-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez N.E., Zhang Z., Madamanchi A., Boyd K.L., O’Rear L.D., Nashabi A., Li Z., Dupont W.D., Zijlstra A., and Zutter M.M.. 2011. The α2β1 integrin is a metastasis suppressor in mouse models and human cancer. J. Clin. Invest. 121:226–237 10.1172/JCI42328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razidlo G.L., Schroeder B., Chen J., Billadeau D.D., and McNiven M.A.. 2014. Vav1 as a central regulator of invadopodia assembly. Curr. Biol. 24:86–93 10.1016/j.cub.2013.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci E., Mattei E., Dumontet C., Eaton C.L., Hamdy F., van der Pluije G., Cecchini M., Thalmann G., Clezardin P., and Colombel M.. 2013. Increased expression of putative cancer stem cell markers in the bone marrow of prostate cancer patients is associated with bone metastasis progression. Prostate. 73:1738–1746 10.1002/pros.22689 [DOI] [PubMed] [Google Scholar]
- Rigbolt K.T., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., and Blagoev B.. 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4:rs3. [DOI] [PubMed] [Google Scholar]
- Roeder B.A., Kokini K., Sturgis J.E., Robinson J.P., and Voytik-Harbin S.L.. 2002. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J. Biomech. Eng. 124:214–222 10.1115/1.1449904 [DOI] [PubMed] [Google Scholar]
- Sameni M., Dosescu J., Yamada K.M., Sloane B.F., and Cavallo-Medved D.. 2008. Functional live-cell imaging demonstrates that beta1-integrin promotes type IV collagen degradation by breast and prostate cancer cells. Mol. Imaging. 7:199–213. [PMC free article] [PubMed] [Google Scholar]
- Savoy R.M., and Ghosh P.M.. 2013. The dual role of filamin A in cancer: can’t live with (too much of) it, can’t live without it. Endocr. Relat. Cancer. 20:R341–R356 10.1530/ERC-13-0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtner H., Calaminus S.D., Sinclair A., Monypenny J., Blundell M.P., Leon C., Holyoake T.L., Thrasher A.J., Michie A.M., Vukovic M., et al. 2013. Megakaryocytes assemble podosomes that degrade matrix and protrude through basement membrane. Blood. 121:2542–2552 10.1182/blood-2012-07-443457 [DOI] [PubMed] [Google Scholar]
- Schiller H.B., Hermann M.R., Polleux J., Vignaud T., Zanivan S., Friedel C.C., Sun Z., Raducanu A., Gottschalk K.E., Théry M., et al. 2013. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15:625–636 10.1038/ncb2747 [DOI] [PubMed] [Google Scholar]
- Schmidt S., Nakchbandi I., Ruppert R., Kawelke N., Hess M.W., Pfaller K., Jurdic P., Fässler R., and Moser M.. 2011. Kindlin-3–mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J. Cell Biol. 192:883–897 10.1083/jcb.201007141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., and Vignjevic D.M.. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz G.S., Davidson J.M., Kirsner R.S., Bornstein P., and Herman I.M.. 2011. Dynamic reciprocity in the wound microenvironment. Wound Repair Regen. 19:134–148 10.1111/j.1524-475X.2011.00673.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.A., and Ginsberg M.H.. 2002. Networks and crosstalk: integrin signalling spreads. Nat. Cell Biol. 4:E65–E68 10.1038/ncb0402-e65 [DOI] [PubMed] [Google Scholar]
- Sharma V.P., Eddy R., Entenberg D., Kai M., Gertler F.B., and Condeelis J.. 2013. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr. Biol. 23:2079–2089 10.1016/j.cub.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Ye Y., Dong L., Vainionpää S., Mustonen H., Puolakkainen P., and Wang S.. 2012. Kindlin-2: a novel adhesion protein related to tumor invasion, lymph node metastasis, and patient outcome in gastric cancer. Am. J. Surg. 203:222–229 10.1016/j.amjsurg.2011.06.050 [DOI] [PubMed] [Google Scholar]
- Shields M.A., Dangi-Garimella S., Redig A.J., and Munshi H.G.. 2012. Biochemical role of the collagen-rich tumour microenvironment in pancreatic cancer progression. Biochem. J. 441:541–552 10.1042/BJ20111240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottnik J.L., Daignault-Newton S., Zhang X., Morrissey C., Hussain M.H., Keller E.T., and Hall C.L.. 2013. Integrin alpha2beta 1 (α2β1) promotes prostate cancer skeletal metastasis. Clin. Exp. Metastasis. 30:569–578 10.1007/s10585-012-9561-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Rowe R.G., Botvinick E.L., Kurup A., Putnam A.J., Seiki M., Weaver V.M., Keller E.T., Goldstein S., Dai J., et al. 2013. MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev. Cell. 25:402–416 10.1016/j.devcel.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T., Barlow B., O’Rear L., Jarvis B., Li Z., Dickeson K., Dupont W., and Zutter M.. 2011. Loss of the α2β1 integrin alters human papilloma virus-induced squamous carcinoma progression in vivo and in vitro. PLoS ONE. 6:e26858 10.1371/journal.pone.0026858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Wu S., Shi X., Chen K., and Wu C.. 2003. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 113:37–47 10.1016/S0092-8674(03)00163-6 [DOI] [PubMed] [Google Scholar]
- van Kempen L.C., Rijntjes J., Mamor-Cornelissen I., Vincent-Naulleau S., Gerritsen M.J., Ruiter D.J., van Dijk M.C., Geffrotin C., and van Muijen G.N.. 2008. Type I collagen expression contributes to angiogenesis and the development of deeply invasive cutaneous melanoma. Int. J. Cancer. 122:1019–1029 10.1002/ijc.23147 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., and Horwitz A.R.. 2011. Adhesion dynamics at a glance. J. Cell Sci. 124:3923–3927 10.1242/jcs.095653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A.M., Page J.M., Guelcher S.A., and Parekh A.. 2013. Synthetic and tissue-derived models for studying rigidity effects on invadopodia activity. Methods Mol. Biol. 1046:171–189 10.1007/978-1-62703-538-5_10 [DOI] [PubMed] [Google Scholar]
- Williams K.C., McNeilly R.E., and Coppolino M.G.. 2014. SNAP23, Syntaxin4, and vesicle-associated membrane protein 7 (VAMP7) mediate trafficking of membrane type 1-matrix metalloproteinase (MT1-MMP) during invadopodium formation and tumor cell invasion. Mol. Biol. Cell. 25:2061–2070 10.1091/mbc.E13-10-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Yoshida N., Takanashi M., Ito Y., Fukami K., Yanagihara K., Yashiro M., and Sakai R.. 2014. Stromal fibroblasts mediate extracellular matrix remodeling and invasion of scirrhous gastric carcinoma cells. PLoS ONE. 9:e85485 10.1371/journal.pone.0085485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G.G., Risteli L., Mäkinen M., Risteli J., Kauppila A., and Stenbäck F.. 1995. Immunohistochemical study of type I collagen and type I pN-collagen in benign and malignant ovarian neoplasms. Cancer. 75:1010–1017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.