In yeast, the protein kinase Gin4 locally controls plasma membrane lipid asymmetry, which is necessary for optimal cytokinesis.
Abstract
Plasma membrane function requires distinct leaflet lipid compositions. Two of the P-type ATPases (flippases) in yeast, Dnf1 and Dnf2, translocate aminoglycerophospholipids from the outer to the inner leaflet, stimulated via phosphorylation by cortically localized protein kinase Fpk1. By monitoring Fpk1 activity in vivo, we found that Fpk1 was hyperactive in cells lacking Gin4, a protein kinase previously implicated in septin collar assembly. Gin4 colocalized with Fpk1 at the cortical site of future bud emergence and phosphorylated Fpk1 at multiple sites, which we mapped. As judged by biochemical and phenotypic criteria, a mutant (Fpk111A), in which 11 sites were mutated to Ala, was hyperactive, causing increased inward transport of phosphatidylethanolamine. Thus, Gin4 is a negative regulator of Fpk1 and therefore an indirect negative regulator of flippase function. Moreover, we found that decreasing flippase function rescued the growth deficiency of four different cytokinesis mutants, which suggests that the primary function of Gin4 is highly localized control of membrane lipid asymmetry and is necessary for optimal cytokinesis.
Introduction
A eukaryotic plasma membrane (PM) is a complex structure in which a multitude of lipid species are arranged in a spatially defined manner. PM lipids are organized laterally in the plane of the membrane into microdomains (Lingwood and Simons, 2010) and also transversely across the membrane, such that each leaflet of the bilayer has a distinct lipid composition (Fadeel and Xue, 2009). The marked difference in leaflet lipid content (referred to as bilayer asymmetry) was first noted in the erythrocyte PM (Gordesky and Marinetti, 1973), but is characteristic of the PM in all cell types (Devaux, 1991; van Meer, 2011). The exocellular leaflet is enriched in phosphatidylcholine, sphingolipids, and glycolipids, whereas the inner leaflet is enriched in phosphatidylethanolamine (PtdEth), phosphatidylserine (PtdSer), phosphatidylinositol (PtdIns), and derived phosphoinositides (e.g., PtdIns4,5P2; Devaux, 1991; Fadeel and Xue, 2009).
Bilayer asymmetry does not arise de novo during PM biogenesis, but is generated, in part, by active translocation of PtdEth and PtdSer inwards (“flipping”) along with similar translocation outwards of exoleaflet lipids (“flopping”; Daleke, 2003; van Meer, 2011). Maintenance of bilayer asymmetry is necessary in the face of the PM remodeling that results from continual exocytic vesicle insertion and endocytic vesicle removal, which would otherwise scramble leaflet lipid content. In eukaryotes, inward translocation of PtdEth and PtdSer is catalyzed by a subfamily (class 4) of P-type ATPases, dubbed flippases (Daleke, 2007; Lenoir et al., 2007; Tanaka et al., 2011; Sebastian et al., 2012). In budding yeast, there are five flippases: Dnf1, Dnf2, Dnf3, Drs2, and Neo1 (Catty et al., 1997). Dnf1 and Dnf2 localize primarily in the PM, whereas Dnf3, Drs2, and Neo1 are mainly confined to intracellular membranes (Daleke, 2007). Exit of Dnf1 (1,571 residues) and Dnf2 (1,612 residues) from the ER and their insertion and function in the PM requires their association with a smaller escort protein, Lem3/Ros3 (414 residues; Kato et al., 2002; Noji et al., 2006). Genetic analysis in yeast has implicated Dnf1 and Dnf2, and the other flippases—and thus membrane asymmetry—in endocytosis, protein trafficking, and vesicle formation (Chen et al., 1999; Gall et al., 2002; Hua et al., 2002; Pomorski et al., 2003; Liu et al., 2007; Natarajan et al., 2009; Hachiro et al., 2013) and in establishment of cell polarity (Iwamoto et al., 2004; Saito et al., 2007; Fairn et al., 2011; Das et al., 2012). Mutations in the human homologue of Dnf1 and Dnf2, ATP8B1, result in progressive familial intrahepatic cholestasis (Byler disease), as well as benign recurrent intrahepatic cholestasis and intrahepatic cholestasis in pregnancy (van der Mark et al., 2013).
The role of flippases in polarized growth is particularly intriguing. PtdEth is enriched in the exocellular leaflet at sites of polarized growth (Iwamoto et al., 2004; Saito et al., 2004), which suggests that flippase function must be temporally down-regulated during early bud formation when highly directional growth is required, but then reactivated when isotropic growth needs to resume. Until recently, there was little understanding about whether and how flippase function is regulated. For Drs2, specifically, it has been reported that PtdIns4P binds to its C-terminal tail and is required for its activity (Natarajan et al., 2009). The first clue about how Dnf1 and Dnf2 might be regulated came when it was found that loss-of-function mutations in FPK1, encoding a Ser/Thr protein kinase, were synthetically lethal with a deletion of CDC50, which encodes a Lem3 paralogue that associates with and is required uniquely for the function of flippase Drs2 (Nakano et al., 2008), which suggests that Fpk1 action was needed for optimal activity of the remaining flippases. Indeed, yeast cells lacking Fpk1 and its paralogue Fpk2/Kin82 did not have any change in flippase abundance or localization, yet showed a reduced ability to internalize fluorescently labeled PtdEth and PtdSer derivatives and exhibited an elongated bud phenotype with prolonged exocellular PtdEth exposure (Nakano et al., 2008). Moreover, purified Fpk1 was capable of direct phosphorylation of all yeast flippases (except Neo1), but with a marked preference for Dnf1 and Dnf2 (Nakano et al., 2008). Together, these results implicate Fpk1-mediated phosphorylation as a primary stimulus of Dnf1 and Dnf2 activity, providing the first mechanistic insight about cellular control of membrane asymmetry.
Subsequently, we enlarged this regulatory circuitry by showing that Fpk1 (the closest mammalian homologue is p70S6K; Hunter and Plowman, 1997) is, in turn, phosphorylated and inactivated by Ypk1 (Roelants et al., 2010), a protein kinase whose function is up-regulated in response to membrane stress (Roelants et al., 2011) and which we previously showed is the sequence homologue and functional orthologue of mammalian serum- and glucocorticoid-inducible kinase (SGK1; Casamayor et al., 1999). Additionally, we showed that a complex sphingolipid species, mannosyl-inositol-phosphorylceramide (MIPC), is required for Fpk1 activity (Roelants et al., 2010). Although these observations uncovered novel inputs into Fpk1 regulation, they did not explain how Fpk1-dependent regulation of flippases is controlled spatially and temporally during passage through the cell division cycle to control the budding process. By the means described here, we found that a septin-associated protein kinase, Gin4, phosphorylates and negatively regulates Fpk1, allowing us to demonstrate that Gin4 is responsible for the negative regulation of aminophospholipid flipping at the incipient bud site. Moreover, Gin4-mediated inhibition of Fpk1 action appears to be necessary for efficient cytokinesis.
Results
Screening for novel Fpk1 regulators
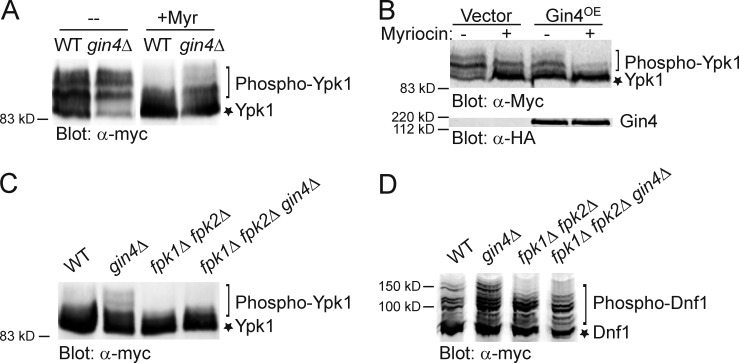
Flippase function requires activating phosphorylation by Fpk1 (Nakano et al., 2008), a protein kinase located at the bud cortex and under negative control by protein kinase Ypk1 (Roelants et al., 2010). To identify additional potential regulators of aminophospholipid flipping, we asked whether any other protein kinase might affect Fpk1 activity. Fpk1 not only phosphorylates flippases (large proteins hard to extract from the PM), but also feedback phosphorylates Ypk1 at S51 and/or S71 (or both), leading to multiple, slower mobility Ypk1 phospho-isoforms resolvable by SDS-PAGE (Roelants et al., 2010). We have demonstrated previously that a Ypk1 mobility shift provides a convenient and reliable assay for Fpk1 activity in vivo (Roelants et al., 2010). Hence, to identify other protein kinases that may influence Fpk1 function, we expressed Ypk1-myc in a collection of strains each deleted for a nonessential protein kinase gene. Strikingly, Ypk1 phospho-isoforms were substantially increased in gin4Δ cells (Fig. 1 A), which suggests that Gin4 may negatively regulate Fpk1. Complex sphingolipid MIPC promotes Fpk1 activity in vivo (Roelants et al., 2010). Accordingly, treatment of cells with myriocin (Myr), a drug that potently inhibits all sphingolipid biosynthesis by blocking the first enzyme (l-serine:palmitoyl-CoA C-palmitoyltransferase; SPT) in the pathway, greatly reduced the Ypk1 mobility shift. However, in cells lacking Gin4, the effect of reducing MIPC-dependent stimulation of Fpk1 was partially alleviated (Fig. 1 A), which indicates that the down-regulation of Fpk1 activity by Gin4 is not mediated through any effect on the MIPC level.
Figure 1.
Gin4 negatively regulates Fpk1-dependent phosphorylation of Ypk1 and Dnf1. (A) Wild-type (WT) strain (BY4741) or an isogenic gin4Δ mutant (YAT100) expressing Ypk1-myc from the GAL1 promoter (pAM76) were grown to mid-exponential phase, then either mock-treated or treated with Myr (1.25 µM) as indicated, collected, and lysed. The resulting extracts were resolved by SDS-PAGE and analyzed by immunoblotting with anti–c-myc mAb 9E10. (B) As in A, except that wild-type cells (Y258) carried both a plasmid expressing Ypk1-myc from the GAL1 promoter (pAM54) and either an empty vector (BG1805) or the same vector expressing Gin4 (pGin4-zz) from the GAL1 promoter. Gin4-zz was detected via its zz tag using an anti-HA mAb as a source of IgG. (C) As in A, except that a wild-type strain (BY4741) or isogenic gin4Δ mutant (YAT100), fpk1Δ fpk2Δ double mutant (YFR205), or fpk1Δ fpk2Δ gin4Δ triple mutant (YFR278) were used and only the Myr-treated cells are shown. (D) Same as in C, except that the cells expressed Dnf1(1403–1571)-myc from plasmid pES10 and the extract was analyzed using a Phos-tag gel.
Conversely, and in agreement with an independent role for Gin4 action in down-regulating Fpk1 activity, Gin4 overexpression markedly decreased the Ypk1 mobility shift and was additive with the effect of Myr (Fig. 1 B). Moreover, this effect was specific to Gin4 because elevated expression of neither its closest paralogue (Kcc4) nor its next closest relative (Hsl1), both septin-associated protein kinases (Barral et al., 1999; Shulewitz et al., 1999), like Gin4 (Longtine et al., 1998), had no effect on Ypk1 phospho-isoforms (Fig. S1). Furthermore, the increase in slower mobility Ypk1 isoforms seen in the absence of Gin4 was totally dependent on the presence of active Fpk1 and Fpk2 (Fig. 1 C), confirming that a primary role of Gin4 is to inhibit Fpk1. Although the pattern of phosphorylation of the PM flippases is much more complicated, we verified that loss of Gin4 also increased Fpk1-mediated phosphorylation of Dnf1, especially the appearance of the slowest mobility phospho-isoforms (Fig. 1 D), which further corroborates negative regulation of Fpk1 by Gin4.
Gin4 phosphorylates Fpk1
Gin4-dependent modification of Fpk1 could be direct or indirect, and could impede Fpk1 function by affecting its localization, level, or catalytic activity. We showed previously that Fpk1 localizes to the cell cortex in young buds and to the bud neck in large budded cells (Roelants et al., 2010). Compared with GIN4 cells, the pattern of Fpk1 localization was not detectably different in cells lacking Gin4 or overexpressing Gin4 (Fig. S2 A). Likewise, compared with GIN4 cells, the steady-state level of Fpk1 was not detectably different in cells lacking or overexpressing Gin4 (Fig. S2 B). Together, these results suggested that the primary effect of Gin4 might be to control the activity of Fpk1.
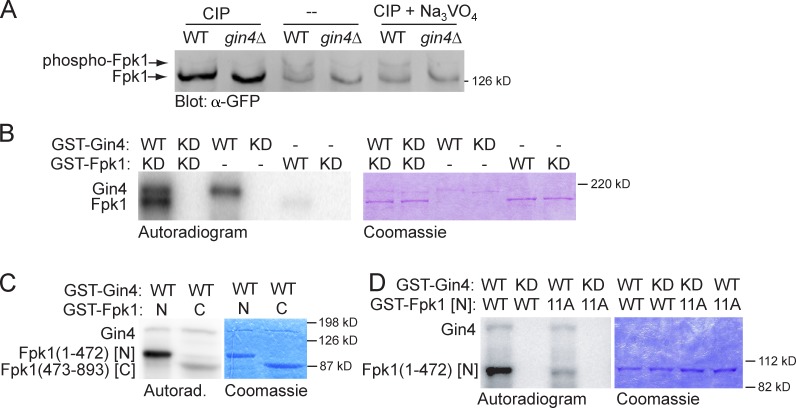
To assess whether phosphorylation of Fpk1 is Gin4 dependent in vivo, we first analyzed the migration pattern of Fpk1 fused to GFP using phosphate affinity (Phos-tag) gels (Kinoshita et al., 2009) and found a readily detectable level of a slower mobility Fpk1-GFP species whose appearance required Gin4 (Fig. 2 A, middle). This isoform was indeed due to phosphorylation because it was eliminated by treatment of the samples with calf intestinal phosphatase (CIP; Fig. 2 A, left), but persisted in the presence of CIP and the phosphatase inhibitor Na3VO4 (Fig. 2 A, right). To determine whether the Gin4-dependent phosphorylation of Fpk1-GFP observed in vivo may be direct, we tested whether Fpk1 serves as a substrate of Gin4 in vitro. To avoid any possibility of self-phosphorylation, catalytically inactive GST-Fpk1(D621A) (Roelants et al., 2010) purified from Escherichia coli was incubated with purified recombinant GST-Gin4 or an equivalent amount of a catalytically inactive mutant, GST-Gin4(K48A). We found that Fpk1 was readily phosphorylated by Gin4 (Fig. 2 B). Furthermore, when fused to GST, the N-terminal noncatalytic domain of Fpk1 was a much more robust substrate for Gin4 than its C-terminal (kinase) domain (Fig. 2 C). After exhaustive in vitro phosphorylation of Fpk1(1–472) by Gin4 in vitro, the sites of phosphorylation were mapped by mass spectrometry. This analysis revealed that 17 Ser or Thr residues were detectably modified, the majority of which fit the consensus (-R/KxxS-; Fig. S2 C), in accord with the phosphoacceptor site motif preference of Gin4 determined using synthetic peptide arrays (Mok et al., 2010). 11 of the most efficiently phosphorylated sites in Fpk1(1–472) were mutated to Ala, which decreased phosphorylation to a near-background level (Fig. 2 D), confirming that these sites were Gin4 targets.
Figure 2.
Gin4 phosphorylates Fpk1 in vivo and in vitro. (A) Wild-type strain (YFR221, WT) or an isogenic gin4Δ (YFR224) expressing Fpk1-GFP were grown to mid-exponential phase and lysed. The resulting extracts were either not treated or treated with CIP or CIP + Na3VO4, resolved on a Phos-tag gel, and analyzed by immunoblotting with anti-GFP antibody. (B) GST-Gin4 (pAB1, WT) or catalytically inactive (“kinase-dead,” KD) mutant, GST-Gin4(K48A) (pAT103), were purified from E. coli, incubated with γ-[32P]ATP and either GST-Fpk1 (pFR143, WT) or catalytically inactive (KD) mutant GST-Fpk1(D621A) (pFR144), also purified from E. coli. Resulting products were resolved by SDS-PAGE and analyzed by autoradiography (left) and staining with Coomassie dye (right). (C) GST-Gin4 (pAB1, WT) was purified from E. coli, incubated with γ-[32P]ATP and either GST-Fpk1(1–472) (pBS1, N) or catalytically inactive GST-Fpk1(473–893; D621A) (pBS2, C), which were also purified from E. coli, and the products were analyzed as in B. (D) As in C, except that either GST-Gin4 (pAB1, WT) or catalytically inactive (KD) GST-Gin4(K48A) (pAT103) were incubated with either GST-Fpk1(1–472) (pBS1, WT) or GST-Fpk1(1–472; 11A) in which 11 Gin4 phosphorylation sites were mutated to Ala (pJW2).
Gin4-mediated phosphorylation down-regulates Fpk1 activity
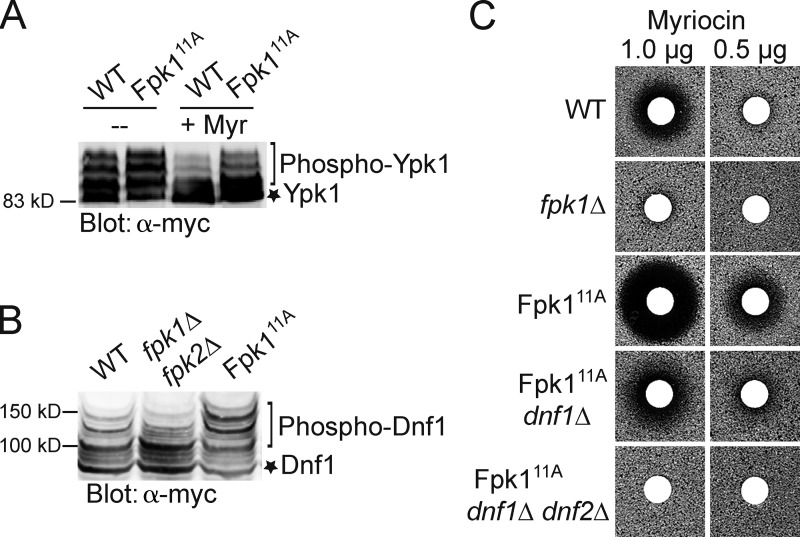
To determine whether a lack of Gin4-mediated phosphorylation enhances Fpk1 activity, we first examined by SDS-PAGE the mobility of Ypk1 isolated from wild-type cells or an isogenic strain in which the endogenous FPK1 locus was replaced by an allele (Fpk111A) in which the 11 Gin4 sites in full-length Fpk1 were mutated to Ala. We found that Fpk111A greatly enhanced formation of the slower mobility phospho-isoforms compared with the control cells, even when the levels of MIPC were reduced by Myr treatment (Fig. 3 A), and this increase was comparable to that observed in gin4Δ cells (Fig. 1 A). Likewise, compared with control cells, the appearance of the slowest mobility Fpk1-dependent isoforms of the flippase Dnf1 were also markedly increased in the cells expressing Fpk111A (Fig. 3 B). Thus, absence of Gin4 phosphorylation increased the specific activity of Fpk1 in vivo on both substrates. As a third criterion to assess its level of activity, we examined the Myr sensitivity conferred by Fpk111A compared with wild-type Fpk1 because we have shown previously that cells lacking Fpk1 are quite resistant to Myr (Roelants et al., 2010). Consistent with the conclusion that Fpk111A is hyperactive, the cells expressing Fpk111A cells were markedly more sensitive to Myr than cells expressing wild-type Fpk1 (Fig. 3 C). Moreover, in agreement that this phenotype was exerted through Fpk1-mediated stimulation of the flippases, the elevated drug sensitivity caused by Fpk111A was almost eliminated in cells lacking Dnf1 and totally abrogated in cells lacking both Dnf1 and Dnf2 (Fig. 3 C). Indeed, in agreement with the observation that flippases regulate the permeability of the yeast PM to xenobiotics (Yamane-Sando et al., 2014), we found that 400 ng/ml of Myr drastically reduced formation of complex sphingolipids in wild-type cells, whereas even 600 ng/ml had no effect in dnf1Δ dnf2Δ cells (Fig. S3 A).
Figure 3.
Phosphorylation by Gin4 down-regulates Fpk1 activity. (A) Cells expressing either wild-type Fpk1 (BY4741, WT) or Fpk111A (YJW2) and also expressing Ypk1-myc from the GAL1 promoter (pAM76) were grown to mid-exponential phase, untreated or treated with Myr (1.25 μM), and lysed. The resulting extracts were resolved by SDS-PAGE and analyzed by immunoblotting with anti–c–myc mAb 9E10. (B) Cells expressing wild-type Fpk1 (BY4741, WT), an isogenic strain lacking both Fpk1 and Fpk2 (YFR205, fpk1Δ fpk2Δ) or expressing Fpk111A (YJW2) and also expressing Dnf1(1403–1571)–myc from the GAL1 promoter (pES10) were grown to mid–exponential phase and lysed. The resulting extracts were resolved on a Phos-tag gel and analyzed by immunoblotting with anti–c–myc mAb 9E10. (C) Wild-type (BY4742, WT), fpk1Δ (JTY6581), Fpk111A (YFR307), Fpk111A dnf1Δ (YFR308), or Fpk111A dnf1Δ dnf2Δ (YFR312) strains, as indicated, were plated as a lawn on YPD plates and then overlaid immediately at the center with a sterile filter paper disk onto which a solution containing the indicated amounts of Myr had been spotted. After incubation at 30°C for 2 d, plates were photographed.
Furthermore, the elevated Myr sensitivity displayed by Fpk111A cells was not attributable to enhanced Fpk1-mediated feedback inhibition of Ypk1. Ypk1 phosphorylates and inactivates Orm1 and Orm2 (Roelants et al., 2011), two major negative regulators of SPT (Breslow et al., 2010); hence, elevated Fpk1-mediated feedback inhibition of Ypk1 would be expected to impede all sphingolipid biosynthesis. However, total sphingolipid levels were not significantly altered in cells expressing Fpk111A compared with wild-type or fpk1Δ fpk2Δ cells (Fig. S3 B).
Gin4 phosphorylation of Fpk1 down-regulates flippase function
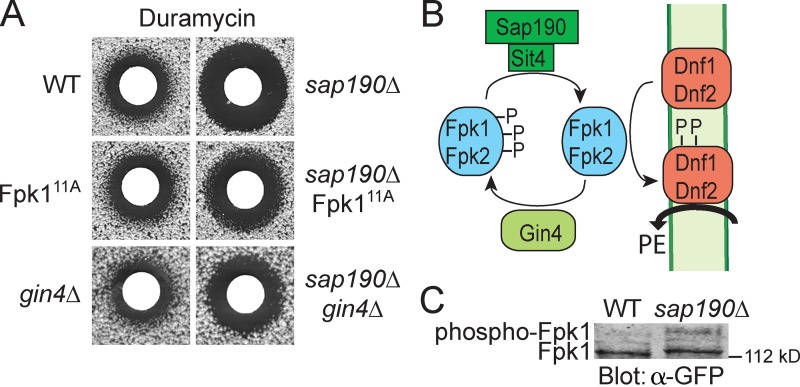
Collectively, our results indicate that modification by Gin4 inhibits Fpk1 function and that a major physiological consequence of this regulation is inhibition of flippase function. As further confirmation of this conclusion, we took advantage of the fact that the efficiency of flippase action can be monitored by assessing the sensitivity of cells to duramycin, a drug that binds to PtdEth in the outer leaflet of the PM and kills cells (Zhao, 2011). Because Fpk1 is a flippase activator, Gin4-mediated inhibition of Fpk1 should decrease flippase function, leaving more PtdEth on the cell surface and making cells more susceptible to duramycin, whereas cells expressing Fpk111A, which is largely “immune” to Gin4 phosphorylation, or cells lacking Gin4 altogether, should have enhanced flippase function, less PtdEth in the outer leaflet, and greater resistance to duramycin. However, any differences in duramycin sensitivity between wild-type, Fpk111A, and gin4Δ cells were too subtle to reliably detect (Fig. 4 A, left). We reasoned that we might be able to “trap” more wild-type Fpk1 in its Gin4-phosphorylated state (Fig. 4 B), but not Fpk111A or Fpk1 in gin4Δ cells, and thus enhance differences in duramycin sensitivity, if we removed the phosphatase responsible for reversing Gin4-mediated phosphorylation of Fpk1. To pinpoint any phosphatase that contributes to dephosphorylation of Fpk1, the duramycin sensitivity of a collection of deletion strains lacking each of the nonessential protein phosphatase genes or their associated factors was assessed. We found that cells lacking the phosphoprotein phosphatase 2A–related phosphatase Sit4 (Sutton et al., 1991; unpublished data) or one member of the family of Sit4-associated proteins, Sap190 (Luke et al., 1996; Fig. 4 A, right), exhibited a marked increase in sensitivity to duramycin, which suggests that the Sit4–Sap190 complex specifically dephosphorylates Fpk1. Indeed, in sap190Δ cells, there was a significant increase in the level of phosphorylated Fpk1 (Fig. 4 C).
Figure 4.
Gin4 phosphorylation of Fpk1 results in down-regulation of flippase activity. (A) Wild-type (BY4741, WT), Fpk111A (YJW2), or gin4Δ (YAT100) cells, or isogenic derivatives of the same strains carrying a sap190Δ mutation (sap190Δ [YFR320], sap190Δ Fpk111A [YFR323], and sap190Δ gin4Δ [YFR326]), were plated as a lawn on YPD plates and then overlaid immediately at the center with a sterile filter paper disk onto which 10 µl of a stock solution of duramycin (8 mM) had been spotted. After incubation at 30°C for 3 d, plates were photographed. (B) Model for control of Fpk1 (and Fpk2)-dependent activation of flippases Dnf1 and Dnf2 by Gin4-mediated phosphorylation and Sit4-Sap190 dephosphorylation of Fpk1. Trapping of Fpk1 (and Fpk2) in their Gin4-phosphorylated state prevents optimal flippase function, increasing the level of PtdEth in the outer leaflet of the PM, thereby conferring greater sensitivity to duramycin. (C) Either wild-type cells (YFR221, WT) or an isogenic sap190Δ mutant (YFR328) expressing Fpk1-GFP were grown to mid-exponential phase and lysed. The resulting extracts were resolved on a Phos-tag gel and analyzed by immunoblotting with anti-GFP antibody.
Using this sensitized sap190Δ background, it was clear that either Fpk111A or the absence of Gin4 enhanced flippase function to a greater extent than wild-type Fpk1 because the cells became reproducibly less sensitive to duramycin (Fig. 4 A, right). Thus, Fpk111A (or absence of Gin4) allows Fpk1 to be more active, stimulating flippase activity, reducing PtdEth in the outside leaflet, and conferring greater duramycin resistance (Fig. 4 B).
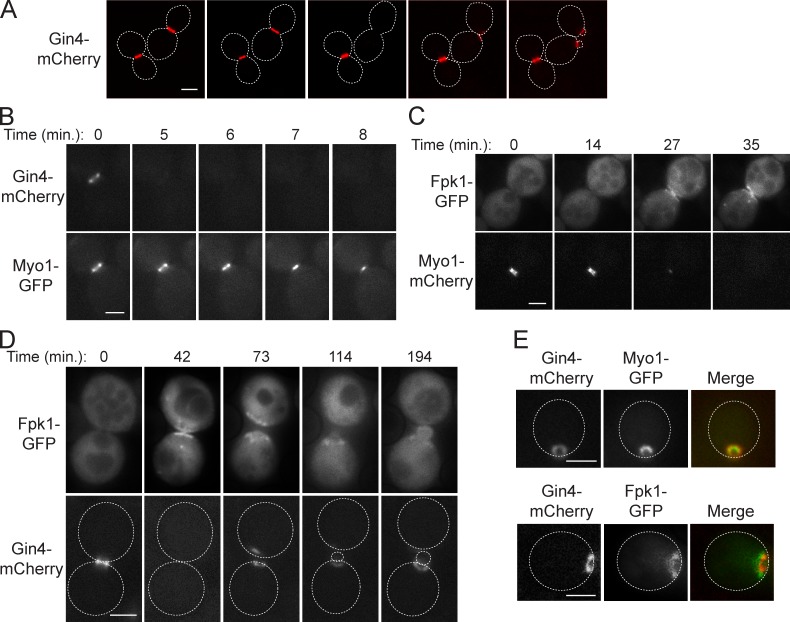
Gin4 and Fpk1 localization indicate potential roles in cytokinesis and bud emergence
To gain insight into the biological processes regulated by Gin4-mediated inhibition of Fpk1, we first investigated the dynamics of both Gin4 and Fpk1 localization during the cell cycle. Gin4 localizes at the bud site just at bud emergence and remains at the bud neck through M-phase, then disappears before cytokinesis (Okuzaki et al., 1997; Longtine et al., 1998; Fig. 5 A). Detailed observation revealed, first, that Gin4 disappears from the bud neck before the onset of actomyosin ring (AMR) contraction, as judged by the timing of ingression of the type II myosin heavy chain (Myo1; Fig. 5 B). By the same criterion, Fpk1 accumulates at the bud neck only at the end of AMR contraction (Fig. 5 C). This reciprocal behavior suggests that increased Dnf1- and Dnf2-mediated inward translocation of PtdEth may be important at the completion of cytokinesis. These observations further suggest that a function of Gin4 is to ensure complete suppression of any Fpk1-dependent flippase activation before AMR contraction and, therefore, that preventing inward PtdEth translocation is important for establishing the conditions for cytokinesis.
Figure 5.
Gin4 and Fpk1 colocalize prominently at the site of bud emergence. (A) Cells expressing Gin4-mCherry (YFR355) were viewed by time-lapse fluorescence microscopy at ∼30-min intervals. (B) As in A, for cells coexpressing Gin4-mCherry and Myo1-GFP (YFR388) at the indicated times. (C) As in B, for cells coexpressing Myo1-mCherry (YFR385) and Fpk1-GFP from the TPI1 promoter on a CEN plasmid (pFR150). (D) As in B, for cells coexpressing Fpk1-GFP (pFR150) and Gin4-mCherry (YFR355). (E, top) Same cells as in B. (E, bottom) Same cells as in D. Broken lines represent the perimeter of the indicated cells. Bars, 2 µm.
Furthermore, we found that Gin4 reappears at the cortical site of future bud emergence concomitant with Myo1 (Fig. S4 A), raising the possibility that Gin4 down-regulation of Fpk1 plays a role at that stage. Indeed, Fpk1 is also present at the new polarization site (Fig. 5 D), associated mainly with the perimeter of the ring-like structure containing Gin4 and Myo1 (Fig. 5 E). To confirm that Gin4-dependent inhibition of Fpk1 requires Gin4 association with the PM, we overexpressed Gin4(Δ1026–1125), a mutant lacking its C-terminal kinase associated-1 (KA1) domain, which binds to anionic phospholipids and is required for PM and bud neck localization of Gin4 (Moravcevic et al., 2010). In contrast to overexpressed full-length Gin4 (1,142 residues), an equivalent level of Gin4(Δ1026–1125) did not inhibit Fpk1-mediated phosphorylation of Ypk1 (Fig. S4 B). Given that Dnf1 and Dnf2 are found at the incipient bud site (Hua et al., 2002; Pomorski et al., 2003; Saito et al., 2004), our observations suggest that, at bud emergence, Gin4 is in a position to down-regulate Fpk1 and impose a reduction in flippase activity that may be important for proper bud polarization.
Control of leaflet lipid composition is important for cytokinesis
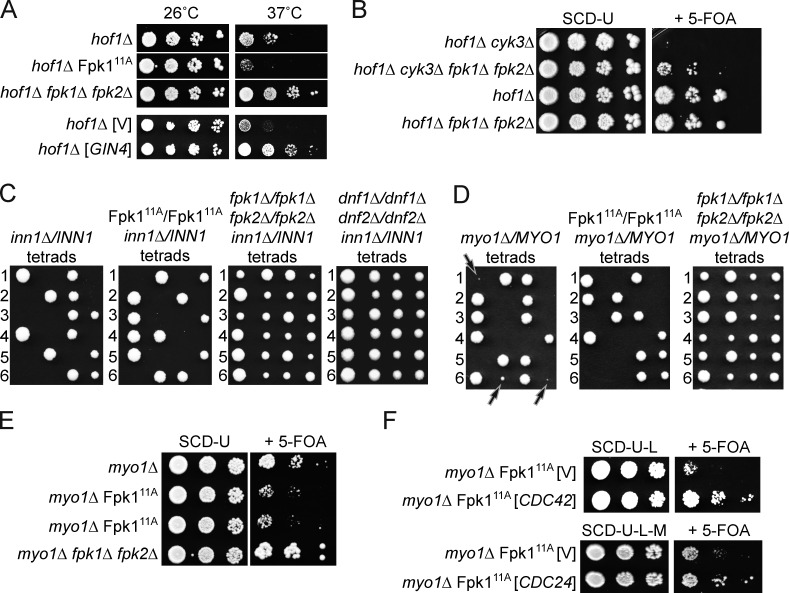
The accumulation of Fpk1 at the bud neck at the end of AMR contraction (Fig. 5 C) prompted us to look at the potential role of Fpk1 in cytokinesis. Interestingly, in mammalian cells, there is evidence that PtdEth plays some critical role in contractile ring disassembly and completion of cytokinesis (Emoto and Umeda, 2001), whereas, in S. pombe, PtdEth has been implicated in the execution of cytokinesis (Luo et al., 2009). In budding yeast, it has been reported that a gin4Δ mutation (but not a kcc4Δ or hsl1Δ mutation) is synthetically lethal with the absence of Hof1, an SH3 domain–containing bud neck–localized protein required for cytokinesis (Atkins et al., 2013; Meitinger et al., 2013). Based on the findings we describe here, gin4Δ cells should have elevated Fpk1 activity and, hence, elevated flippase function. Consistent with increased flippase function being responsible for the observed lethal phenotype of gin4Δ hof1Δ cells, we found that Fpk111A, which we showed has elevated PtdEth flipping (Fig. 4), exacerbated the temperature-sensitive growth defect of hof1Δ cells, and, conversely, elimination of both of the flippase-activating protein kinases Fpk1 and Fpk2 almost completely rescued the growth defect of hof1Δ cells at 37°C (Fig. 6 A, top). Completely consistent with the same conclusions, increased expression of Gin4 itself potently suppressed the temperature-sensitive growth defect of hof1Δ cells (Fig. 6 A, bottom).
Figure 6.
Decreasing flippase function rescues the growth defect of cytokinesis mutants. (A) Cultures of isogenic hof1Δ cells (YFR386), hof1Δ Fpk111A cells (YFR396), and hof1Δ fpk1Δ fpk2Δ (YFR398) cells, each carrying pRS316-HOF1, and hof1Δ cells (YFR386) carrying both an empty vector (YCpLG, V) or the same vector expressing GIN4 (pJT5241) and pRS316-HOF1, were streaked onto plates containing 5-fluoro-orotic acid (5-FOA) medium (Boeke et al., 1984) to select against the presence of the URA3-marked pRS316-HOF1 vector. Colonies arising were streaked on a second set of 5-FOA plates, and the resulting colonies were grown to mid-exponential phase in SCD. Serial 10-fold dilutions were then spotted on SCD plates. After incubation at 26°C or 37°C, as indicated, for 3 d, the plates were photographed. (B) Strains hof1Δ cyk3Δ (YFR422), hof1Δ cyk3Δ fpk1Δ fpk2Δ (YFR425), hof1Δ (YFR387), and hof1Δ fpk1Δ fpk2Δ (YFR424), all carrying pRS316-HOF1, were grown to mid-exponential phase in SCD-U. Serial 10-fold dilutions were spotted on SCD-U plates and plates containing 5-FOA. After incubation at 26°C for 3 d, the plates were photographed. (C) Tetrad analysis of isogenic inn1Δ/INN1 heterozygous diploids that were homozygous for wild-type FPK1 (YFR421), Fpk111A (YFR419), for both fpk1Δ and fpk2Δ (YFR414), or for both dnf1Δ and dnf2Δ (YFR434). (D) Tetrad analysis of isogenic myo1Δ/MYO1 heterozygous diploids that were homozygous for wild-type FPK1 (YFR420) or Fpk111A (YFR412), or for both fpk1Δ and fpk2Δ (YFR413). Arrows indicate microcolonies formed by myo1Δ spores expressing wild-type Fpk1 (left), which are inviable when Fpk111A is present (middle) and, conversely, grow much more robustly when both Fpk1 and Fpk2 are absent (right). (E) The same strains as in D were transformed with pRS316-MYO1 and dissected on SCD-U. The resulting haploid myo1Δ (YFR438), myo1Δ Fpk111A (YFR439; shown in duplicate), and myo1Δ fpk1Δ fpk2Δ (YFR440) strains each carrying pRS316-MYO1 were grown to mid-exponential phase, and serial 10-fold dilutions were spotted on SCD-U plates and plates containing 5-FOA to select against the presence of the pRS316-MYO1 vector. After incubation at 30°C for 3 d, the plates were photographed. (F) A myo1Δ/MYO1 heterozygous diploid that was homozygous for Fpk111A (YFR443) carrying an empty vector (YCpLG, V) or the same vector expressing CDC42 (PB3050), and the same strain carrying an empty vector (pRS315, V) or the same vector expressing CDC24 from the MET25 promoter (pJT4350) were transformed with pRS316-MYO1 and dissected on SCD-U-L (for the CDC42-expressing vector) or SCD-U-L-M (for the CDC24-expressing vector). The resulting myo1Δ Fpk111A strains (YFR448, YFR449, YFR451, and YFR452) were treated as in E.
Cyk3 is another SH3 domain–containing protein that interacts at the bud neck with Hof1. Although hof1Δ cyk3Δ double mutants are inviable even at 30°C (Korinek et al., 2000), we found that absence of Fpk1 and Fpk2 partially rescued this lethality (Fig. 6 B). Likewise, absence of Inn1, a protein required for primary septum (PS) ingression at the bud neck during cytokinesis, is lethal (Nishihama et al., 2009); yet, removal of both Fpk1 and Fpk2, or removal of both flippases Dnf1 and Dnf2, rescued the inviability of inn1Δ cells (Fig. 6 C).
Hof1, Cyk3, and Inn1 are all needed for efficient cell division because they each participate, directly or indirectly, in activation of Chs2, the chitin synthase isoform required for PS formation (Nishihama et al., 2009; Devrekanli et al., 2012). To determine whether the suppression observed was specific to mutants defective in PS formation or would also affect mutants defective in AMR contraction, we tested myo1Δ cells, which are viable but grow very slowly (Bi et al., 1998). Remarkably, expression of Fpk111A greatly exacerbated the growth defect of cells lacking Myo1, and, conversely, elimination of Fpk1 and Fpk2 markedly improved the growth of myo1Δ cells (Fig. 6, D and E). These results indicate that a decrease in the rate of PtdEth flipping is beneficial for cells impaired in either AMR contraction or PS formation, which suggests that during execution of normal cytokinesis and cell division, the dynamic movement of lipids in the PM needs to be properly coordinated with protein-driven events.
One predicted consequence of a decrease in Fpk1-stimulated and Dnf1- and Dnf2-catalyzed PtdEth flipping should be an increase in the activity of Cdc42, a key PM-associated regulator of cell polarization and cytokinesis (Tong et al., 2007; Atkins et al., 2013; Lo et al., 2013; Onishi et al., 2013), because inner leaflet PtdEth is required for activation of the GTPase-activating proteins (GAPs), Rga1 and Rga2, that down-regulate Cdc42-GTP (Saito et al., 2007), and/or for the Rdi-mediated dissociation of Cdc42 from the PM (Das et al., 2012). Consistent with the idea that a contributing factor to the beneficial effect of decreased flippase function in cells impaired for cytokinesis is due to enhanced Cdc42 function, a modest increase in the expression of either Cdc42 or Cdc24 (Cdc42-specific guanine nucleotide exchange factor) rescued the severe growth debility of myo1Δ Fpk111A cells (Fig. 6 F).
High-level overexpression of Cdc42 is reportedly not grossly deleterious to normal cells (Johnson and Pringle, 1990), but kills cytokinesis mutants defective in either AMR assembly or PS formation (Atkins et al., 2013); yet, as we observed for myo1Δ Fpk111A cells, a modest elevation in Cdc42 level markedly suppressed the temperature-sensitive growth defect of hof1Δ cells (Fig. S5).
Collectively, our results suggest that the role of the Gin4–Fpk1–flippase pathway is to properly adjust the inner leaflet PtdEth concentration as a means to fine-tune the level of active Cdc42 at the cell cortex. The dramatic departure of Gin4 from the bud neck that occurs just before AMR contraction would allow for Sit4-Sap190–mediated reactivation of the Fpk1 present immediately thereafter, stimulating influx of PtdEth, thereby promoting near-complete Cdc42-GTP inactivation. Consistent with this scenario, several groups have reported that a sharp drop in Cdc42-GTP occurs at the end of cytokinesis (Atkins et al., 2013; Okada et al., 2013; Kang et al., 2014).
Discussion
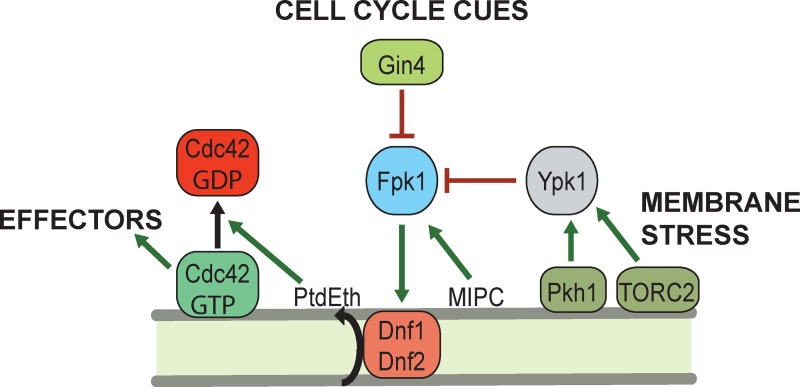
We have demonstrated previously (Roelants et al., 2010) that the flippase-activating protein kinase Fpk1 (and its paralogue Fpk2) is subject to two physiologically important levels of control: via inhibitory phosphorylation by the membrane-stress activated protein kinase Ypk1 (and its paralogue Ypk2); and via stimulation specifically by the complex sphingolipid MIPC (by a mechanism not yet understood). As we show here, an equally important new component of this regulatory network is inhibitory phosphorylation by the septin-associated and cell cycle–regulated protein kinase Gin4 (Fig. 7). This specific function is a previously unrecognized role for Gin4, which was thought to be involved only in proper septin collar formation via direct phosphorylation of septin Shs1 (Mortensen et al., 2002; Asano et al., 2006). It has been suggested that this modification promotes formation of the septin collar that demarcates the bud neck during bud emergence, thereby generating a stable scaffold that recruits other proteins needed for bud growth and then cytokinesis (McMurray and Thorner, 2009; Oh and Bi, 2011). Consistent with such a function, after mitosis, Shs1 is dephosphorylated by the Rts1-bound isoform of phosphoprotein phosphatase 2A, concomitant with splitting of the septin collar into two discrete rings at cytokinesis (Dobbelaere et al., 2003). However, aberrant septin structure at the bud neck is displayed by only a very small fraction of the cells in a culture of a gin4Δ mutant (Longtine et al., 1998). Moreover, time-lapse microscopy of gin4Δ cells expressing a GFP-tagged septin revealed that, in the majority of these rare anomalous cells, the abnormal septin morphology eventually resolves into a uniform collar and the cell then divides (McMurray and Thorner, 2009). The findings we have presented here demonstrate that Gin4 control of Fpk1 governs the level of PtdEth in the inner leaflet of the PM and argue that a fundamentally important function of Gin4 is its role in regulation of PM bilayer asymmetry.
Figure 7.
Gin4 controls inner leaflet PtdEth and Cdc42-GTP level. During progression through the cell cycle, Gin4 localizes at the incipient bud site, and phosphorylates and inhibits Fpk1 at the same location, preventing flippase activation and keeping PtdEth level on the inner leaflet low. Because inner leaflet PtdEth promotes Cdc42-GTP inactivation (Saito et al., 2007; Das et al., 2012), Gin4-imposed reduction in PtdEth allows for buildup of Cdc42-GTP to stimulate recruitment and function of effectors, like formin Bni1, that promote bud emergence. Further accumulation of Gin4 at the septin collar will continue to suppress the inner leaflet PtdEth level, allowing for localized protection of Cdc42-GTP, perhaps facilitating recruitment and activation of effectors required for efficient cytokinesis. Despite a normally beneficial role, flippase function also permits entry of toxic xenobiotics. To help prevent an influx of such noxious agents when the PM barrier is already compromised by heat stress or other membrane-perturbing or -damaging agents, an independent stress response mediated by protein kinase Ypk1 also down-regulates flippase function by inhibiting Fpk1. Full Ypk1 activity requires its stimulation by two PM-associated protein kinases: eisosome-bound Pkh1 and the TORC2 complex.
In this regard, it must be appreciated that reversal of Gin4-dependent phosphorylation of Fpk1 may also be under tight control. Although the responsible phosphatase Sit4 associates with Sap4, Sap155, Sap185, and Sap190 (Luke et al., 1996), we found that only sap190Δ mutations, and not sap4Δ, sap155Δ, or sap185Δ mutations (unpublished data), exhibited an increase in duramycin sensitivity, which indicates that reactivation of Fpk1 is a function specific to only the Sap190-bound form of Sit4.
We found that, in contrast to their spatiotemporal partitioning during cytokinesis, Gin4 and Fpk1 colocalize at the incipient bud site. Given that inner leaflet PtdEth promotes inactivation of Cdc42-GTP by its GAPs and/or GDI (Cherfils and Zeghouf, 2013), our results connect Gin4 action to modulation of the level of active Cdc42, an essential and pivotal regulator of cell polarity and morphogenesis (Etienne-Manneville, 2004; Bi and Park, 2012). Gin4, by negatively regulating Fpk1, inhibits the flippases under Fpk1 control, lowers inner leaflet PtdEth content, thus reducing PtdEth-stimulated GAPCdc42 (Rga1 and Rga2; Saito et al., 2007) and/or Rho GDI (Rdi1; Das et al., 2012) function, thereby maintaining or increasing the concentration of PM-associated GTP-bound Cdc42 (Fig. 7). An important aspect of the production of Cdc42-GTP above a certain threshold at this cell cycle stage is to trigger actin cable formation for bud emergence, mediated by the formin Bni1 (Chen et al., 2012). As a result of their colocalization, Gin4-mediated phosphorylation and inhibition of Fpk1 and the ensuing reduced level of inner leaflet PtdEth would allow for sufficient Cdc42-GTP at the incipient bud site for activation of this and other effectors.
Recent work has proposed a model wherein active Cdc42 recruits septins, which, upon assembly, inhibit Cdc42 in a negative feedback loop via the bud neck–localized Cdc42-GAPs (Okada et al., 2013). However, our findings provide deeper insight into the dynamics of this process. Assembled septins also recruit Gin4, which, by phosphorylating and inhibiting Fpk1 in its immediate vicinity, will result in down-regulation of the flippases and lowering of inner leaflet PtdEth, thereby reducing Cdc42-GAP and/or Cdc42 GDI activity, thus locally maintaining Cdc42-GTP within a tightly delimited site at the bud cortex to support bud emergence. Moreover, our visualization of these proteins showed that, at the site of bud emergence, some Fpk1 localizes in a concentric ring around the periphery of where Gin4 localizes (Fig. 5 E); if that more distant pool of Fpk1 escapes Gin4-imposed inhibition, it would be able to activate flippase function, raise inner leaflet PtdEth, stimulate Cdc42-GAP and/or Cdc42-GDI activity, and thereby delineate a rather sharp boundary outside of which Cdc42 cannot be active. Showing that there is an inverse correlation between inner leaflet PtdEth content and the concentration of active Cdc42 is not now technically feasible because the only known PtdEth-specific probe is a fluorescently tagged derivative of a tetracyclic, disulfide-bonded peptidic antibiotic (Ro09-0198/cinnamycin) that is only able to bind this lipid when it is present in the outer leaflet of the PM (Aoki et al., 1994; Iwamoto et al., 2004). Moreover, this agent blocks PtdEth flipping and, hence, can only report the total amount of PtdEth that has been delivered to the PM.
If colocalization of Gin4 with Fpk1 suppresses flippase function during polarity establishment and bud site selection, their dramatic separation just before AMR contraction (Fig. 5) presumably achieves the opposite. Viewed in this way, Gin4 and Fpk1 provide an example of how differential spatial and temporal reorganization of components of the same signaling modality can allow for rewiring of pathway connectivity to accomplish distinct outputs during cell cycle progression. Up to the onset of cytokinesis, Gin4 is maintained at the bud neck, which, based on our data, would ensure that any residual flippase function is totally suppressed, allowing for Cdc42-GTP–stimulated processes at the bud neck, possibly Bnr1-promoted actin filament formation for AMR assembly (Vallen et al., 2000; Buttery et al., 2012). Just before initiation of AMR ingression, Gin4 rapidly disappears from the bud neck, whereas Fpk1 accumulates at the bud neck as AMR contraction is completed, which would allow for reactivation of flippases, buildup of inner leaflet PtdEth, and the down-regulation of active Cdc42 that occurs at the end of cell division (Atkins et al., 2013; Okada et al., 2013; Kang et al., 2014).
Although we do not have direct evidence for this sort of regulation of Cdc42 during normal cytokinesis because roles for Cdc42 at this stage of the cell cycle have only recently started to emerge, our results indicate that regulation of Cdc42 exerted by the Gin4–Fpk1–flippase pathways plays a crucial role in AMR-independent cytokinesis. In the absence of Myo1, no AMR can form and PS formation is also defective, yet cytokinesis can be completed by abnormal deposition of an irregular, chitinaceous secondary septum, referred to as the remedial septum (Schmidt et al., 2002), that allows for cell separation and survival (Bi et al., 1998). We clearly show that lack of the flippase activators Fpk1 and Fpk2 ameliorates the growth defect of myo1Δ cells and that the hyperactive Fpk111A allele worsens the growth defect of myo1Δ cells (Fig. 6, D and E). Moreover, elimination of Fpk1 and Fpk2, or of the PM flippases they control, also ameliorated the growth defects of three different mutants (hof1Δ, hof1Δ cyk3Δ, and inn1Δ) that prevent PS formation (Fig. 6, A, B, and C). Consistent with this suppression arising from increased Cdc42 activity due to lower inner leaflet PtdEth, modest elevation of total Cdc42 or of its GEF (Cdc24) also ameliorated the growth defect of both myo1Δ cells (Fig. 6 F) and hof1Δ cells (Fig. S5). Therefore, in our hands, we find that Cdc42 action somehow promotes formation of this remedial septum. It is also possible that inner leaflet PtdEth influences other Rho GTPases whose overexpression has been shown to suppress the growth defect of cytokinesis mutants (Dong et al., 2003; Yoshida et al., 2009; Onishi et al., 2013).
If, in normal cells, activation of flippases and subsequent inhibition of Cdc42 is important for the completion of cytokinesis, one might wonder why the lack of flippase activity, and thus activation of Cdc42, is beneficial for all the mutants we tested that are defective in cytokinesis? Atkins et al. (2013) proposed that inappropriate Cdc42 activation during mitotic exit impairs cytokinesis via mislocalization of Inn1 and the IQGAP orthologue Iqg1, which are important for PS formation. However, in the absence of AMR or PS formation, activation of Cdc42 cannot have this deleterious effect because the proteins that are mislocalized when Cdc42 is activated are not present in the first place. Thus, the mechanism that cells use to complete cytokinesis under such conditions is different. Hence, although activation of Cdc42 may be detrimental to AMR and PS formation, our results suggest that it is beneficial with regard to remedial septum formation.
Given the central role we have uncovered for Gin4 in controlling the PtdEth content of the PM inner leaflet and, consequently, Cdc42-GTP level, one important mechanistic question to address in the immediate future is what element within Gin4 is responsible for its specific association with septin-containing structures, how it may be controlled, and, conversely, which septins or aspect of septin organization are recognized by Gin4. Another critical question is what inputs are responsible for the dramatic changes in level and localization of Gin4 during progression through the cell cycle. Gin4 has been shown to be activated by the upstream septin-associated protein kinase Elm1 (Asano et al., 2006), most likely by phosphorylation of Thr189 in its activation loop (related kinase Hsl1 is phosphorylated by Elm1 at the corresponding site, Thr273; Szkotnicki et al., 2008). Gin4 activity and localization also appear to be modulated by Cdk1/Cdc28 (Okuzaki et al., 1997; Mortensen et al., 2002; Holt et al., 2009). In Candida albicans, Gin4 is also phosphorylated and activated by Cdk1, and it has been shown that forced activation of Cdc42 restores defects in bud neck formation observed in gin4Δ mutants (Li et al., 2012), placing Cdc42 downstream of the Gin4–Fpk1–flippase pathway, which is consistent with our findings. Interestingly, there is some evidence that Cdc42 itself is required in vivo for the mitosis-specific activation of Gin4 and the role it plays in polarized bud growth (Tjandra et al., 1998), which may represent an elegant feedback loop for regulation of the Gin4–Fpk1–flippase pathway.
Materials and methods
Strains and growth conditions
Yeast strains used in this study (Table S1) were grown routinely at 30°C. Standard rich (YP) and defined minimal (SC) media (Sherman et al., 1986), containing either 2% glucose (Glc), 2% raffinose, and 0.2% sucrose (Raf-Suc) or 2% galactose (Gal) as the carbon source, as indicated, and supplemented with appropriate nutrients to maintain selection for plasmids, were used for yeast cultivation. In particular, YFR433 and YFR434 (Table S1) were derived from a strain provided by T.R. Graham (Vanderbilt University, Nashville, TN). For gene induction from the GAL1 promoter, cells were pregrown to mid-exponential phase in SC+Raf-Suc medium, Gal was added (2% final concentration), and incubation was continued for 3 h. When cells were treated with Myr (Sigma-Aldrich) or phytosphingosine (PHS; Avanti Polar Lipids, Inc.), the cultures were grown to mid-exponential phase, induced with Gal for 1 h, and the compounds were added at the final concentrations indicated (Myr, 1.25 µM; PHS, 5 or 10 µM) and incubation was continued for an additional 2 h. Standard yeast genetic techniques were performed according to Sherman et al. (1986).
Plasmids and recombinant DNA methods
Plasmids used in this study (Table S2) were constructed using standard procedures (Sambrook et al., 1989) in E. coli strain DH5α. In particular, plasmid PB3050 (Table S2) was provided by D. Pellman (Harvard Medical School, Boston, MA). Fidelity of all constructs was verified by nucleotide sequence analysis. All PCR reactions were performed using Phusion DNA polymerase (Finnzymes; Thermo Fisher Scientific). Site-directed mutagenesis using appropriate mismatch oligonucleotide primers was conducted using the QuikChange method and Pfu Turbo DNA polymerase (Agilent Technologies).
Preparation of cell extracts and immunoblotting
The cells in samples (1.5 ml) of an exponentially growing culture (A600nm = 0.6) were collected by brief centrifugation, immediately frozen in liquid N2, and then lysed by resuspension in 150 µl of 1.85 M NaOH, 7.4% β-mercaptoethanol. Protein in the resulting lysate was precipitated by the addition of 150 µl of 50% trichloroacetic acid on ice. After 10 min, the resulting denatured protein was collected by centrifugation, washed twice with acetone, and solubilized by resuspension in 80 µl of 0.1 M Tris, 5% SDS. Then, 20 µl of a 5× stock of SDS-PAGE sample buffer was added. After boiling for 5 min, portions (15 µl) of the samples containing Ypk1-myc were resolved by SDS-PAGE (8% acrylamide, 75:1 monomer-to-cross-linker), and samples containing Dnf1(1403–1571)-myc were resolved on a Phos-tag gel (8% acrylamide, 29:1 monomer-to-cross-linker, 30 µM Phos-tag reagent; Wako Pure Chemical Industries), transferred to nitrocellulose, incubated with appropriate primary antibodies in Odyssey buffer (LI-COR Biosciences), washed, incubated with appropriate secondary antibodies conjugated to infrared fluorophores, and visualized using an Odyssey infrared imaging system (LI-COR Biosciences). The antibodies used in this work were: tissue culture medium containing mouse anti–c-myc mAb 9E10, raised against a synthetic peptide corresponding to the C-terminal residues (408–432) of human c-myc (AEEQKLISEEDLLRKRREQLKHKLE; epitope underlined) conjugated to keyhole limpet hemocyanin (1:100; Monoclonal Antibody Facility, Cancer Research Laboratory, University of California, Berkeley), mouse anti-HA mAb (1:1,000, Covance), mouse anti-GFP mAb (1:1,000, Roche), and rabbit polyclonal anti-Gin4 antibodies, raised against a purified bacterially expressed (His)6-tagged C-terminal fragment (residues 870–1,142; 1:1,000, a gift of D. Kellogg, University of California, Santa Cruz, Santa Cruz, CA).
Protein kinase assay
GST-Gin4 or a kinase-dead mutant (K68A) were expressed and purified from E. coli and incubated at 30°C in protein kinase assay buffer (20 mM Tris-HCl, pH 7.2, 125 mM potassium acetate, 12 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 2 mM DTT, 1% glycerol, 0.02% BSA, 25 mM β-glycerol phosphate, and 1 mM sodium orthovanadate) with 100 µM γ-[32P]ATP (∼5 × 105 cpm/nmol) and 0.5 µg of GST-fused substrate protein (which also were prepared by expression in and purification from E. coli, as described in the following section). After 30 min, reactions were terminated by the addition of SDS-PAGE sample buffer containing 6% SDS followed by boiling for 5 min. Labeled proteins were resolved by SDS-PAGE and analyzed by autoradiography using a PhosphorImager (Molecular Dynamics Division, GE Healthcare).
Purification of GST fusion proteins
Freshly transformed BL21(DE3) cells carrying a plasmid expressing the desired GST fusion protein were grown to A600nm = 0.6, and expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (final concentrations: 0.2 mM for GST-Fpk1 and 0.5 mM for GST-Gin4). After vigorous aeration for 8 h at 26°C (for GST-Fpk1) or 4 h at 30°C (for GST-Gin4), cells were harvested and the GST fusion protein was purified by column chromatography on glutathione-agarose beads using standard procedures.
Analysis of complex sphingolipids
Cells were adjusted to A600nm = 1.0. 2-ml cultures were labeled with 100 µCi of [32P] H2PO4 and cells were allowed to grow for 3 h. Myr was added to the overnight cultures at the indicated final concentrations from a stock of 0.625 mM (250 µg/ml) in methanol. An equal volume of methanol was added to the negative culture. Lipids were extracted and resolved as described previously (Momoi et al., 2004) with minor modifications. The cell pellet was washed twice with 2 ml water and treated with 5% trichloroacetic acid for 20 min on ice. Pellets were extracted twice with ethanol/water/diethyl ether/pyridine/NH4OH (15:15:5:1:0.018) at 60°C for 1 h. The combined extracts were treated with 0.1 N KOH in methanol for 1 h at 50°C to hydrolyze glycerophospholipids. Sphingolipids then were extracted with 3 ml chloroform, 1 ml methanol, and 4.5 ml of 0.5 N NH4OH, and the chloroform layer was washed three times with alkaline water (0.05 N NH4OH in water). For some samples, 1 ml of 4 M NaCl was added during chloroform extraction to facilitate separation of the aqueous and organic phases. The organic layer was dried in a Speedvac and resuspended in 50 µl of chloroform/methanol/water (16:16:5), spotted at the origin of silica gel TLC plates, and resolved by ascending chromatography using chloroform/methanol/4.2 N NH4OH (9:7:2) as the solvent. Radioactive species on the TLC plate was visualized using a phosphorimaging screen and a Typhoon imaging system.
Determination of total sphingolipids
Overnight cultures were adjusted to A600nm = 0.2 and allowed to grow to an A600nm = 1.0. The cell pellet from 10 ml of such cultures was used for analysis. Lipids were extracted, as described in the previous section, and total lipid extract was hydrolyzed with 0.5 M HCl in acetonitrile-water for 4 h at 70°C. After hydrolysis, the acid was neutralized by 0.2 ml NH4OH, 1 ml CHCl3 and 1 ml of water were added, solutions were mixed, phases were separated by centrifugation, and the organic layer was removed and dried in a Speedvac (Thermo Fisher Scientific). Dried lipids were resuspended in 0.3 ml methanol and derivatized with 0.1 ml o-phthalaldehyde, as described previously (Merrill et al., 2000). After incubation for 30 min, the reaction was centrifuged to remove particulate matter and 20–50 µl of the supernatant was analyzed by HPLC performed using an Eclipse Plus C18 column (4.6 × 150 mm, 5 µm) on an HPLC device (120 Infinity; Agilent) equipped with a UV detector. Samples were resolved using a mobile phase gradient of methanol/5 mM potassium phosphate, pH 7.0, from 50:50 (vol/vol) to 95:5 (vol/vol) over a 7-min elution. PHS, both pure standard (Avanti Polar Lipids, Inc.) and in yeast samples, was reproducibly observed as a sharp peak at 2.9 min with a >1,000-fold signal-to-noise ratio. PHS standard was serially diluted and resolved by the same method to determine the linear range of the analysis.
Subcellular localization by fluorescence microscopy
Yeast proteins fused to either GFP (Tsien, 1998) or mCherry (Shaner et al., 2004) were constructed by in-frame integration at the corresponding chromosomal locus and expressed from the endogenous promoter, except Fpk1-GFP that was expressed from the TPI1 promoter on a CEN plasmid, which, as we demonstrated previously, displays a pattern of localization brighter than, but indistinguishable from, that exhibited by integrated Fpk1-GFP expressed from its endogenous promoter (Roelants et al., 2010). All such fusions were functional, as judged by complementation tests, i.e., the ability of an integrated construct to confer a normal phenotype or a plasmid-borne construct to restore a normal phenotype to the corresponding null mutant. To visualize yeast proteins tagged with GFP or mCherry, cells were grown to mid-exponential phase, spotted on 1% agarose pads on a glass slide at room temperature, and viewed directly under an epifluorescence microscope (model BH-2; Olympus) using a 100× objective lens (D plan Apochromat 100, 1.3 numerical aperture) equipped with appropriate band-pass filters (Chroma Technology Corp.) and illuminated with a SOLA SE Light Engine (Lumencor, Inc.) for excitation. Images were collected using a charged-couple device camera (CoolSNAP MYO; Photometrics), and processed with Micro-Manager imaging software (Edelstein et al., 2010) and Photoshop (Adobe Systems).
Analysis of phosphorylation sites by mass spectrometry
GST-Fpk1(1–472) that was expressed and purified from E. coli was incubated exhaustively with excess Mg-ATP and GST-Gin4 that was also expressed and purified from E. coli in a standard protein kinase assay buffer (see the “Protein kinase assay” section). Portions of the phosphorylated GST-Fpk1 were denatured in 8 M urea, 100 mM Tris-HCl, pH 8.5, reduced with 5 mM tris-(2-carboxyethyl)-phosphine, amidomethylated at its sole Cys (C173) by incubation in the dark with 10 mM freshly prepared iodoacetamide, and then concentrated fivefold using a Microcon 50 centrifugal filtration device (EMD Millipore). Portions of the resulting protein were diluted fourfold into 1 mM CaCl2 and 100 mM Tris-HCl, pH 8.5, and digested with at final concentration of 1.5 µg/ml sequencing grade trypsin (Promega) or endoprotease Glu-C (Staphylococcus aureus V8 protease) at 37°C for at least 8 h. Reactions were terminated by the addition of 5% formic acid (final concentration). The resulting peptides were desalted using a C18 spin tip (OMIX; Varian/Agilent Technologies). A nano LC column (consisting of 10 cm of C18 5-µm packing material; Polariz, Varian/Agilent Technologies) was packed in a 100-µm inner diameter glass capillary with an emitter tip, loaded with the sample by use of a pressure bomb, and washed extensively with Buffer A (5% acetonitrile, 0.02% heptafluorobutyric acid). The column was then directly coupled to an electrospray ionization source mounted on an LTQ XL linear ion trap mass spectrometer (Thermo Fisher Scientific). An HPLC (Model 1200; Agilent Technology) equipped with a split line, so as to deliver a flow rate of 300 nl/min, was used for elution using a linear gradient from 100% Buffer A to 60% Buffer B (80% acetonitrile, 0.02% heptafluorobutyric acid). The programs SEQUEST and DTASELECT (Eng et al., 1994; Tabb et al., 2002) were used to identify peptides from the S. cerevisiae database, supplemented with a database of common contaminants. Spectra identified by SEQUEST as phosphopeptides were inspected manually to further assess the quality of the match.
Online supplemental material
Fig. S1 shows that Kcc4 and Hsl1 do not affect Fpk1-dependent phosphorylation of Ypk1. Fig. S2 shows that Gin4 does not affect Fpk1 localization or level and the Gin4 phosphorylation sites in Fpk1(1–472). Fig. S3 shows that PM flippase activity is required for susceptibility to Myr, but does not affect sphingolipid levels. Fig. S4 shows that the KA1 domain of Gin4 is required for its negative regulation of Fpk1 activity. Fig. S5 shows that modest elevation of Cdc42 rescues hof1Δ cells, whereas galactose-driven overexpression is toxic. Tables S1 and S2 list the yeast strains and plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410076/DC1.
Supplementary Material
Acknowledgments
We thank Todd Graham for the dnf1Δ dnf2Δ dnf3Δ strain, Douglas Kellogg for anti-Gin4 antibodies, David Pellman for plasmid PB3050, Lori Kohlstaedt for expert help with mass spectrometry, Thorner laboratory members Anna Roelants and Hannah Thorner for technical assistance, and Alexander Muir, Alexander Baltz, Gregory Finnigan, and Jesse Patterson for reagents, plasmids, and useful discussions.
This work was supported by National Institutes of Health (NIH) R01 Research grant GM21841 (to J. Thorner), by NIH Ruth L. Kirschstein National Research Service Award Postdoctoral Fellowship GM77886 (to A.E. Trott), and, in part, by NIH S10 Instrumentation grant RR025622 to the Vincent Coates Proteomics/Mass Spectrometry Facility at the University of California, Berkeley.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- 5-FOA
- 5-fluoro-orotic acid
- AMR
- actomyosin ring
- CIP
- calf intestinal phosphatase
- GAP
- GTPase-activating protein
- MIPC
- mannosyl-inositol-phosphorylceramide
- Myr
- myriocin
- PHS
- phytosphingosine
- PM
- plasma membrane
- PS
- primary septum
- PtdEth
- phosphatidylethanolamine
- PtdIns
- phosphatidylinositol
- PtdSer
- phosphatidylserine
References
- Aoki Y., Uenaka T., Aoki J., Umeda M., and Inoue K.. 1994. A novel peptide probe for studying the transbilayer movement of phosphatidylethanolamine. J. Biochem. 116:291–297. [DOI] [PubMed] [Google Scholar]
- Asano S., Park J.E., Yu L.R., Zhou M., Sakchaisri K., Park C.J., Kang Y.H., Thorner J., Veenstra T.D., and Lee K.S.. 2006. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J. Biol. Chem. 281:27090–27098 10.1074/jbc.M601483200 [DOI] [PubMed] [Google Scholar]
- Atkins B.D., Yoshida S., Saito K., Wu C.F., Lew D.J., and Pellman D.. 2013. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J. Cell Biol. 202:231–240 10.1083/jcb.201301090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y., Parra M., Bidlingmaier S., and Snyder M.. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176–187 10.1101/gad.13.2.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., and Park H.O.. 2012. Cell polarization and cytokinesis in budding yeast. Genetics. 191:347–387 10.1534/genetics.111.132886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., and Pringle J.R.. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301–1312 10.1083/jcb.142.5.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., LaCroute F., and Fink G.R.. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345–346 10.1007/BF00330984 [DOI] [PubMed] [Google Scholar]
- Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C.S., and Weissman J.S.. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463:1048–1053 10.1038/nature08787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery S.M., Kono K., Stokasimov E., and Pellman D.. 2012. Regulation of the formin Bnr1 by septins and a MARK/Par1-family septin-associated kinase. Mol. Biol. Cell. 23:4041–4053 10.1091/mbc.E12-05-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A., Torrance P.D., Kobayashi T., Thorner J., and Alessi D.R.. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9:186–197 10.1016/S0960-9822(99)80088-8 [DOI] [PubMed] [Google Scholar]
- Catty P., de Kerchove d’Exaerde A., and Goffeau A.. 1997. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 409:325–332 10.1016/S0014-5793(97)00446-8 [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Ingram M.F., Rosal P.H., and Graham T.R.. 1999. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147:1223–1236 10.1083/jcb.147.6.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Kuo C.C., Kang H., Howell A.S., Zyla T.R., Jin M., and Lew D.J.. 2012. Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p. Mol. Biol. Cell. 23:3814–3826 10.1091/mbc.E12-05-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J., and Zeghouf M.. 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93:269–309 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- Daleke D.L.2003. Regulation of transbilayer plasma membrane phospholipid asymmetry. J. Lipid Res. 44:233–242 10.1194/jlr.R200019-JLR200 [DOI] [PubMed] [Google Scholar]
- Daleke D.L.2007. Phospholipid flippases. J. Biol. Chem. 282:821–825 10.1074/jbc.R600035200 [DOI] [PubMed] [Google Scholar]
- Das A., Slaughter B.D., Unruh J.R., Bradford W.D., Alexander R., Rubinstein B., and Li R.. 2012. Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat. Cell Biol. 14:304–310 10.1038/ncb2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P.F.1991. Static and dynamic lipid asymmetry in cell membranes. Biochemistry. 30:1163–1173 10.1021/bi00219a001 [DOI] [PubMed] [Google Scholar]
- Devrekanli A., Foltman M., Roncero C., Sanchez-Diaz A., and Labib K.. 2012. Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J. Cell Sci. 125:5453–5466 10.1242/jcs.109157 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Gentry M.S., Hallberg R.L., and Barral Y.. 2003. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell. 4:345–357 10.1016/S1534-5807(03)00061-3 [DOI] [PubMed] [Google Scholar]
- Dong Y., Pruyne D., and Bretscher A.. 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161:1081–1092 10.1083/jcb.200212040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K., and Umeda M.. 2001. Membrane lipid control of cytokinesis. Cell Struct. Funct. 26:659–665 10.1247/csf.26.659 [DOI] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., and Yates J.R.. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S.2004. Cdc42—the centre of polarity. J. Cell Sci. 117:1291–1300 10.1242/jcs.01115 [DOI] [PubMed] [Google Scholar]
- Fadeel B., and Xue D.. 2009. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit. Rev. Biochem. Mol. Biol. 44:264–277 10.1080/10409230903193307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn G.D., Hermansson M., Somerharju P., and Grinstein S.. 2011. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat. Cell Biol. 13:1424–1430 10.1038/ncb2351 [DOI] [PubMed] [Google Scholar]
- Gall W.E., Geething N.C., Hua Z., Ingram M.F., Liu K., Chen S.I., and Graham T.R.. 2002. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 12:1623–1627 10.1016/S0960-9822(02)01148-X [DOI] [PubMed] [Google Scholar]
- Gordesky S.E., and Marinetti G.V.. 1973. The asymetric arrangement of phospholipids in the human erythrocyte membrane. Biochem. Biophys. Res. Commun. 50:1027–1031 10.1016/0006-291X(73)91509-X [DOI] [PubMed] [Google Scholar]
- Hachiro T., Yamamoto T., Nakano K., and Tanaka K.. 2013. Phospholipid flippases Lem3p-Dnf1p and Lem3p-Dnf2p are involved in the sorting of the tryptophan permease Tat2p in yeast. J. Biol. Chem. 288:3594–3608 10.1074/jbc.M112.416263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., and Morgan D.O.. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 325:1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Fatheddin P., and Graham T.R.. 2002. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell. 13:3162–3177 10.1091/mbc.E02-03-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., and Plowman G.D.. 1997. The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22:18–22 10.1016/S0968-0004(96)10068-2 [DOI] [PubMed] [Google Scholar]
- Iwamoto K., Kobayashi S., Fukuda R., Umeda M., Kobayashi T., and Ohta A.. 2004. Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells. 9:891–903 10.1111/j.1365-2443.2004.00782.x [DOI] [PubMed] [Google Scholar]
- Johnson D.I., and Pringle J.R.. 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111:143–152 10.1083/jcb.111.1.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P.J., Lee M.E., and Park H.O.. 2014. Bud3 activates Cdc42 to establish a proper growth site in budding yeast. J. Cell Biol. 206:19–28 10.1083/jcb.201402040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato U., Emoto K., Fredriksson C., Nakamura H., Ohta A., Kobayashi T., Murakami-Murofushi K., Kobayashi T., and Umeda M.. 2002. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 277:37855–37862 10.1074/jbc.M205564200 [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., and Koike T.. 2009. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4:1513–1521 10.1038/nprot.2009.154 [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Bi E., Epp J.A., Wang L., Ho J., and Chant J.. 2000. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 10:947–950 10.1016/S0960-9822(00)00626-6 [DOI] [PubMed] [Google Scholar]
- Lenoir G., Williamson P., and Holthuis J.C.. 2007. On the origin of lipid asymmetry: the flip side of ion transport. Curr. Opin. Chem. Biol. 11:654–661 10.1016/j.cbpa.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Li C.R., Au Yong J.Y., Wang Y.M., and Wang Y.. 2012. CDK regulates septin organization through cell-cycle-dependent phosphorylation of the Nim1-related kinase Gin4. J. Cell Sci. 125:2533–2543 10.1242/jcs.104497 [DOI] [PubMed] [Google Scholar]
- Lingwood D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327:46–50 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Liu K., Hua Z., Nepute J.A., and Graham T.R.. 2007. Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol. Biol. Cell. 18:487–500 10.1091/mbc.E06-07-0592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.C., Lee M.E., Narayan M., Chou C.S., and Park H.O.. 2013. Polarization of diploid daughter cells directed by spatial cues and GTP hydrolysis of Cdc42 budding yeast. PLoS ONE. 8:e56665 10.1371/journal.pone.0056665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., Fares H., and Pringle J.R.. 1998. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143:719–736 10.1083/jcb.143.3.719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke M.M., Della Seta F., Di Como C.J., Sugimoto H., Kobayashi R., and Arndt K.T.. 1996. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16:2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Matsuo Y., Gulis G., Hinz H., Patton-Vogt J., and Marcus S.. 2009. Phosphatidylethanolamine is required for normal cell morphology and cytokinesis in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell. 8:790–799 10.1128/EC.00029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray M.A., and Thorner J.. 2009. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 4:18 10.1186/1747-1028-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F., Palani S., Hub B., and Pereira G.. 2013. Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol. Biol. Cell. 24:1290–1304 10.1091/mbc.E12-08-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill A.H., Caligan T.B., Wang E., Peters K., and Ou J.. 2000. Analysis of sphingoid bases and sphingoid base 1-phosphates by high-performance liquid chromatography. Methods Enzymol. 312:3–9 10.1016/S0076-6879(00)12894-0 [DOI] [PubMed] [Google Scholar]
- Mok J., Kim P.M., Lam H.Y., Piccirillo S., Zhou X., Jeschke G.R., Sheridan D.L., Parker S.A., Desai V., Jwa M., et al. . 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3:ra12 10.1126/scisignal.2000482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momoi M., Tanoue D., Sun Y., Takematsu H., Suzuki Y., Suzuki M., Suzuki A., Fujita T., and Kozutsumi Y.. 2004. SLI1 (YGR212W) is a major gene conferring resistance to the sphingolipid biosynthesis inhibitor ISP-1, and encodes an ISP-1 N-acetyltransferase in yeast. Biochem. J. 381:321–328 10.1042/BJ20040108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K., Mendrola J.M., Schmitz K.R., Wang Y.H., Slochower D., Janmey P.A., and Lemmon M.A.. 2010. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 143:966–977 10.1016/j.cell.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen E.M., McDonald H., Yates J. III, and Kellogg D.R.. 2002. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell. 13:2091–2105 10.1091/mbc.01-10-0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Yamamoto T., Kishimoto T., Noji T., and Tanaka K.. 2008. Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol. Biol. Cell. 19:1783–1797 10.1091/mbc.E07-07-0646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P., Liu K., Patil D.V., Sciorra V.A., Jackson C.L., and Graham T.R.. 2009. Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat. Cell Biol. 11:1421–1426 10.1038/ncb1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R., Schreiter J.H., Onishi M., Vallen E.A., Hanna J., Moravcevic K., Lippincott M.F., Han H., Lemmon M.A., Pringle J.R., and Bi E.. 2009. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 185:995–1012 10.1083/jcb.200903125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji T., Yamamoto T., Saito K., Fujimura-Kamada K., Kondo S., and Tanaka K.. 2006. Mutational analysis of the Lem3p-Dnf1p putative phospholipid-translocating P-type ATPase reveals novel regulatory roles for Lem3p and a carboxyl-terminal region of Dnf1p independent of the phospholipid-translocating activity of Dnf1p in yeast. Biochem. Biophys. Res. Commun. 344:323–331 10.1016/j.bbrc.2006.03.095 [DOI] [PubMed] [Google Scholar]
- Oh Y., and Bi E.. 2011. Septin structure and function in yeast and beyond. Trends Cell Biol. 21:141–148 10.1016/j.tcb.2010.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Leda M., Hanna J., Savage N.S., Bi E., and Goryachev A.B.. 2013. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell. 26:148–161 10.1016/j.devcel.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzaki D., Tanaka S., Kanazawa H., and Nojima H.. 1997. Gin4 of S. cerevisiae is a bud neck protein that interacts with the Cdc28 complex. Genes Cells. 2:753–770 10.1046/j.1365-2443.1997.1590358.x [DOI] [PubMed] [Google Scholar]
- Onishi M., Ko N., Nishihama R., and Pringle J.R.. 2013. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J. Cell Biol. 202:311–329 10.1083/jcb.201302001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorski T., Lombardi R., Riezman H., Devaux P.F., van Meer G., and Holthuis J.C.. 2003. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell. 14:1240–1254 10.1091/mbc.E02-08-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Baltz A.G., Trott A.E., Fereres S., and Thorner J.. 2010. A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. USA. 107:34–39 10.1073/pnas.0912497106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Breslow D.K., Muir A., Weissman J.S., and Thorner J.. 2011. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 108:19222–19227 10.1073/pnas.1116948108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., and Tanaka K.. 2004. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:3418–3432 10.1091/mbc.E03-11-0829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Hanamatsu H., Kato U., Umeda M., Kozminski K.G., and Tanaka K.. 2007. Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev. Cell. 13:743–751 10.1016/j.devcel.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsh E.F., and Maniatis T.. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Lab Press, Cold Spring Harbor, NY: 1626 pp. [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D.H., and Cabib E.. 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115:293–302. [DOI] [PubMed] [Google Scholar]
- Sebastian T.T., Baldridge R.D., Xu P., and Graham T.R.. 2012. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta. 1821:1068–1077 10.1016/j.bbalip.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., and Tsien R.Y.. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., and Hicks J.B.. 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Lab Press, Cold Spring Harbor, NY: 186 pp. [Google Scholar]
- Shulewitz M.J., Inouye C.J., and Thorner J.. 1999. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Immanuel D., and Arndt K.T.. 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11:2133–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkotnicki L., Crutchley J.M., Zyla T.R., Bardes E.S., and Lew D.J.. 2008. The checkpoint kinase Hsl1p is activated by Elm1p-dependent phosphorylation. Mol. Biol. Cell. 19:4675–4686 10.1091/mbc.E08-06-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D.L., McDonald W.H., and Yates J.R. III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26 10.1021/pr015504q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Fujimura-Kamada K., and Yamamoto T.. 2011. Functions of phospholipid flippases. J. Biochem. 149:131–143 10.1093/jb/mvq140 [DOI] [PubMed] [Google Scholar]
- Tjandra H., Compton J., and Kellogg D.. 1998. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr. Biol. 8:991–1000 10.1016/S0960-9822(07)00419-8 [DOI] [PubMed] [Google Scholar]
- Tong Z., Gao X.D., Howell A.S., Bose I., Lew D.J., and Bi E.. 2007. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J. Cell Biol. 179:1375–1384 10.1083/jcb.200705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R.Y.1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509–544 10.1146/annurev.biochem.67.1.509 [DOI] [PubMed] [Google Scholar]
- Vallen E.A., Caviston J., and Bi E.. 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:593–611 10.1091/mbc.11.2.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Mark V.A., Elferink R.P., and Paulusma C.C.. 2013. P4 ATPases: flippases in health and disease. Int. J. Mol. Sci. 14:7897–7922 10.3390/ijms14047897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G.2011. Dynamic transbilayer lipid asymmetry. Cold Spring Harb. Perspect. Biol. 3:a004671 10.1101/cshperspect.a004671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane-Sando Y., Shimobayashi E., Shimobayashi M., Kozutsumi Y., Oka S., and Takematsu H.. 2014. Fpk1/2 kinases regulate cellular sphingoid long-chain base abundance and alter cellular resistance to LCB elevation or depletion. Microbiologyopen. 3:196–212 10.1002/mbo3.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Bartolini S., and Pellman D.. 2009. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 23:810–823 10.1101/gad.1785209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.2011. Lantibiotics as probes for phosphatidylethanolamine. Amino Acids. 41:1071–1079 10.1007/s00726-009-0386-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.