Abstract
Mitosis entails global alterations to chromosome structure and nuclear architecture, concomitant with transient silencing of transcription. How cells transmit transcriptional states through mitosis remains incompletely understood. While many nuclear factors dissociate from mitotic chromosomes, the observation that certain nuclear factors and chromatin features remain associated with individual loci during mitosis originated the hypothesis that such mitotically retained molecular signatures could provide transcriptional memory through mitosis. To understand the role of chromatin structure in mitotic memory, we performed the first genome-wide comparison of DNase I sensitivity of chromatin in mitosis and interphase, using a murine erythroblast model. Despite chromosome condensation during mitosis visible by microscopy, the landscape of chromatin accessibility at the macromolecular level is largely unaltered. However, mitotic chromatin accessibility is locally dynamic, with individual loci maintaining none, some, or all of their interphase accessibility. Mitotic reduction in accessibility occurs primarily within narrow, highly DNase hypersensitive sites that frequently coincide with transcription factor binding sites, whereas broader domains of moderate accessibility tend to be more stable. In mitosis, proximal promoters generally maintain their accessibility more strongly, whereas distal regulatory elements tend to lose accessibility. Large domains of DNA hypomethylation mark a subset of promoters that retain accessibility during mitosis and across many cell types in interphase. Erythroid transcription factor GATA1 exerts site-specific changes in interphase accessibility that are most pronounced at distal regulatory elements, but has little influence on mitotic accessibility. We conclude that features of open chromatin are remarkably stable through mitosis, but are modulated at the level of individual genes and regulatory elements.
Condensation of chromosomes during mitosis gives rise to microscopic structures that have been recognizable to biologists for centuries. Numerous studies have investigated the structural properties of mitotic chromosomes using imaging, biochemical, and biophysical approaches (Vagnarelli 2013), but details of their internal organization remain largely mysterious. A recent study showed that long-range interphase chromatin interactions ranging from hundreds of kilobases to megabases are obscured during mitosis (Naumova et al. 2013), but the influence of mitosis on chromatin structure at finer genomic resolutions remained unresolved.
At the level most directly relevant for gene regulation—individual genes and cis-regulatory modules (CRMs)—the structural configuration of the mitotic genome is of tremendous interest to the study of epigenetics. Mitosis, concomitant with altering chromosome structure, disassembles the metazoan nucleus (Güttinger et al. 2009), silences transcription (Prescott and Bender 1962), and evicts many components of the general transcription machinery (Gottesfeld and Forbes 1997; Akoulitchev and Reinberg 1998; Prasanth et al. 2003) and sequence-specific transcription factors from chromatin (Hershkovitz and Riggs 1995; Martínez-Balbás et al. 1995; Kadauke and Blobel 2013). How cells transmit gene regulatory signals through mitosis remains a major frontier in understanding cellular memory. Studies have proposed that the physical basis for mitotic memory includes molecular entities coupled to DNA, covalently or noncovalently, during mitosis, which may store gene regulatory information locally at individual loci to direct appropriate transcriptional control upon genome reactivation during G1 entry. Such proposed memory signatures, often referred to as mitotic “bookmarks,” can include (1) DNA methylation patterns, which are presumably unaltered through mitosis, (2) mitotically stable histone modifications and variants (Kelly et al. 2010; Varier et al. 2010; Wang and Higgins 2012), (3) a growing list of transcription regulators that are partially or fully retained on mitotic chromatin, usually at a minority of their interphase occupancy sites, including general factors (Dey et al. 2000; Christova and Oelgeschläger 2001; Blobel et al. 2009; Follmer et al. 2012) and sequence-specific transcription factors (Raff et al. 1994; Zaidi et al. 2003; Delcuve et al. 2008; Egli et al. 2008; Yang et al. 2008; Kadauke et al. 2012; Caravaca et al. 2013; Kadauke and Blobel 2013; Yang et al. 2013), and (4) structural properties of chromatin, such as nucleosome architecture and DNA topology (Kuo et al. 1982; Martínez-Balbás et al. 1995; Michelotti et al. 1997; Kadauke et al. 2012), that are maintained in or unique to mitosis.
The influence of each of these proposed types of mitotic bookmarks on gene regulation remains largely unclear. Chromatin structure deserves special attention, as it remains unknown to what extent mitotic chromatin condensation sterically hinders macromolecular access during mitosis and how that might contribute to mitotic eviction of factors and transcriptional silencing. A number of studies have used the DNase I sensitivity assay (Weintraub and Groudine 1976; Wu et al. 1979a,b) to probe the accessibility of mitotic chromatin; they found that DNase sensitivity is maintained in bulk on metaphase spreads (Gazit et al. 1982; Kerem et al. 1983) and at a few individual loci, such as at the Gapdh locus (Kuo et al. 1982) and Hsp70 promoter (Martínez-Balbás et al. 1995), by Southern blot detection. In the case of the Hsp70 promoter, DNase I sensitivity in the general region is preserved in mitosis despite loss of in vivo footprints of DNA-binding factors (Martínez-Balbás et al. 1995). More recently, we found that a number of regulatory regions in the murine erythroid genome preserve most of their accessibility in mitosis by DNase-qPCR measurements (Kadauke et al. 2012), and others have observed some level of mitotic DNase sensitivity at cohesin binding sites in mitosis (Yan et al. 2013). Biophysical measurements of transcription factor mobility on mitotic chromatin (Chen 2004; Caravaca et al. 2013) also support the permissiveness of mitotic chromatin to macromolecular access. Despite providing important insights, these previous studies were limited to examining subsets of genomic sites or behaviors of individual proteins. Thus, the field has lacked an unbiased genome-wide framework for understanding the influence of mitosis on chromatin accessibility.
In this study, we applied the DNase I sensitivity assay coupled to high-throughput sequencing (DNase-seq) to map mitotic chromatin accessibility genome-wide. We performed DNase-seq on cells in mitosis versus interphase using a rapidly dividing murine erythroblast cell line, G1E. G1E cells are null for the erythroid master regulator GATA1 and arrested in their maturation at the pro-erythroblast stage (Weiss et al. 1997). Restoration of GATA1 activity via stable expression of a GATA1-estrogen receptor fusion protein (hereafter referred to as “G1E + GATA1”) enables estradiol-inducible erythroid maturation (Weiss et al. 1997). Using this system, we studied the dynamics of chromatin accessibility of the interphase and mitotic genome across two erythroid maturation stages and dissected the potential interplay between chromatin accessibility and occupancy by GATA1, a transcription factor known to bind mitotic chromatin (Kadauke et al. 2012).
We found that, despite dramatic alterations to chromosome morphology at the microscopic level, preservation of DNase sensitivity during mitosis is widespread, although diverse site-specific patterns exist. An overall mild reduction in accessibility during mitosis is concentrated among the crop of narrow, highly hypersensitive sites (DNase-sensitive “peaks”), which often coincide with transcription factor binding sites. This contrasts with broader regions of sensitivity (DNase-sensitive “hotspots”) that are very stable through mitosis. Importantly, susceptibility to mitotic perturbation varies across classes of CRMs: Distal CRMs are more prone to losing accessibility during mitosis than promoters. A subset of promoters that is accessible across many cell or tissue types maintains accessibility during mitosis, and is marked by large domains of low DNA methylation. GATA1 exerts effects on chromatin accessibility mostly in interphase, triggering site-specific alterations that are most pronounced at distal CRMs.
Our data provide the first detailed accessibility map of the mitotic genome, revealing transcriptional regulatory signatures that remain widely visible within mitotic chromatin structure. The observation that structural changes of chromatin during mitosis are distinct for promoter-proximal and distal gene regulatory elements indicates mitosis-specific regulation at the level of individual loci. These findings have broad implications for transcriptional memory in dividing cells.
Results
DNase-seq analysis of pure mitotic erythroid cells at distinct maturation stages
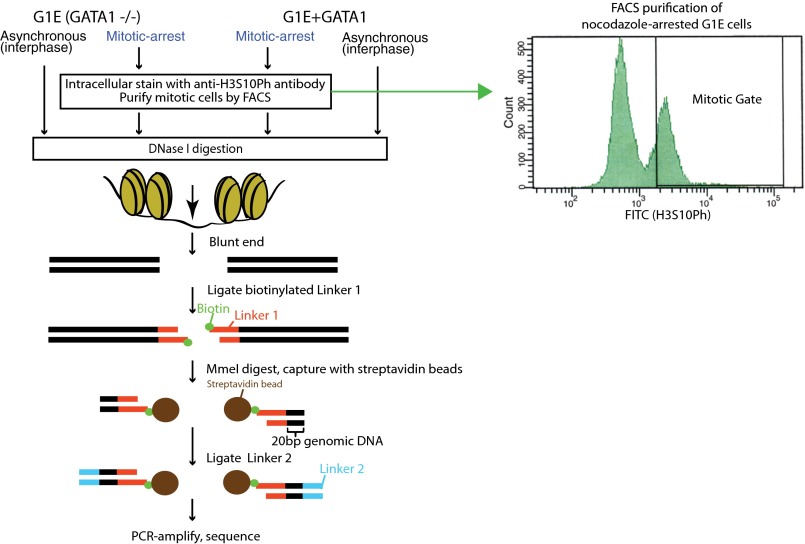
A number of recent studies have used DNase-seq to study asynchronous cells and tissues (Boyle et al. 2008a; Wu et al. 2011; Degner et al. 2012; Neph et al. 2012; Thurman et al. 2012). Applying DNase-seq to study the mitotic genome of suspension cells requires isolating mitotic populations at high purity, because contaminating interphase cells could contribute to apparent DNase sensitivity signals that do not actually reflect the configuration of the mitotic genome. Similar to many other cell types, enrichment of mitotic G1E cells by nocodazole arrest alone typically yields a relatively low mitotic purity of ∼55%. We thus applied a previously established protocol to purify mitotic cells from the nocodazole-treated population by intracellular staining of cells mildly fixed with 0.1% formaldehyde using an antibody against the mitosis-specific histone 3 Ser10-phospho (H3Ser10Ph) epitope (Fig. 1), followed by fluorescence-activated cell sorting (FACS) of the H3Ser10Ph-positive cells to obtain a mitotic population at >98% purity (Follmer and Francis 2012; Kadauke et al. 2012; Campbell et al. 2014). Chromatin was digested with DNase I, and the resulting fragments isolated, amplified, and sequenced using methods based on Song and Crawford (2010) (Fig. 1). Mild formaldehyde fixation of chromatin does not noticeably alter DNase sensitivity (Supplemental Fig. S1). DNase-seq libraries were generated in biological triplicates for asynchronous (containing ∼97% interphase cells, hereafter referred to as interphase) and purified mitotic cells from G1E and G1E + GATA1, yielding 219–266 million total mapped reads from the biological triplicates combined for each sample (Table 1). The biological triplicates show strong pairwise concordance (Pearson correlation coefficient of read densities ranging from 0.71 to 0.93; Supplemental Fig. S2). Thus, in the main text and figures we present results from analyses performed on the reads pooled from biological triplicates, with the experimental variance obtained from library-size normalized read densities of individual biological replicates indicated where appropriate for quantitative comparisons. We discuss additional considerations for normalization and quantitative interpretation in further detail in the Supplemental Methods.
Figure 1.
Experimental strategy for performing DNase-seq. G1E and G1E + GATA1 cells were grown asynchronously or arrested in mitosis by nocodazole treatment. Mitotic populations of nocodazole-treated cells were isolated by FACS using intracellular antibody staining of H3S10Ph. All samples were subjected to DNase digestion, followed by affinity capture of cleaved fragments, library preparation, and sequencing.
Table 1.
Summary of DNase-seq libraries and their overlapping previously known transcription factor binding sites and erythroid CRMs

We note that applying DNase-seq to mitotic cells requires special consideration of potential global differences from interphase cells that are unrelated to chromatin configuration, such as the lack of nuclear-cytoplasmic compartmentalization during mitosis, and which might have unknown effects on DNase sensitivity. Such intrinsic differences, if they exist, are challenging to control for. Thus, DNase-seq read density reveals the DNase sensitivity of a given site relative to other regions in the same experimental condition. Any potential global scaling differences across cellular states would not be expected to produce differing behaviors between genomic elements, and thus we focus the majority of our analyses on these types of site-specific changes.
Importantly, our algorithm defined 4.4% of the mappable mouse genome as DNase-sensitive regions (DNase “hotspots,” described in the next section and in Supplemental Methods). These regions overlap 66.5%–80.6% of previously identified binding sites of transcription factors GATA1 and TAL1 in the corresponding cell conditions. Moreover, these hotspots across all experimental conditions cover 94% of 286 experimentally validated erythroid CRMs manually curated from the literature (Table 1). Since DNase sensitivity is known to often coincide with transcription factor binding sites and CRMs, these results support the validity of our DNase-seq data sets.
Chromatin accessibility is widely preserved during mitosis, with diverse locally specified patterns
DNase sensitivity profiles exhibit distinct spatial patterns (Fig. 2A). Often, promoters and known transcription factor binding sites coincide with narrow hypersensitive regions, such as at the Klf13, +5 kb, and +42 kb distal CRMs, and at the Gata2 promoter, −8 kb, and +9 kb distal CRMs (Fig. 2B). In some cases, these hypersensitive sites are surrounded by relatively broad regions of moderately increased DNase cut density that mark domains of sensitivity in the range of kilobases, such as that coinciding with the entire gene body of Gata2 (Fig. 2B) and Myc (Supplemental Fig. S3). The sharp increases in DNase cut density likely reflect sites commonly referred to in the literature as “hypersensitive,” and often colocalize quite precisely with the binding sites of transcription factors (as shown for the Klf13 locus in Fig. 2B). To distinguish broad moderately sensitive domains from the narrow hypersensitive sites systematically, we defined them as “hotspots” and “peaks,” respectively, modified from previously published definitions (Baek et al. 2011). Hotspots are contiguous >250-bp regions significantly enriched in DNase cut density relative to the 200-kb surrounding background, as well as ranking among the top 100,000 most DNase-sensitive regions in the mappable genome (the fulfillment of these criteria is based on applying two independent algorithms for calling enrichments in DNase sensitivity, DNase2Hotspots and F-seq; see Supplemental Methods for details). The median width of hotspots is ∼650 bp, but the largest extends to ∼15 kb. Within hotspots, 150-bp regions that are further enriched in DNase cut density over the surrounding hotspot are defined as peaks, and a given hotspot may contain any number of peaks, or none. Our algorithm for hotspot and peak detection distinguishes the broad and narrow patterns, respectively, of DNase sensitivity that we aimed to capture (Fig. 2; Supplemental Fig. S3).
Figure 2.
Individual sites display diverse patterns of interphase-to-mitosis dynamics in chromatin accessibility. (A) Example of DNase hotspots and peaks. (B) G1E + GATA1 DNase cut density profiles at the Gata2, Slc8b1, and Klf13 loci are shown to illustrate their spatial patterns. Broad versus narrow sensitivity patterns are captured by the hotspots (brown bars) and peaks (orange bars), respectively, as defined in the main text and in Supplemental Methods. Note that individual sites can retain very little (green boxes), or virtually all (red boxes) of their accessibility in mitosis. A number of different patterns are also shown for the Klf13 locus, for which ChIP-seq tracks for GATA1 (interphase and mitosis), CTCF (Wu et al. 2011), and TAL1 are also shown to illustrate colocalization of their binding sites with DNase peaks.
Given the striking morphologic alterations of chromosomes associated with the known dissociation of many factors during mitosis, a widely held assumption is that mitosis limits macromolecular access to chromatin by increasing steric hindrance. On the contrary, we observed that at individual sites, DNase sensitivity can range from being virtually eliminated to partially or fully preserved during mitosis. For example, the hotspot covering the Gata2 gene body (Fig. 2B) is relatively stable through mitosis, as is the peak at the promoter region; in contrast, levels of DNase sensitivity at the −8 kb and +9 kb distal CRMs are partially or completely diminished during mitosis. To illustrate the variety of interphase-to-mitosis dynamics possible with respect to hotspots and peaks, other types of patterns are shown for the Slc8b1 (near complete loss of mitotic accessibility at promoter) and Klf13 (mitotic accessibility well-preserved at promoter, but largely eliminated at distal sites) loci in Figure 2B.
As illustrated by Figure 2B, interphase-to-mitosis transitions in DNase sensitivity are mostly gradual, rather than binary. Hence, while categorizing DNase cut density as either DNase “sensitive” or “insensitive” facilitates systematic analysis of genomic regions, doing so requires setting thresholds that can distort interpretations at sites where the DNase cut density is close to the threshold. For example, some hotspots can have similar levels of DNase cut density in interphase and mitosis, but happen to pass the threshold for our algorithm in only the mitotic sample. This can lead to an overestimate of “mitosis-only” hotspots (Supplemental Fig. S4), when in fact hotspots in this group mostly display similar levels of interphase and mitotic DNase sensitivity (Supplemental Fig. S5). We mitigated these thresholding effects by analyzing regions defined by the union of all hotspots or peaks present in any one of the four samples (G1E interphase or mitosis, G1E + GATA1 interphase or mitosis). Hereafter, “hotspots” and “peaks” refer to regions defined by their respective unions across samples. Using these final sets of 123,674 hotspots and 27,978 peaks enabled us to investigate quantitative changes in read densities within them across experimental conditions.
To examine the global effect of mitosis on the accessibility of the 4.4% of the mappable genome covered by hotspots, we calculated the fraction of total reads in the library mapped within all hotspots in interphase versus mitosis. By this measure, the aggregate accessibility of hotspots decreases from 11.3% in interphase to 8.3% in mitosis for G1E, and 14.0% in interphase to 12.3% in mitosis for G1E + GATA1 (Table 1). These results demonstrate that aggregate changes of accessible sites, relative to inaccessible background regions, are small during mitosis.
In the context of this mild change in aggregate accessibility among hotspots during mitosis, most individual hotspots and peaks show extensive preservation of accessibility during mitosis (Fig. 3A for G1E + GATA1; Supplemental Fig. S6 for G1E). This observation is consistent with absolute measurements of DNase sensitivity by qPCR at a number of individual sites performed in our previous study (Kadauke et al. 2012), reproduced in Supplemental Figure S7 for comparison, indicating that sequencing did not introduce large global scaling differences between mitotic and interphase measurements. Notably, the reduction in accessibility during mitosis is more pronounced among peaks than hotspots—a difference that is visible across a wide range of interphase accessibility (Fig. 3B for G1E + GATA1; Supplemental Fig. S6 for G1E). Using the mitosis-to-interphase ratio of read densities as a metric, hotspots preserve a median of 69% (G1E) and 93.3% (G1E + GATA1) of their interphase accessibility during mitosis; in contrast, peaks retain only a median of 46.9% (G1E) and 52.4% (G1E + GATA1) of their interphase accessibility in mitosis (Fig. 3C). Thus, the mechanisms that underlie the presence of hotspots and peaks are differentially susceptible to mitotic perturbation. This difference reveals that changes in mitotic chromatin accessibility preferentially occur in a spatially confined manner within DNase peaks. Such patterns are likely triggered by reduced affinity of trans-acting factors to specific sites that lead to local changes in nucleosome positioning, rather than by a large-scale increase in steric hindrance that would be expected to affect broad genomic regions relatively evenly.
Figure 3.
Chromatin accessibility is widely preserved during mitosis, with reductions occurring preferentially within narrow, hypersensitive DNase peaks. (A) After obtaining the union of the regions defined by hotspots/peaks across all samples, the mitotic versus interphase library size-normalized read densities were obtained from reads pooled from biological triplicates. Shown are mitotic versus interphase scatter plots (binned 2D density plots) for G1E + GATA1 cells. The color scale indicates the density of data points within each bin. The dashed diagonal line marks where mitotic and interphase read densities are equal. The overall trend is summarized by the moving mean (curve overlaid on plot) obtained from dividing the x-axis into bins consisting of ∼1000 hotspots or peaks. Gray dotted horizontal and vertical lines mark the estimated read density of inaccessible background regions, defined as all regions outside of hotspots. Data points corresponding to individual promoter peaks shown in Figure 2B are highlighted. The same graphs for G1E cells are shown in Supplemental Figure S6. (B) A zoomed-in view of the juxtaposition of moving means of hotspots and peaks in A. Error bars denote SEM of read densities from individual biological replicates (n = 3). (C) Box plot summaries of the mitosis-to-interphase ratio of the library size-normalized read densities for hotspots and peaks in G1E and G1E + GATA1, using reads pooled from biological triplicates. The horizontal dashed line marks where mitotic read density is equal to interphase read density.
Promoters preserve accessibility in mitosis more than distal CRMs
Cis-regulatory elements are important platforms upon which trans-acting factors assemble to regulate transcription, yet the degree to which these genomic elements remain accessible to transcription regulators during mitosis has remained largely unknown. Figure 2B demonstrates that for a given locus, such as Gata2 or Klf13, the promoter region can fully retain accessibility during mitosis, but the nearby distal hotspots and peaks can lose mitotic accessibility. We tested whether there are systematic differences in mitotic preservation of accessibility between proximal and distal CRMs. To identify regulatory regions, we intersected our DNase sensitivity map with a nine-state chromatin annotation for G1E and G1E + GATA1 derived from ChromHMM (Ernst and Kellis 2012), a genome segmentation program based on a multivariate hidden Markov model learned jointly from H3K4me1, H3K4me3, H3K36me3, H3K9me3, and H3K27me3 ChIP-seq data sets (Wu et al. 2011; Ernst and Kellis 2012). Since the profiles of histone lysine methylation modifications are overall similar in G1E and G1E + GATA1 (Wu et al. 2011), we defined promoter hotspots or peaks as those that either overlap an annotated transcriptional start site (TSS), and/or are covered predominantly by H3K4me3 (which also captures unannotated potential TSSs) in either G1E or G1E + GATA1 (Supplemental Fig. S8). We defined predicted distal CRM hotspots or peaks as those that do not overlap an annotated TSS and are mostly marked with H3K4me1 in either G1E or G1E + GATA1 (Supplemental Fig. S8). Based on these criteria, of the 123,674 hotspots across all four experimental conditions, 13% correspond to promoters and 26.2% are predicted distal CRMs. Moreover, our distal CRM detection algorithm correctly identified all 191 of the erythroid distal CRMs that overlapped hotspots and have been experimentally confirmed in the literature (Wu et al. 2011).
Strikingly, promoters as a group are the most accessible sites in the genome in both interphase and mitosis (Fig. 4A), revealing that the hierarchy of accessibility between classes of CRMs is well preserved in mitosis. Given that the degree of mitotic accessibility strongly correlates with interphase accessibility, we asked whether high mitotic accessibility of promoters is entirely explained by their high interphase accessibility, or if other unknown properties unique to promoters might also contribute to this pattern. Even when matched for interphase accessibility, the average mitotic accessibility of hotspots is still markedly higher at promoters than distal CRMs across nearly the full range of interphase accessibility (Fig. 4B). These trends apply to both G1E and G1E + GATA1 and show similar results when the same analyses are performed on peaks (Fig. 4C; Supplemental Fig. S9). These results point to promoter-specific mechanisms that enable them to preserve overall a larger fraction of their interphase accessibility during mitosis.
Figure 4.

Maintenance of mitotic accessibility at promoters exceeds that of distal CRMs. (A) Box plots of read densities of peaks in interphase and mitosis color-coded by their classification as either promoter, distal CRMs (see main text and Supplemental Methods for detailed definitions), or “other” regions that do not correspond to these defined categories. Gray dotted horizontal lines mark the global background estimate of read density in all regions outside of hotspots. (B, top) Scatter plots (binned 2D density, produced similarly as in Fig. 3A) of mitosis versus interphase read density at the union of peaks across all samples, grouped by promoter versus distal CRMs peaks. (Bottom row) A zoomed-in view of the juxtaposition of the moving means for promoter and distal CRM peaks, with error bars denoting SEM from biological replicates (n = 3). Size of circles conveys the number of promoter or distal CRM peaks within each bin. (C) Box plot summaries of the mitosis-to-interphase ratio of read densities for hotspots and peaks. Horizontal dashed line marks location on y-axis where interphase read density is equal to mitosis read density.
Promoters that are accessible across many murine tissues are exceptionally accessible in mitosis and marked by large conserved domains of DNA hypomethylation
While promoters overall preserve chromatin accessibility during mitosis quite well, a wide range of mitotic accessibility exists (Fig. 4B). To explore potential predictors of promoter hotspots with exceptionally well-preserved mitotic accessibility, we examined the tissue distribution of the accessibility of our erythroid hotspots by intersecting them with the available interphase DNase hypersensitive sites (DHS) from across 45 murine tissue or cell types from the Mouse ENCODE Consortium (Vierstra et al. 2014). We found that preservation of accessibility across multiple cell or tissue types is strongly indicative of high mitosis-to-interphase accessibility ratio (Fig. 5A, top). This correlation is mostly restricted to promoters, because distal CRMs show a much more tissue-specific distribution of accessibility (Fig. 5A, bottom).
Figure 5.
Promoters that are accessible across many murine tissues are marked by high mitotic accessibility and large DNA hypomethylation domains. (A) Promoter and distal CRM hotspots in G1E + GATA1 are shown in scatterplots (binned 2D density plot) of mitosis-to-interphase accessibility ratio versus the number of murine tissues in which the promoter hotspot overlaps at least one DNase hypersensitive site (DHS). Dashed horizontal line marks where mitotic and interphase DNase sensitivities are equal. (B) The fraction of promoter hotspots at genes encoding for sequence-specific transcription factor genes is specifically enriched by applying each of the two criteria (mitosis-to-interphase accessibility ratio and tissue preservation of hotspot) individually and together. (C) Myc is an example of a locus demarcated by a DNA methylation canyon. Shown are interphase and mitotic DNase accessibility profiles from G1E + GATA1, DNA methylation ratios in mouse HSCs, and DNase accessibility profiles from across Mouse ENCODE cell or tissue types, with dark bands representing DNase-sensitive regions. (D) G1E + GATA1 promoter hotspots are shown in scatterplots (binned 2D density plot) of mitosis-to-interphase accessibility ratio versus the number of murine cell or tissue (out of 45) DHS that is present. The panels are divided into promoter hotspots that overlap UMRs of the indicated size ranges. (E) Mitosis versus interphase DNase read densities of G1E + GATA1 promoter hotspots are shown as moving means color-coded by overlap with UMRs of the indicated size ranges, with error bars denoting SEM from biological replicates (n = 3). Error bars for the UMRs ≥1 kb and <3.5 kb (blue line) are omitted to avoid obscuring the difference between the red and green curves.
Gene ontology (GO) analysis showed that, compared to all promoter hotspots as background, the 5059 promoter hotspots with >85% mitosis-to-interphase ratio are mildly enriched for a mix of molecular function categories consisting of cell surface proteins and transcription regulatory proteins (Fig. S10). Figure 5B quantifies these enrichments for two GO terms that encompass these two distinct functional gene categories (“sequence-specific DNA-binding transcription factor activity” and “signaling receptor activity”). In contrast, the 1698 promoter hotspots that are preserved in ≥10 cell or tissue types are enriched specifically for molecular functions involving transcriptional regulation by 2.2-fold over all promoter hotspots, without any enrichment for the surface receptor GO terms (Fig. 5B; Supplemental Fig. S10). Of note, 15.7% of the 945 promoter hotspots that meet the dual criteria of >85% mitosis-to-interphase ratio and preserved across ≥10 cell or tissue types belong to sequence-specific transcription factor genes, representing a 2.7-fold enrichment over all promoter hotspots that is higher than applying either criteria alone (Fig. 5B).
The tissue-invariant patterns of accessibility and GO enrichment of this subset of promoters are reminiscent of the properties of another recently discovered chromatin feature. By examining genome-wide DNA methylation profiles encompassing a large range of cell types and species, several studies (Long et al. 2013; Xie et al. 2013; Jeong et al. 2014) described the existence of very large hypomethylation regions spanning multiple kilobases. These large domains of DNA hypomethylation—referred to as “broad nonmethylated islands” (Long et al. 2013), DNA methylation “valleys” (Xie et al. 2013), or “canyons” (Jeong et al. 2014)—are distinct from smaller hypomethylated regions in that the large domains are maintained across many tissues (Xie et al. 2013; Jeong et al. 2014) and can be evolutionarily conserved at individual loci (Long et al. 2013). These large hypomethylation domains tend to demarcate genes involved in transcriptional regulation (such as genes encoding members of the HOX, FOX, ZIC, GATA, and KLF protein families) and developmental signaling pathways (Long et al. 2013; Xie et al. 2013; Jeong et al. 2014). Many of these genes, especially the transcription regulator genes, are also the ones we found in our DNase analysis to be among those whose promoters are accessible across many tissues and in mitosis. Thus, large DNA hypomethylation domains and a subset of our promoter hotspots share characteristics—identified independently—of relatively ubiquitous tissue distribution and a propensity to demarcate transcription regulator genes.
Given these shared characteristics, we examined the degree of genomic overlap between large DNA hypomethylation domains previously obtained from mouse hematopoietic stem cells (HSCs) (Jeong et al. 2014) and the DNase hotspots from this study. Of the 13,579 undermethylated regions (UMRs) ≥1 kb detected in HSCs, 90.8% overlap with our DNase hotspots in G1E or G1E+GATA1. Among the UMRs, 1104 were previously defined as methylation canyons based on a size threshold of ≥3.5 kb; 97.6% of the canyons overlap a promoter hotspot in our DNase data sets. Given that ∼70%–90% of the HSC methylation canyons are shared by a large number of diverse cell types previously examined (Jeong et al. 2014), the two chromatin features can be compared across different hematopoietic cell types. Where DNase hotspots and methylation canyons overlap, they very often approximate each other’s borders, usually spanning the promoter proximal regions or an entire gene, such as at the Myc (Fig. 5C), Foxa1, Uncx, and Gata2 loci (Supplemental Fig. S11). Importantly, among the 189 promoter hotspots preserved across ≥15 cell or tissue types, 98.9% overlap the larger UMRs (≥1 kb) and 50.8% overlap the largest UMRs (≥3.5 kb, or canyons) (Fig. 5D). Moreover, promoter hotspots demarcated by methylation canyons are overall significantly higher in mitotic accessibility than other promoter hotspots matched for their levels of interphase accessibility in both G1E + GATA1 (Fig. 5E) and G1E (Supplemental Fig. S12).
Together, these findings implicate a role for large DNA hypomethylation domains in contributing to exceptional promoter accessibility in mitosis and across many tissues. Of note, genes overlapping DNase hotspots and DNA methylation canyons can exhibit any level of expression (Supplemental Fig. S13), including some that are silent in HSCs and G1E cell types, such as the hepatocyte transcription factor Foxa1 (Supplemental Fig. S11). These results indicate that maintenance of genes in hypomethylated and mitotically accessible chromatin can be uncoupled from active RNA synthesis, consistent with previous analyses of methylation canyons and gene expression in HSCs (Jeong et al. 2014).
GATA1-driven erythroid maturation exerts site-specific alterations to interphase chromatin accessibility that are most pronounced at distal CRMs, but little effect on mitotic accessibility
Chromatin accessibility is closely related to trans-acting factor binding, but the exact nature of this relationship is often unknown for individual factors. Genetic complementation of GATA1 in the G1E cell differentiation model enabled us to test for direct effects of GATA1 occupancy on chromatin accessibility in interphase and mitosis, as well as indirect influences resulting from cell maturation. Recent mRNA measurements normalized to spike-in controls revealed that restoration of GATA1 function represses >5000 genes and induces only about 200 genes (A Stonestrom, S Hsu, K Jahn, P Huang, S Kadauke, A Campbell, R Hardison, and G Blobel, in prep.). A previous study showed that even promoters of genes that alter expression drastically show minimal change in accessibility (Wu et al. 2011). Figure 6A illustrates several of such repressed (Kit, 33.5-fold repressed) and induced genes (Slc4a1, 849-fold induced), where the changes in promoter interphase DNase sensitivity are very mild between the presence and absence of GATA1, compared to the large differential expression in mRNA measured by RNA-seq (T Mishra, C Morrissey, C Keller, B Giardine, E Heuston, S Anderson, V Paralkar, M Pimkin, M Weiss, D Bodine, et al., in prep.; GEO accession number GSE40522). At select distal CRMs, significant changes in interphase DNase sensitivity can be observed in the direction consistent with expression changes (such as Kit -114 kb and to a lesser extent Slc4a1 + 9.9 kb in Fig. 6A).
Figure 6.

Dynamics of interphase and mitotic chromatin accessibility during GATA1-driven erythroid maturation. (A) Browser track views of DNase accessibility and GATA1 ChIP-seq profiles in interphase, with the fold change in mRNA levels from G1E to G1E + GATA1 indicated at the top. (B) Interphase accessibility dynamics of DNase peaks in G1E versus G1E + GATA1 are presented as scatterplots (binned 2D density plots), grouped by promoter versus distal CRMs, and by overlap with GATA1 binding sites. Graphing conventions are the same as in Figure 3, with error bars denoting SEM of biological replicates (n = 3) for moving means. Promoters and distal regulatory sites associated with GATA1-repressed (Kit) and GATA1-induced Slc4a1 loci shown in A are highlighted. (C) Scatterplots (binned 2D density plots) of G1E mitotic accessibility versus G1E + GATA1 mitotic accessibility of DNase peaks are shown, grouped by overlap with GATA1 binding sites in interphase only (I-GATA1), interphase and mitosis (IM-GATA1), or mitosis only (M-GATA1). Graph conventions are similar to B.
We extended the results from Wu et al. (2011) on promoter interphase accessibility by examining promoters and distal CRMs in detail in the new data sets presented here. Consistent with Wu et al. (2011), we found that overall the levels of interphase accessibility of promoter peaks exhibit little change between the G1E and G1E + GATA1 states; furthermore, this trend is largely unchanged regardless of whether the promoter coincides with one of GATA1’s 10,460 binding sites (Fig. 6B, left two panels), suggesting that GATA1 occupancy does not contribute significantly to variations in promoter accessibility. Of the top 100 most up-regulated and top 100 most down-regulated genes from G1E to G1E + GATA1, only some are associated with mild site-specific increases and decreases, respectively, in interphase promoter accessibility (Supplemental Fig. S14), including Kit and Slc4a1 (Fig. 6A; highlighted in Fig. 6B). There is no correlation between preservation of promoter accessibility during mitosis and the extent of differential expression from G1E to G1E + GATA1 (Supplemental Fig. S15).
In contrast, the dynamics of distal CRMs’ interphase accessibility upon restoration of GATA1 function are more site-specific and pronounced. Individual distal CRMs can increase, decrease, or maintain the same DNase cut densities (Fig. 6B, right two panels; Slc4a1 + 9.9 kb and Kit-144 from Fig. 6A are highlighted). Importantly, at distal CRMs bound by GATA1, there is a general shift toward reduced interphase accessibility, compared to those not bound by GATA1 (Fig. 6B, right two panels). This finding suggests that GATA1 and its cofactors function as repressors at the majority of distal CRMs, including the Kit -114-kb regulatory region as previously described (Jing et al. 2008). However, site-specific behaviors likely depend on the activating or repressive cofactor complexes present at individual loci. The observation that GATA1-driven maturation reduces the interphase accessibility of most distal CRMs is consistent with the observation that genes repressed by GATA1 function vastly outnumber the activated genes (A Stonestrom, S Hsu, K Jahn, P Huang, S Kadauke, A Campbell, R Hardison, and G Blobel, in prep.). We assigned distal CRMs to their nearest genes and found that changes in the interphase accessibility of distal CRMs are not associated with alterations in expression of the nearest gene (Supplemental Fig. S14); however, the true association of distal CRM accessibility with expression of the correct target genes is likely stronger, as this method of pairing distal CRMs with genes discounts the fact that many distal CRMs regulate genes far away.
In the context of mitotic chromatin, GATA1-induced erythroid maturation results in a slight global increase in accessibility at promoters (Fig. 6C, left), but not at distal CRMs (Fig. 6C, right). We next examined whether mitotic GATA1 chromatin occupancy influences chromatin accessibility. GATA1 binding sites can be divided into 8831 interphase-only (I-GATA1), 527 interphase-and-mitosis (IM-GATA1), and 1102 mitosis-only (M-GATA1) occupancy sites (previously categorized based on the presence or absence of ChIP-seq peak calls [Kadauke et al. 2012], which are generally accurate for GATA1’s sharp, well-defined ChIP peaks). The majority of these subcategories of GATA1 binding sites overlap DNase hotspots (Supplemental Fig. S16). Examples of these patterns of GATA1 mitotic binding are shown in Supplemental Figure S3. These subcategories of GATA1 binding sites showed no significant differences in their distributions of G1E and G1E + GATA1 mitotic chromatin accessibility patterns, suggesting that promoter mitotic accessibility differences between the two maturation states are not a direct result of differential GATA1 mitotic binding (Fig. 6C). Moreover, in G1E + GATA1 cells, sites bound by GATA1 during mitosis show similar distributions of mitotic accessibility maintenance as sites bound by GATA1 only in interphase (Supplemental Fig. S17), suggesting that GATA1 binding does not contribute significantly to site-specific variations in mitotic chromatin accessibility.
Together, these results indicate that GATA1 clearly influences chromatin accessibility in interphase, especially at distal CRMs, likely in part via the action of cofactor complexes. In contrast, GATA1 mitotic occupancy does not contribute significantly to variations in preservation of accessibility in mitosis. These findings implicate yet unknown GATA1-independent mechanisms that regulate mitotic chromatin dynamics.
Discussion
This study provides a detailed DNase accessibility map of the mitotic genome, lending insights into structural principles and their relationship to gene regulation. Our finding that the genome retains significant DNase sensitivity during mitosis establishes a genome-wide framework for previous reports of DNase sensitivity during mitosis for select loci and mitotic occupancy of individual factors. These results are consistent with other studies using microscopic volume measurements of an artificial genomic array (Li et al. 1998) and whole chromosomes (Martin and Cardoso 2010; Vagnarelli 2012), as well as FRET-based assays of histone–histone interactions (Llères et al. 2009), that demonstrate only two to threefold condensation of chromosomes during mitosis compared to interphase. Thus, we conclude that condensation of chromosomes during mitosis, relative to interphase, is not as extreme as commonly assumed, and is unlikely to be sufficient for displacing many chromatin regulators from mitotic chromatin.
We uncovered several novel trends that distinguish sites favoring open versus closed chromatin configurations in mitosis. First, reduction in accessibility during mitosis occurs preferentially among DNase peaks (Fig. 3A,B). This observation narrows the likely mechanisms underlying mitotic changes in chromatin accessibility; specifically, large-scale, indiscriminate steric occlusion is unlikely to produce such site-specific and spatially confined changes in accessibility. Rather, site specificity is more likely explained by the binding of sequence-specific transcription factors and their cofactors. We speculate that transcription factor binding could generate a narrow DNase peak by evicting the nucleosomes in the vicinity of the binding site, while also recruiting factors capable of spreading along and remodeling chromatin to generate the broader accessibility pattern of a DNase hotspot. Loss of trans-acting factor affinity for chromatin during mitosis, perhaps due to mitosis-specific phosphorylation (Rizkallah et al. 2011), could explain the preferential loss of DNase sensitivity at peaks. In contrast, patterns of generalized accessibility across hotspots, such as at the Gata2 (Fig. 2B) and Myc (Fig. 5C) loci, are likely attributable to mitotically stable chromatin features, with DNA methylation patterns being a potential candidate responsible at a subset of genes (Supplemental Fig. S11).
While the model described above is likely generally applicable, our findings for GATA1 show that the influence of transcription factor binding on chromatin accessibility must be tested on a case-by-case basis, and can be related to whether the factor is involved in activation or repression of a given locus. Thus, while it may appear counterintuitive that GATA1 binding is associated with a pronounced decrease in interphase accessibility at most distal CRMs (Fig. 6B), this observation is consistent with its predominantly repressive role on the majority of its target genes (A Stonestrom, S Hsu, K Jahn, P Huang, S Kadauke, A Campbell, R Hardison, and G Blobel, in prep.). This result is also consistent with the ability of GATA1 to bind regions with relatively high nucleosome occupancy (Hu et al. 2011). Despite GATA1’s ability to bind mitotic chromatin at a subset of its interphase occupancy sites, GATA1 mitotic occupancy does not measurably alter mitotic chromatin accessibility (Fig. 6C). This result might be accounted for by the lack of nucleosome remodeling activity intrinsic to GATA1 itself, and the absence of all tested GATA1 cofactors from GATA1 mitotic occupancy sites (Kadauke et al. 2012). Thus, transcription factor occupancy is not necessarily positively correlated with interphase or mitotic DNase sensitivity, and the precise relationship between the two can be specific to the cofactor milieu.
Providing additional support for local modulation of mitotic chromatin accessibility, we found that promoters tend to preserve mitotic accessibility to a greater extent than distal CRMs, though a wide variation exists within each category (Fig. 4). The mechanisms underlying these general patterns are unknown. It remains unclear how other chromatin features known to be associated with promoter regions in mitosis, including trans-acting factor binding (Kadauke and Blobel 2013), certain histone modifications (Wang and Higgins 2012), global shifts in the positioning of histone variants (Kelly et al. 2010), and increased single-strandedness of DNA (Michelotti et al. 1997), could potentially contribute to general maintenance of promoter accessibility in mitosis. For the group of promoter hotspots that are accessible across many tissues, large domains of tissue-invariant DNA hypomethylation likely contribute to maintaining an open chromatin configuration at these sites during mitosis; however, definitive support for hypotheses regarding the causal role of a given mark will require experimental evidence beyond correlations. While we are not aware of studies directly examining DNA methylation patterns specifically in mitosis, given its well-established role in epigenetic memory on the time scales of organismal development (Bartolomei and Ferguson-Smith 2011), it is reasonable to assume that DNA methylation is unaltered during mitosis and thus might contribute to one aspect of mitotic memory. A conundrum from our analyses of the promoter DNase hotspots associated with large DNA hypomethylation domains is that some genes in these regions are silent. We speculate that maintenance of open chromatin configuration stably through cell divisions might ensure that even these silent genes, including those important for developmental regulation, are permissive in receiving regulatory signals for transcriptional activation or other chromatin transactions.
From the perspective of cell-fate maintenance and reprogramming, perhaps the most intriguing finding is that mitosis preferentially disrupts the accessibility of most distal CRMs (Fig. 4). Given that enhancers play important roles in driving tissue-specific gene expression, what is the consequence of preferential reduction in their mitotic accessibility? Presumably, this loss of accessibility could reflect dissociation of enhancer-binding proteins, and by extension, dissolution of long-range enhancer–gene interactions. Does the preferential loss of distal CRMs’ accessibility during mitosis lead to a transient absence of normal enhancer regulation immediately post-mitosis? If so, this might present a window of transient instability in tissue-specific gene regulation. We envision that the insights provided in this study will lead to testable hypotheses that address these and other fundamental questions about how gene expression contends with mitotic division.
Methods
DNase I digestion of asynchronous and purified mitotic cells
G1E and the G1E-ER4 sublines were cultured as described in Weiss et al. (1997). Experiments were performed in biological triplicates as follows. G1E-ER4 cells were treated with 100 nM estradiol for 22 h prior to harvest (referred to as “G1E + GATA1”). Both G1E and G1E-ER4 cells were treated with nocodazole (200 ng/mL) for 7 h prior to harvest. At harvest, nocodazole-treated and asynchronous cells were cross-linked with 0.1% formaldehyde at room temperature for 10 min, then quenched with 1 M glycine. Fixed cells were washed with PBS and resuspended in 1× Cell Lysis Buffer (60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 10 mM Tris at pH 7.4, 300 mM sucrose, 0.1 mM EGTA, 0.5 mM PMSF, 0.1% NP-40, and 2 μL/mL protease inhibitor cocktail [Sigma]). For mitotic samples, cells were then stained with anti-H3S10Phos antibody (Millipore 04-817) and Dy488 F(ab′)2 anti-rabbit IgG secondary antibody (Jackson 711-485-152), and sorted by FACS for H3S10Phos-positive cells as shown in Figure 1.
DNase I digestion was performed based on protocol from Cockerill (1999) with details outlined as follows: 2–10 million asynchronous cells (in Cell Lysis Buffer) or 2 million mitotic cells (collected from the FACS machine in PBS) were resuspended in 50 μL Nuclei Lysis Buffer (300 mM sodium acetate, 5 mM EDTA at pH 7.4, 0.5% SDS), added to 5 μL of 100 mM CaCl2, equilibrated at room temperature for 10 min. A range of units of DNase I were added (see DNase-seq library preparation below for the range of units selected for sequencing) and the digestion reaction proceeded for 10 min at room temperature, then terminated by adding 350 μL of 0.1 mg/mL proteinase K in Nuclei Lysis Buffer. Samples were gently mixed by inversion, incubated at 55°C for 5 min, then overnight at 65°C for reversal of formaldehyde crosslinks. Additional proteinase K was added to a final concentration of 0.1 mg/mL and incubated at 55°C for 1 h. DNA fragments were isolated by phenol-chloroform extraction and ethanol precipitation.
DNase-seq library generation
DNase-seq library construction was performed as described in Song and Crawford (2010) with the following modifications. Standard 0.8% agarose gels were run for 2 h at 80V with 5 μL of each sample and stained with ethidium bromide to check the extent of chromatin digestion. A range of three different DNase I concentrations were chosen for each condition that best matched digestion patterns between conditions. For mitotic samples, this was 2, 4, and 8 units of DNase. For asynchronous samples, this was between 4 and 40 units, adjusted proportionally to the number of cells in the sample. A total of 70 μL of each sample (for each DNase I concentration) was subjected to blunt-end reaction containing 20 μL NEB Buffer 2, 7 μL 10 mM dNTP, 6 μL T4 DNA polymerase (NEB M0203), 2 μL BSA (100×), 4 μL 50 mM MgCl2, and 95 μL dH2O, incubated at room temperature for 3.25 h. Two-hundred microliters of TE buffer was added and samples placed at 65°C for 15 min to deactivate enzyme. Reactions were cleaned up by phenol-chloroform extraction and DNA was resuspended in 30 μL 10 mM Tris-Cl. The samples corresponding to the three DNase I concentrations chosen for each condition were measured by Nanodrop and pooled at equimolar concentrations into a single tube for overnight ligations to the first DNase-seq linker, as per Song and Crawford (2010). The one modification from Song and Crawford (2010) is that we added a 5′ phosphate to linker 1 to increase ligation efficiency. Replicate 1 was sequenced in one lane using Illumina HiSeq 2000. Replicates 2 and 3 were sequenced using Illumina Genome Analyzer IIx.
Bioinformatic analysis
Reads from DNase-seq libraries were trimmed to the first 20 bp that corresponded to genomic DNA and then mapped to mouse mm9 genome using Bowtie (Langmead et al. 2009), allowing mapping to at most four locations but reporting only the single best alignment. Mapped reads pooled from three biological replicates were used to call hotspots and peaks using DNase2Hotspots, as described in Baek et al. (2011). We additionally required that hotspots must also overlap the top 100,000 read-enriched regions called by F-seq (Boyle et al. 2008b). A final set of hotspots and peaks was defined as the union across each experimental condition. From these hotspots and peaks, smoothed signals from F-seq (proportional to library size-normalized read densities) were obtained for each of the biological replicates for quantitative comparisons. As described in the main text, hotspots and peaks were intersected with histone lysine methylation states obtained from segmenting the genome using ChromHMM (Ernst and Kellis 2012); a master list of DHSs from 45 cell or tissue types from the Mouse ENCODE Consortium (Vierstra et al. 2014); and previously identified GATA1 (Kadauke et al. 2012) and TAL1 (Wu et al. 2011) binding sites defined by MACS (Zhang et al. 2008). GO analyses were performed using Gene Regions Enrichment of Annotations Tool (GREAT 2.0.2) (McLean et al. (2010)). Additional details and rationale for our bioinformatic methods can be found in the Supplemental Methods.
Data access
Raw and processed data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE61885. DNase-seq signal tracks can also be viewed on the PSU Genome Browser at http://main.genome-browser.bx.psu.edu/cgi-bin/hgTrackUi?hgsid=192185_VwI1eSEbwN9drvE7zCtOYiqES0IB&c=chr7&g=chopDnase2. Processed data in the form of a table of DNase hotspots and peaks, containing read densities and intersection with other data sets used in this study, are also available as a Supplemental File (HotspotPeakTables.xls).
Supplementary Material
Acknowledgments
We especially thank Gautham P. Nair for extensive advice on data analysis and comments on the manuscript. We also thank Jennifer E. Phillips-Cremins, Kenneth Zaret, Caleb Ng, and Caroline Bartman for comments on the manuscript, and Margaret Goodell, the Raj, Hardison, Weiss, Tong, and Blobel laboratories for discussions. This work was supported by NIH T32GM008216 to C.C.H., the Intramural Research Program of NCI at the National Institutes of Health (M.H.S. and S.B.), NIH R01 DK065806 and NIH RC2 HG005573 to R.C.H., and NIH RO1 DK054937 to G.A.B.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.180646.114.
References
- Akoulitchev S, Reinberg D. 1998. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev 12: 3541–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S, Sung M-H, Hager GL. 2011. Quantitative analysis of genome-wide chromatin remodeling, pp. 433–441. Humana Press, New Jersey. [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. 2009. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell 36: 970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. 2008a. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS. 2008b. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24: 2537–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Hsiung CC-S, Blobel GA. 2014. Comparative analysis of mitosis-specific antibodies for bulk purification of mitotic populations by fluorescence-activated cell sorting. BioTechniques 56: 90-1–93-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. 2013. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev 27: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. 2004. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol 168: 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Oelgeschläger T. 2001. Association of human TFIID–promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol 4: 79–82. [DOI] [PubMed] [Google Scholar]
- Cockerill PN. 1999. Identification of DNaseI hypersensitive sites within nuclei, pp. 29–46. Humana Press, New Jersey. [Google Scholar]
- Degner JF, Pai AA, Pique-Regi R, Veyrieras J-B, Gaffney DJ, Pickrell JK, De Leon S, Michelini K, Lewellen N, Crawford GE, et al. 2012. DNaseI sensitivity QTLs are a major determinant of human expression variation. Nature 482: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcuve GP, He S, Davie JR. 2008. Mitotic partitioning of transcription factors. J Cell Biochem 105: 1–8. [DOI] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol Cell Biol 20: 6537–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Birkhoff G, Eggan K. 2008. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol 9: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follmer NE, Francis NJ. 2012. Preparation of Drosophila tissue culture cells from different stages of the cell cycle for chromatin immunoprecipitation using centrifugal counterflow elutriation and fluorescence-activated cell sorting, 1st ed., Vol. 513. Elsevier, NY. [Google Scholar]
- Follmer NE, Wani AH, Francis NJ. 2012. A polycomb group protein is retained at specific sites on chromatin in mitosis. PLoS Genet 8: e1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit B, Cedar H, Lerer I, Voss R. 1982. Active genes are sensitive to deoxyribonuclease I during metaphase. Science 217: 648–650. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem Sci 22: 197–202. [DOI] [PubMed] [Google Scholar]
- Güttinger S, Laurell E, Kutay U. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 10: 178–191. [DOI] [PubMed] [Google Scholar]
- Hershkovitz M, Riggs AD. 1995. Metaphase chromosome analysis by ligation-mediated PCR: heritable chromatin structure and a comparison of active and inactive X chromosomes. Proc Natl Acad Sci 92: 2379–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K, et al. 2011. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 21: 1650–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al. 2014. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet 46: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. 2008. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA. 2013. Mitotic bookmarking by transcription factors. Epigenetics Chromatin doi: 10.1186/1756-8935-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. 2012. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA. 2010. H2A.Z Maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell 39: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem BS, Goitein R, Richler C, Marcus M, Cedar H. 1983. In situ nick-translation distinguishes between active and inactive X chromosomes. Nature 304: 88–90. [DOI] [PubMed] [Google Scholar]
- Kuo MT, Iyer B, Schwarz RJ. 1982. Condensation of chromatin into chromosomes preserves an open configuration but alters the DNase I hypersensitive cleavage sites of the transcribed gene. Nucleic Acids Res 10: 4565–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Sudlow G, Belmont AS. 1998. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J Cell Biol 140: 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llères D, James J, Swift S, Norman DG, Lamond AI. 2009. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J Cell Biol 187: 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grützner F, Odom DT, Patient R, Ponting CP, et al. 2013. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. eLife 2: e00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RM, Cardoso MC. 2010. Chromatin condensation modulates access and binding of nuclear proteins. FASEB 24: 1066–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83: 29–38. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D. 1997. Marking of active genes on mitotic chromosomes. Nature 388: 895–899. [DOI] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. 2013. Organization of the mitotic chromosome. Science 342: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. 2012. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. 2003. Sequential entry of components of gene expression machinery into daughter nuclei. Mol Biol Cell 14: 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. 1962. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res 26: 260–268. [DOI] [PubMed] [Google Scholar]
- Raff JW, Kellum R, Alberts B. 1994. The Drosophila GAGA transcription factor is associated with specific regions of heterochromatin throughout the cell cycle. EMBO J 13: 5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizkallah R, Alexander KE, Hurt MM. 2011. Global mitotic phosphorylation of C2H2 zinc finger protein linker peptides. Cell Cycle 10: 3327–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE. 2010DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc 2: doi: 10.1101/pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P. 2012. Mitotic chromosome condensation in vertebrates. Exp Cell Res 318: 1435–1441. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P. 2013. Chromatin reorganization through mitosis. Adv Protein Chem Struct Biol 90: 179–224. [DOI] [PubMed] [Google Scholar]
- Varier RA, Outchkourov NS, de Graaf P, van Schaik FMA, Ensing HJL, Wang F, Higgins JMG, Kops GJPL, Timmers HM. 2010. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J 29: 3967–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Higgins JMG. 2012. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol 23: 175–184. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193: 848–856. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol 17: 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. 1979a. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell 16: 797–806. [DOI] [PubMed] [Google Scholar]
- Wu C, Wong YC, Elgin S, 1979b. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell 16: 807–814. [DOI] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen K-B, Drautz D, et al. 2011. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res 21: 1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. 2013. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154: 801–813. [DOI] [PubMed] [Google Scholar]
- Yang Z, He N, Zhou Q. 2008. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol 28: 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Sung E, Donlin-Asp PG, Corces VG. 2013. A subset of Drosophila Myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat Commun 4: 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SM, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. 2003. Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc Natl Acad Sci 100: 14852–14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.