Abstract
We introduce a method for simultaneous prediction of microRNA–target interactions and their mediated competitive endogenous RNA (ceRNA) interactions. Using high-throughput validation assays in breast cancer cell lines, we show that our integrative approach significantly improves on microRNA–target prediction accuracy as assessed by both mRNA and protein level measurements. Our biochemical assays support nearly 500 microRNA–target interactions with evidence for regulation in breast cancer tumors. Moreover, these assays constitute the most extensive validation platform for computationally inferred networks of microRNA–target interactions in breast cancer tumors, providing a useful benchmark to ascertain future improvements.
MicroRNAs (miRNAs) regulate RNA stability and mRNA translation (Filipowicz et al. 2008) and their dysregulation has been implicated in a wide range of human diseases including cancer (Garzon et al. 2009). Consequently, establishing accurate and comprehensive repertoires of miRNA–target interactions is a necessary step toward elucidating their mechanistic role in pathophysiology. Dissecting miRNA regulation, however, has proven challenging because candidate miRNA binding sites are ubiquitous and their regulatory effects are context specific (Liu et al. 2005; Lu et al. 2005; Mukherji et al. 2011). As a result, and despite their relatively low accuracy, computational prediction methods that incorporate context-specific data are preferred for screening for miRNA–target interactions in tumor contexts (Carroll et al. 2013; Erhard et al. 2014).
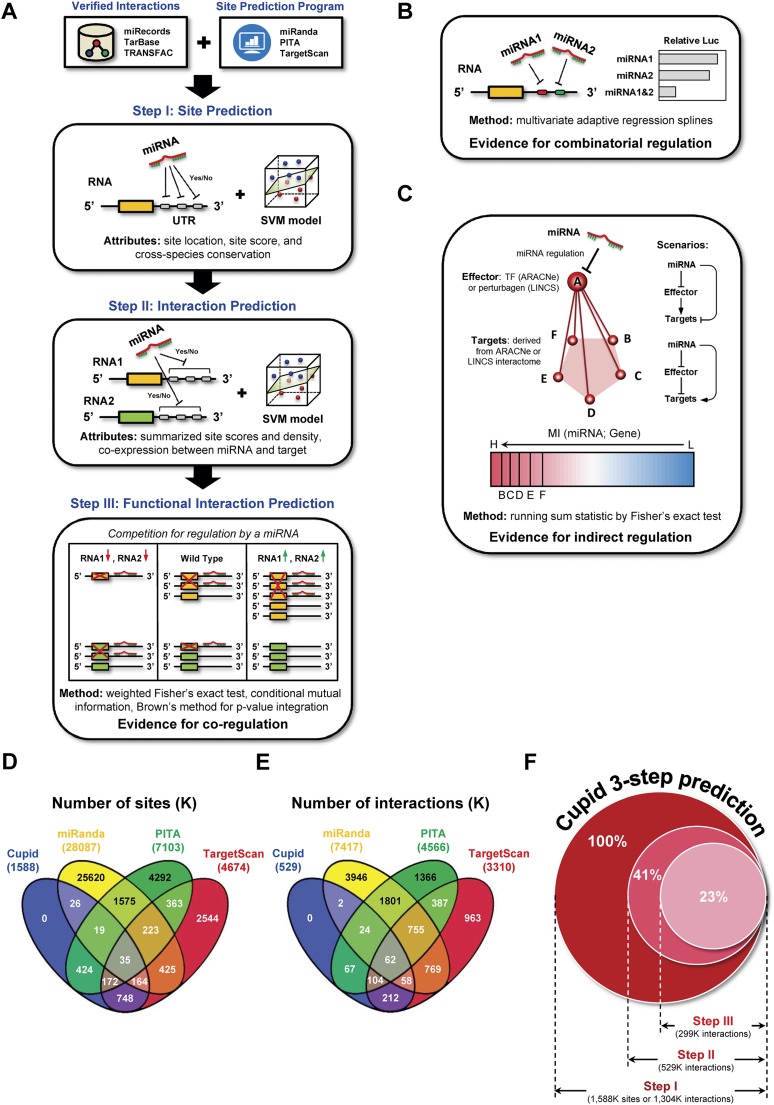
To address these challenges, we introduce Cupid, an integrative framework for the context-specific inference of miRNA targets. Cupid integrates sequence-based evidence and functional clues derived from RNA and miRNA expression analysis, predicting candidate miRNA binding sites and associated target genes using ensemble machine learning classifiers that are trained on validated interactions. Candidate interactions emerging from this step are then refined based on independent, context-specific clues, including their predicted ability to mediate competitive endogenous RNA (ceRNA) interactions, where mRNA compete for shared miRNA regulators (Fig. 1A; Tay et al. 2014). Thus, Cupid simultaneously infers both interaction types (ceRNA and miRNA–target interactions). In addition, we considered evidence for combinatorial regulation by multiple miRNA species (Fig. 1B; Boissonneault et al. 2009; Xu et al. 2011) and for indirect miRNA regulation through effector proteins (Fig. 1C). Taken individually, these clues are predictive of bona fide miRNA–target interactions and can significantly improve the tradeoff between precision and recall.
Figure 1.
Methodology. (A) Cupid first reevaluates sites predicted by TargetScan, miRanda, and PITA, selecting and rescoring each candidate site (Step I). Sites are used to select and score miRNA-target interactions (Step II), which are then examined for evidence for mediating ceRNA interactions (Step III). In addition, to support interaction prediction we considered (B) evidence for combinatorial regulation between miRNAs and (C) evidence for indirect regulation by miRNAs through effectors. (D) The majority of site predictions by TargetScan, miRanda, and PITA are exclusive to a single algorithm; for example, <10% of sites predicted by miRanda are also predicted by another method. (E) Cupid predicted 529K miRNA-target interactions in Step II, excluding 60% of Step I candidate interactions. As a result, it makes considerably fewer predictions than TargetScan, miRanda, and PITA. (F) Cupid Step III predictions include less than a quarter of Step I candidate interactions.
We show that Cupid predictions outperform other leading algorithms, based on multiple experimental assays, including PAR-CLIP data, miRNA perturbation followed by mRNA and protein expression profiles, and 3′ luciferase activity assays. Critically, while Cupid predicts fewer interactions than other methods (Fig. 1D–F), its predictions are much more likely to be consistent with experimental evidence. This is critical since high false-positive prediction rates are a key limitation of current miRNA–target prediction methods.
Results
Algorithm outline and miRNA–target prediction in breast cancer tumors
Cupid is implemented in three sequential steps (Fig. 1A). First, Cupid reevaluates candidate miRNA binding sites in 3′ UTRs, as inferred by TargetScan (Lewis et al. 2005), miRanda (John et al. 2004), and PITA (Kertesz et al. 2007). This is accomplished by integrating their algorithm-specific scores, their location in the 3′ UTR, and their cross-species conservation. Then, miRNA–target interactions are predicted by further integrating information about selected sites, their multiplicity, and the statistical dependency between the expression profiles of miRNA and putative targets.
Likelihoods for each predictive feature are computed based on a positive gold standard set of 588 experimentally confirmed miRNA–target interactions, representing 1481 binding sites in TarBase (Papadopoulos et al. 2009), TRANSFAC (Matys et al. 2006), and miRecords (Xiao et al. 2009). They are then integrated using a support vector machine (SVM) algorithm (Chang and Lin 2011). Finally, Cupid assesses whether inferred targets compete for their predicted miRNA regulators. In the following sections, we discuss results from Cupid-inferred miRNA targets based on gene expression profile data of TCGA breast cancer samples (The Cancer Genome Atlas Network 2012).
Step I (miRNA binding-site analysis)
miRNA binding sites in 3′ UTRs were predicted and scored independently by TargetScan, miRanda, and PITA. Taken together, these algorithms predicted a total of 37M candidate binding sites (Fig. 1D). Each site was associated with the following features: (1) TargetScan, miRanda, and PITA confidence scores, when available; (2) phastCons (Siepel et al. 2005) species-conservation scores; and (3) relative distance from the 3′ and 5′ ends of the target 3′ UTR. These features were used to train a support vector machine (SVM) classifier (Chang and Lin 2011) using 10-fold cross-validation and bootstrap aggregating (bagging), to identify consensus candidate binding sites. In total, out of 37M candidate sites from sequence analysis, Cupid selected ∼1.6M as likely miRNA binding sites (Fig. 1D); predictions and 3′ UTR sequences are provided in Supplemental Table S1; see Supplemental Methods for details.
Step II (miRNA–target interaction refinement)
Candidate binding sites were then used to assess the probability of an interaction between a miRNA and a target 3′ UTR. Predicted interactions were associated with the following features (see Supplemental Methods): (1) their scores from Step I; (2) the total number of miRNA binding sites in a target 3′ UTR (i.e., multiplicity); (3) binding-site density; and (4) the inverse correlation between the expression of the target 3′ UTR gene and that of the miRNA, as measured by signed normalized mutual information (NMI), which was estimated using adaptive partitioning (Darbellay and Vajda 1999) and Spearman correlation. These individual features were used to train an SVM classifier, leading to selection of 529K candidate miRNA-target interactions from 1.6M original binding sites in Step I (Fig. 1E; see Supplemental Methods for details).
Step III (functional evidence analysis)
Finally, candidate interactions from Step II were tested for sequence- and expression-based evidence for 3′ UTR competition for shared miRNA regulators by identifying miRNAs that are also predicted to “mediate” ceRNA interactions. Thus, ceRNA and miRNA–target interactions are simultaneously predicted based on interaction candidates from Step II. We note that when evaluated individually, other lines of functional evidence (Fig. 1B,C) significantly improved prediction accuracy, but their contribution was relatively small; see Supplemental Methods for description and analyses. In total, combined analysis of TCGA breast cancer gene and miRNA expression profiles supported 299K miRNA–target interactions from Cupid Step II (Fig. 1F).
Quality of binding-site selection
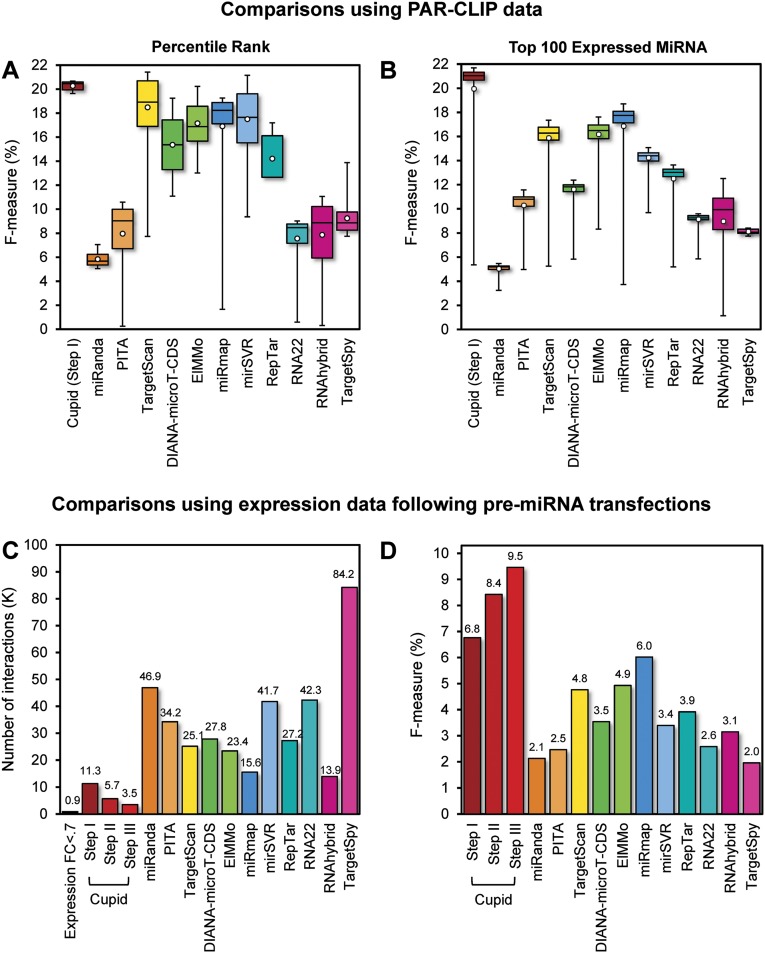
We first compared Cupid Step I performance to that of several published algorithms, including TargetScan (Lewis et al. 2005), miRanda (John et al. 2004), PITA (Kertesz et al. 2007), DIANA-microT-CDS (Reczko et al. 2012), ElMMo (Gaidatzis et al. 2007), miRmap (Vejnar and Zdobnov 2012), mirSVR (Betel et al. 2010), RepTar (Elefant et al. 2011), rna22 (Miranda et al. 2006), RNAhybrid (Rehmsmeier et al. 2004), and TargetSpy (Sturm et al. 2010) by analyzing the overlap of their predictions with experimentally assessed AGO crosslink-centered regions (CCR) in HEK293 cells (Hafner et al. 2010). We tested both the accuracy of binding-site scores (Fig. 2A) and the effects of miRNA expression (Fig. 2B) on AGO localization; note that weakly expressed miRNAs are less likely to be associated with CCRs, and consistently, the 100 most highly expressed miRNAs account for almost all CCRs.
Figure 2.
Site and interaction prediction. (A) miRNA-binding-site predictive accuracy estimates for a panel of target prediction methods are given as cumulative distributions across ranks of predicted miRNA-binding-site scores, starting from the top scoring 1% to all predicted sites. Average, upper, and lower quintiles, and the range of the harmonic mean of precision and recall (F-measure) are shown; precision is the fraction of sites that overlap crosslink-centered regions (CCRs) identified in AGO PAR-CLIP experiments relative to the total number of predicted sites, and recall is the number of CCRs overlapping predicted sites relative to the total number of CCRs. (B) Cumulative distributions across the highest expressed miRNAs, from highest expressed to top 100 highest expressed in HEK293 cells, of predicted binding sites that coincide with CCRs. (C) Number of predicted miRNA–target interactions that were tested through pre-miRNA transfections in MCF7 or MDA-MB-231 for a panel of target-prediction methods, and (D) their predictive ability over target mRNA down-regulation.
Specifically, we computed cumulative F-measure distribution statistics based on the overlap of experimentally assessed CCRs with the 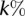 highest confidence targets, as predicted by each algorithm for the most expressed miRNAs in HEK293. The F-metric is defined as the harmonic mean of precision and recall, i.e.,
highest confidence targets, as predicted by each algorithm for the most expressed miRNAs in HEK293. The F-metric is defined as the harmonic mean of precision and recall, i.e., 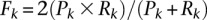 , where
, where 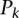 and
and 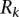 are the method’s precision and recall for the
are the method’s precision and recall for the 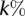 most significant predicted binding sites (Fig. 2A) or for the
most significant predicted binding sites (Fig. 2A) or for the 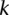 most expressed miRNAs (Fig. 2B). That is,
most expressed miRNAs (Fig. 2B). That is, 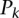 is the frequency with which predicted binding sites overlap CCRs, and
is the frequency with which predicted binding sites overlap CCRs, and 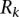 is the frequency with which CCRs overlap predicted binding sites.
is the frequency with which CCRs overlap predicted binding sites.
Results suggest that Cupid consistently finds a good tradeoff between precision and recall for any value of k (Fig. 2A). Similarly, once a sufficiently large number of highly expressed miRNAs are included in the test, Cupid outperforms other site-prediction methods (Fig. 2B). Cupid’s predictive ability peaked at 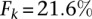 , for the
, for the 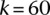 most expressed miRNAs, followed by miRmap and ElMMo with
most expressed miRNAs, followed by miRmap and ElMMo with 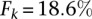 and
and 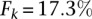 , for
, for 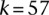 and
and 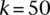 , respectively. Overall, our results suggest that Cupid-predicted binding sites are in better agreement with AGO binding.
, respectively. Overall, our results suggest that Cupid-predicted binding sites are in better agreement with AGO binding.
Quality of interaction prediction in breast cancer cell lines
To assess the algorithm’s ability to predict functional miRNA targets, we used data from three breast cancer-specific studies that provide gene-expression profiles (in duplicates) following transfection of pre-mir-18a, pre-mir-193b, pre-mir-206, pre-mir-302c (Leivonen et al. 2009), pre-mir-101-1 (Frankel et al. 2011), and scrambled controls in MCF7 and pre-mir-145, and control in MDA-MB-231 (Gotte et al. 2010). In total, across the targets predicted for the six miRNAs, we identified 869 down-regulated genes (>30% down-regulation), supporting their role as targets of transfected miRNAs (Fig. 2C; Supplemental Table S3; Guo et al. 2010).
Statistics were obtained separately following each of Cupid’s steps. Specifically, we identified 11.3K (Step I), 5.7K (Step II), and 3.5K (Step III) candidate targets of transfected miRNAs, respectively. We calculated the F-measure for each algorithm under the assumption that false-positive predictions would not be down-regulated following miRNA transfection and that false-negative predictions would be down-regulated but not predicted. Our results suggest that Cupid’s interaction prediction (Step II) and functional-interaction prediction (Step III) significantly improve performance, compared to using only binding-site predictions (Step I) (P < 0.01, FET). Critically, Cupid was significantly more accurate (P < 0.05) than the next best algorithm (miRmap) (Fig. 2D). Thus, Cupid inferred fewer candidate miRNA targets, but with significantly higher F-statistics, suggesting a substantial increase in precision.
Protein expression benchmarks
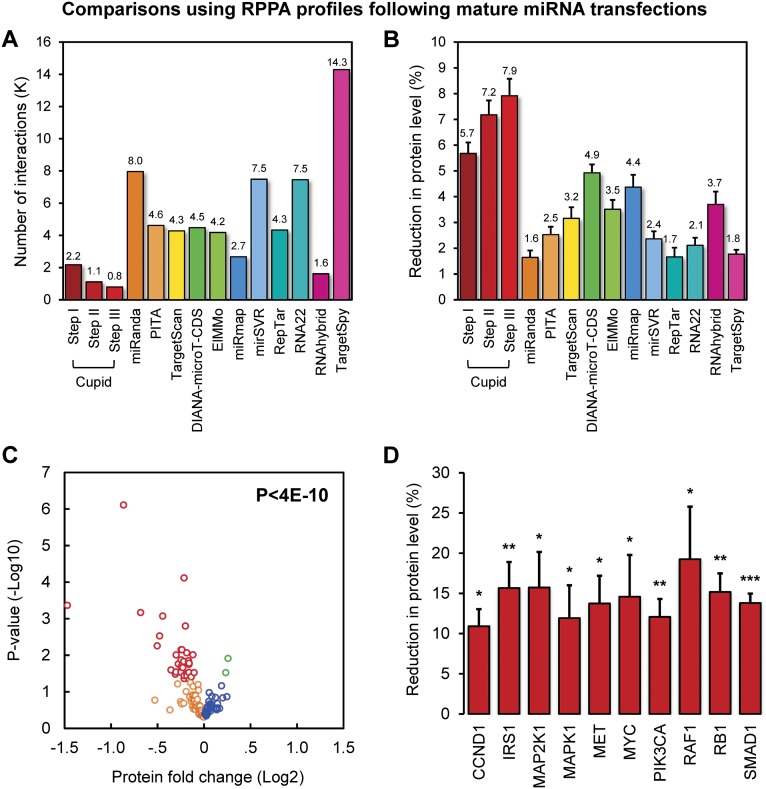
We used RPPA data to measure expression fold reduction of 120 proteins following transfections of 159 miRNA mimics, including four mock controls, in MDA-MB-231 cells. We compared the average fold-expression decrease of protein targets predicted by the various algorithms, relative to mock transfections (Fig. 3B). Of the 158 antibodies included in the RPPA, 117 antibodies (representing 82 predicted miRNA targets) profiled target expression of at least one of 127 transfected Cupid Step III-predicted miRNA regulators, with no replicates; see Supplemental Table S4. In total, this analysis tested nearly 2200, 1000, and 800 Cupid-inferred interactions from Steps I, II, and III, respectively (Fig. 3A). Cupid Step III predictions provided by far the best overlap with the experimental data, improved over both Steps I and II, and significantly outperformed DIANA-microT-CDS, the next best prediction method (P < 1 × 10−4, by Student’s t-test). We thus focused our detailed analysis using Cupid Step III predictions.
Figure 3.
High-throughput perturbation tests using protein-expression profiling. (A) Number of predicted miRNA–target interactions that were tested through miRNA mimic transfection followed by protein-expression profiling. (B) Average reduction in protein level following transfection of predicted miRNA regulators for a panel of target prediction methods. (C) P-values and average protein-expression fold changes after transfection of Cupid-predicted miRNA regulators. In total, considering expression estimates made with 117 antibodies, 34 reported significant down-regulation P < 0.05, in red), 51 reported down-regulation (orange), 30 reported up-regulation (blue), and two reported significant up-regulation (P < 0.05, in green); a comprehensive significance of P < 4 × 10−10. (D) Estimated average reduction in protein expression levels for known breast cancer regulators from C. Data are represented as mean ± SEM.
To test prediction accuracy, we analyzed each antibody independently and evaluated protein expression fold changes after transfection of their predicted miRNA regulators (Fig. 3C). We plotted average protein expression fold change and associated P-value (from Student’s t-test) following transfections of their predicted miRNA regulators and mock controls. Of the 117 antibodies tested, 85 were down-regulated (34 statistically significantly, P < 0.05) and 32 were up-regulated (two significantly) following transfection of their predicted miRNA regulators. This confirms preferential down-regulation of predicted miRNA targets, P < 4 × 10−10 by one-sample Kolmogorov-Smirnov (K-S) test. Moreover, significantly up- and down-regulated antibodies indicate false- and true-positive rates, respectively, suggesting that Cupid predictions have FDR < 0.056; a comparison between Cupid and the next-best method shows a threefold FDR gap (Supplemental Fig. S2). At the individual interaction level, significant reductions in target protein expression were observed following transfection of miRNA regulators for 237 predicted interactions, while increases were observed for only 76 interactions (P < 6 × 10−84 by K-S test). Ten proteins with an established role in breast cancer tumorigenesis, including CCND1, IRS1, MAP2K1, MAPK1, MET, MYC, PIK3CA, RAF1, RB1, and SMAD1, were consistently reduced by >10% on average by their predicted miRNA regulators (Fig. 3D).
Evidence for competition for miRNA regulation
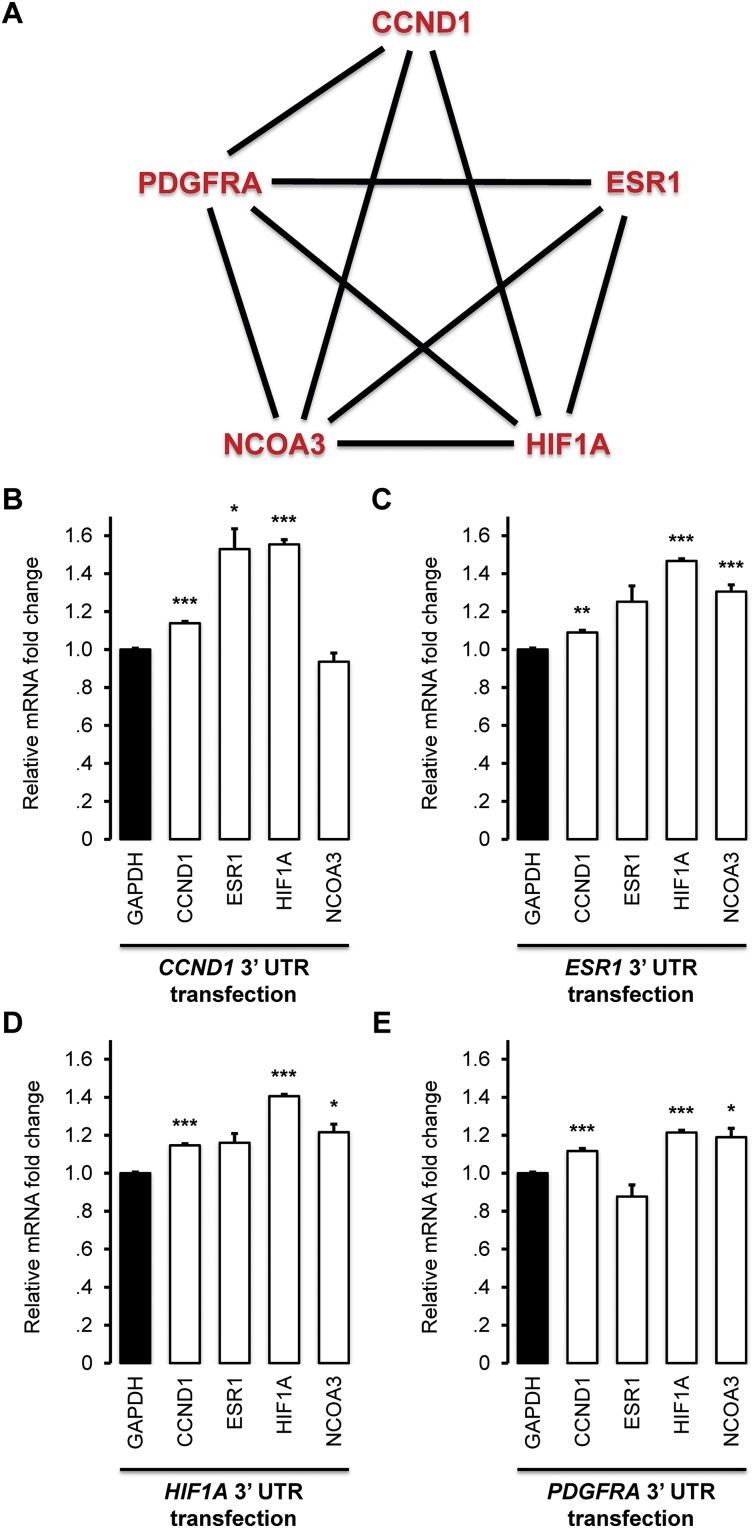
To test predicted miRNA targets using independent evidence for mediating ceRNA interactions, we chose to focus on a fully connected predicted ceRNA network of five established regulators of tumorigenesis in breast cancer (CCND1, ESR1, HIF1A, PDGFRA, and NCOA3) (Fig. 4A; see Supplemental Table S5 for the full data). Specifically, we transfected their 3′ UTRs (except for NCOA3 which could not be cloned) and measured mRNA-expression fold changes of those expressed in MCF7 cells (PDGFRA was not expressed); note that transfection concentrations are not at physiologic levels, but they establish the potential for coregulation by miRNAs. The results confirm the regulatory potential of these 3′ UTR (Fig. 4B–E). In total, eight of 11 of the predicted interactions (Fig. 4A) showed significant up-regulation after transfection of the 3′ UTR of their predicted ceRNA regulators. While this evidence supports ceRNA regulation (Sumazin et al. 2011), it does not identify the miRNAs that mediate the coupling.
Figure 4.
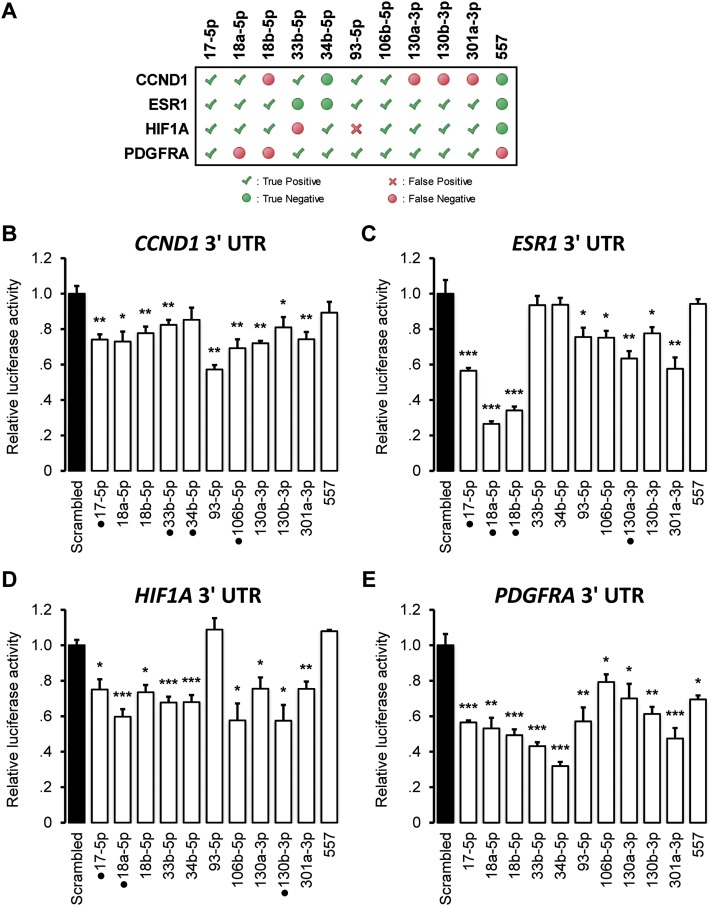
Competition for miRNA regulation. Cupid relies on evidence for competition for miRNA regulation, simultaneously identifying ceRNA and miRNA–target interactions. (A) A ceRNA network of oncogenes implicated in breast cancer regulation. Transfections of the 3′ UTRs of (B) CCND1, (C) ESR1, (D) HIF1A, and (E) PDGFRA in MCF7 up-regulated mRNA expression within the network, as measured by qPCR. Data are represented as mean ± SEM; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
We predicted that these genes compete for several miRNAs, including seven miRNAs predicted to target at least three of the four genes. We used 3′ UTR luciferase reporter assays and miRNA mimic transfections to test whether these miRNAs regulate their predicted 3′ UTR targets. In total, we predicted 30 miRNA-3′ UTR interactions for 10 miRNAs that were predicted to mediate ceRNA interactions in the four-gene subnetwork (Fig. 5). Of particular biological interest, ESR1, HIF1A, and PDGFRA were predicted to compete for miR-17-5p, miR-106b-5p, miR-130a/b-3p, and miR-301a-3p. Our assays tested 44 interactions, including interactions with miR-557 as negative controls. Of the 30 predicted interactions, only regulation of the HIF1A 3′ UTR by miR-93-5p was not supported by the experimental data, suggesting high precision for Cupid’s predictions. The remaining assays tested 14 miRNA-3′ UTR pairs, including four miR-557 targets that were not predicted to interact, and the results suggest that eight of the 14 have regulatory potential.
Figure 5.
Regulatory potential of miRNA mediators. Multiple ceRNA interactions in a network including CCND1, ESR1, HIF1A, and PDGFRA (Fig. 4) were predicted to be mediated by common miRNAs. We tested the regulatory potential of 10 of these miRNAs biochemically, with miR-557 selected as a negative control. (A) Predicted miRNA–target interactions and a summary of biochemical validation, depicting true-positive, true-negative, false-positive, and false-negative predictions; down-regulation of 3′ UTR luciferase activity in response to miRNA-mimic transfection at P < 0.05 was taken as evidence for regulation. Luciferase activity after miRNA mimic transfection relative to transfection of scrambled control is shown for (B) CCND1, (C) ESR1, (D) HIF1A, and (E) PDGFRA 3′ UTRs. Punctuated mimics, for example, miR-17-5p for CCND1, ESR1, HIF1A, but not PDGFRA, correspond to previously validated interactions. Data are represented as mean ± SEM; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Comparing Cupid performance to random selection (same size) from TargetScan, PITA, and miRanda interaction predictions, Cupid had 80% accuracy and an F-measure of 0.87; random-selection had accuracy of 22% and F-measure 0.15. In total, our assays suggest that Cupid predictions have high precision and good, but lower, recall; precision was above 95%, while recall was above 75%. Of the 30 predictions, 10 were previously known (Dweep et al. 2011; Hsu et al. 2011), see Supplemental Table S6, but even after excluding these, Cupid calls were predictive of assay results at P < 0.01 by FET, when comparing true positives and true negatives to false positives and false negatives. Interestingly, one previously reported interaction, CCND1 regulation by miR-34b, was neither predicted nor supported by our luciferase assays.
Other evidence for functional regulation by miRNAs
We tested candidate miRNA–target interactions for evidence for combinatorial regulation by miRNA species, and evidence for indirect regulation through effectors. Evidence for combinatorial regulation is complementary to evidence derived from expression correlation between miRNAs and their targets. Similarly, evidence for indirect regulation by a miRNA examines the correlation between the expression of this miRNA and a set of predicted indirect targets; these were not used to predict direct miRNA–target interactions and are considered complementary evidence. In total, these lines of evidence produced fewer predictions and had weaker predictive ability when compared with evidence for ceRNA regulation. Consequently, we chose to describe them independently. When combined, these lines of evidence support 40,000 predicted interactions that were not included in Step III (an additional 13%); see Supplemental Methods.
ESR1 protein expression is correlated with miRNA regulator expression
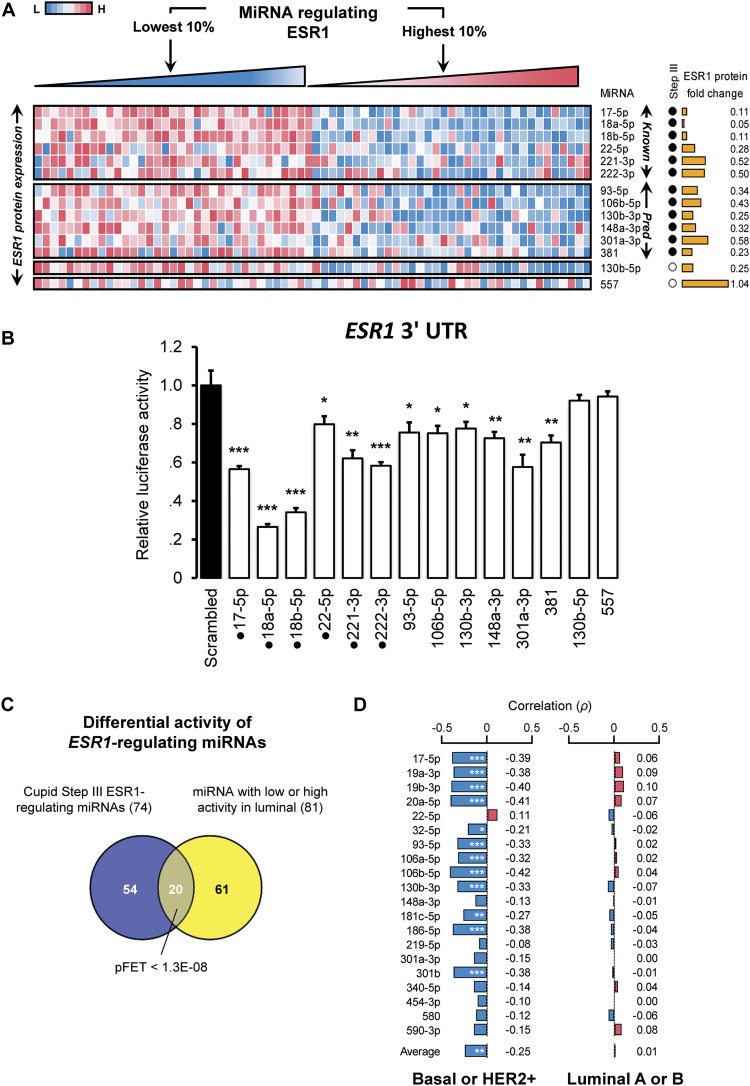
We chose to focus on ESR1 for detailed validation. The analysis of ESR1 protein expression in TCGA breast cancer tumors, profiled by RPPA using the antibody ER.alpha.R.V_GBL.9014870, suggests that ESR1 protein expression is strongly correlated with the expression of predicted miRNA regulators (Fig. 6A). Biochemical validation of select ESR1 miRNA regulators showed significantly reduced ESR1 3′ UTR luciferase activity following transfection of predicted miRNA regulators (Fig. 6B).
Figure 6.
ESR1 expression is anticorrelated with miRNA-regulator expression. (A) ESR1 protein expression in breast cancer tumors is anticorrelated with expression profiles of previously validated (Known) and predicted (Pred) miRNA regulators. ESR1 relative expression in the top and bottom 10% of tumors ranked based on the intensity of the expression of each miRNA; each row was ranked independently. miRNAs predicted to mediate ESR1 ceRNA regulation are marked (Step III); also marked is ESR1 protein expression fold change in tumor samples with low versus high expression for each miRNA. Negative controls include miR-557 and miR-130b-5p; miR-130b-5p expression was anticorrelated with ESR1 expression. (B) 3′ UTR luciferase activity fold changes after miRNA mimic transfections; some data replicated from Figure 5. Punctuated mimics correspond to previously validated interactions. (C) miRNAs with low or high activity in luminal breast cancer tumors are enriched for predicted ESR1 regulators (P < 1.3 × 10−8), and (D) their expression profiles are anticorrelated with ESR1 protein expression in basal-like and HER2-enriched tumors, but not in luminal tumors. Data are represented as mean ± SEM; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Results for 13 selected candidate ESR1 regulators, and miR-557, which was chosen as negative control, are given in Figure 6A and show significant ESR1 protein expression fold change. Eight miRNAs were selected because their effect on ESR1 3′ UTR luciferase activity assays was tested in Figure 5. The other five regulators where chosen at random from Supplemental Figure S4, and include previously validated regulators miR-22-5p, miR-221-3p, and miR-222-3p, as well as previously undescribed ESR1-regulators 381 and 148a-3p; 130b-5p was predicted to regulate ESR1 targets by Cupid Step II but had no evidence for mediating ESR1 ceRNA interactions. Results from biochemical testing of the predicted interactions, including results from assays described in Figure 5, are given in Figure 6B for ease of presentation. All mimic transfections of predicted ESR1 regulators significantly reduced ESR1 3′ UTR luciferase activity. As negative controls, we selected miR-557 and miR-130b-5p, which were not predicted to target ESR1; miR-130b-5p and ESR1 expression were significantly correlated (P < 1 × 10−8) but their interaction was not predicted by Cupid Step III.
In total, 74 miRNAs were predicted to target ESR1 by Cupid Step III. Given that ESR1 mRNA expression is highly subtype specific—its expression is high in luminal A and luminal B breast cancer tumors and very low in basal-like tumors—we set out to test whether miRNAs that target ESR1 may have inferred subtype-specific activity. Our selection criteria for identifying miRNAs with subtype-specific activity included two conditions: (1) miRNA expression must be significantly high or low in one tumor type relative to others (P < 1 × 10−3), and (2) its targets must be enriched for genes with low or high expression in that tumor type (P < 1 × 10−4), respectively, according to running sum statistics; see Supplemental Methods. We also identified ceRNA with subtype activity, and comprehensive lists for both are given in Supplemental Table S11.
In total, we identified 20 miRNAs that are predicted to target ESR1 and have high or low activity in luminal tumors (Fig. 6C); expression profiles of 12 of these miRNAs were significantly anticorrelated with ESR1 protein expression in basal-like and HER2-enriched tumors, but no miRNAs were significantly correlated with ESR1 protein expression in luminal tumors (Fig. 6D). This analysis suggests that ESR1 is regulated by a miRNA program that is specific to basal-like and HER2-enriched tumors and is absent in luminal tumors.
To test the predictive benefit of our compiled evidence for functional regulation, we compared ESR1 protein expression in 352 samples with low (bottom 10%) and high (top 10%) expression of each of the 50 candidate miRNA regulators that were predicted in Cupid Step II and had evidence for indirect regulation through effectors; note that miRNA expression and RPPA data were both available for only 352 of the 728 TCGA breast cancer samples. After removing outliers using the IQR rule for 44 miRNAs within two interquartile ranges from the mean, ESR1 protein expression was 2.8-fold higher in samples with low targeting miRNA expression, on average (Supplemental Fig. S4); only nine of these miRNAs were not predicted by Cupid Step II. This finding is in agreement with genome-wide statistical data that suggests that evidence for functional regulation is significantly predictive of miRNA–target interactions (Supplemental Fig. S4A,B).
Discussion
Identifying and understanding pathological implications due to miRNA dysregulation requires accurate maps of functional miRNA targets in specific disease contexts. We describe systems-biology-based methods that leverage previously validated interactions together with RNA and protein-expression profiles from patient samples to predict miRNA-3′ UTR target interactions with evidence for regulation in these samples. As a proof of principle, we predicted interactions in breast cancer using profiles from TCGA breast cancer tumors together with perturbation data in breast cancer cell lines. A variety of computational and biochemical techniques demonstrated improved fidelity of resulting predictions, including evidence for hundreds of candidate miRNA–target interactions in breast cancer cell lines. Our luciferase reporter assays supported >90% of interactions predicted by Cupid, and we described evidence for functional regulation by miRNAs in breast cancer, including expression-based evidence for ceRNA mediation by nearly 300K miRNA–target interactions. Other lines of evidence that may be useful for building predictive functions in the future failed to significantly improve predictive accuracy.
Focusing on predicted regulators of ESR1, we showed that RPPA data in breast cancer tumors could be used as an effective filter for identifying functional miRNA regulators. To further test the effects of miRNA regulation on a select set of proteins, we profiled protein expression after miRNA perturbation, producing a data set that could be used to compare prediction performance, and identifying breast cancer genes that are particularly amendable to miRNA regulation. In total, we identified nearly 500 miRNA–mRNA interactions with evidence for regulation in breast cancer that were supported by miRNA perturbation assays in breast cancer cell lines.
We observed subtype-specific activity (Supplemental Table S11) and pathway enrichment (Supplemental Table S12) for both miRNA and ceRNA regulators. Our analysis suggests that ESR1 is targeted by miRNA programs that are specific to HER2-enriched and basal-like tumors. We inferred that the PI3K and the p53 signaling pathway are enriched for miR-15, miR-16, and miR-424 targets; the MAPK signaling pathway is enriched for miR-106, miR-20a-5p, and let-7 family targets; the TGF beta signaling pathway is enriched for miR-27b, miR-17-5p, and let-7 targets; and that members of the miR-30 family target the HER2/EGFR pathway. Interestingly, members of the miR-30 family were also identified to have the lowest activity in HER2-enriched breast cancer tumors, suggesting a possible tumor-suppressor role for miR-30 in breast cancer tumors; miR-30 has been recently described as a tumor suppressor in prostate cancer (Kao et al. 2014). We also note that 26 genes were predicted to be regulated by at least 25% of miRNAs tested, and the majority of these 26 are known regulatory factors, including transcription and RNA processing factors.
It is important to note that while our results were based on analyses of reference 3′ UTR targeting, recent work (Sandberg et al. 2008; Brummer and Hausser 2014) suggests that both alternative polyadenylation and miRNA targeting outside of 3′ UTRs may alter miRNA regulation to pathophysiological effects. Incorporating these lines of evidence may help further our understanding of miRNA regulation, and we believe that these challenges can be addressed using current technology. Namely, the effects of alternative polyadenylation can be accounted for through custom analyses of short-RNA libraries, followed by mixture model resolution. Similarly, methods for identifying miRNA binding sites outside of 3′ UTRs can be used to supplement predictions presented here. In addition, the combination of accurate technology for predicting miRNA targets and paired mRNA and protein-expression profiles promises to enable inquiry into miRNA mechanisms of action. For example, statistical evidence suggests that expression profiles of ESR1-regulating miRNAs are predictive of variability in the coupling between ESR1 mRNA and protein-expression profiles. Moreover, a study focusing on the ability of individual miRNA to predict coupling of target mRNA and protein-expression profiles may help identify miRNAs that primarily regulate translation and those that regulate mRNA destabilization (Brummer and Hausser 2014).
In conclusion, the increasing body of molecular profiles in primary disease tissues and in perturbation of disease models presents an opportunity for systems-biology-based approaches to improve the accuracy of regulatory-interaction prediction methods. Our analyses suggest that evidence for regulation by miRNAs in given contexts can significantly improve context-specific miRNA–target predictions, and consequently build better context-specific cellular wiring diagrams. Improving these has implications for efforts to identify and interpret pathologically relevant genomic variants, a key technical challenge in personalized genomics.
Methods
Cupid multistep prediction and other functional evidence for miRNA regulation
Cupid miRNA–target and ceRNA interaction predictions proceed in three sequential steps, as outlined in the Results. A detailed description, including Cupid site prediction (Step I), Cupid interaction prediction (Step II), the predictive features and machine learning processes used in these steps, and methods for identifying candidate miRNA–target interactions with evidence for mediating ceRNA interactions, are presented in the Supplemental Methods. In addition, we used evidence for combinatorial interactions between miRNAs and for indirect miRNA regulation through effectors to support Cupid Step II miRNA–target interaction predictions; methods and results are described in the Supplemental Methods.
Genes-expression profiling following precursor transfection
Gene expression was profiled using Illumina Human-6 Expression BeadChips following transfection of pre-mir-18a, pre-mir-193b, pre-mir-206, pre-mir-302c (Leivonen et al. 2009) (GSE14847), with Affymetrix GeneChip Human Genome U133 Plus 2.0 Array following pre-mir-101-1 and scrambled controls in MCF7 (Frankel et al. 2011) (GSE31397), and pre-miR-145 and control in MDA-MB-231 (Gotte et al. 2010) (GSE19737).
Testing sites and interactions
We used the F-measure to test the ability of binding-site prediction methods to identify in 3′ UTRs with evidence for miRNA binding in HEK293 cells (Hafner et al. 2010), and when testing interactions using gene-expression data after transfection of precursors of predicted miRNA regulators. Detailed methodology is presented in the Supplemental Methods.
3′ UTR cloning, in vitro MIMIC transfection conditions, and luciferase assays
To measure the targeting activity of microRNA MIMICs, the 3′UTRs of specific target genes were cloned downstream from the luciferase reporter in the pMIR-REPORT vector (Life Technologies #AM5795M) by PCR from human genomic DNA using restriction enzymes. 293T cells were plated at 70% confluence in 96-well plates. Twenty-four hours later, cells were transfected with 50 ng of pMIR-REPORT constructs containing the luc-3′-UTR sequences, 50 ng of a Renilla normalization control, and 100 nM of each individual synthetic mirVana miRNA MIMICs (Ambion #4464066) at 100 nM final concentration using the TransIT-LT1 (Mirus Bio #2300A) and TransIT-TKO (Mirus Bio #2150) transfection reagents following the manufacturer’s instructions. After 24 h, relative luciferase units (RLU) were measured using the Dual-Glo Luciferase Assay System (Promega #E2949). Primer sequences are given in the Supplemental Methods.
Forward transfection of 3′ UTRs
Forward transfection of the plasmid was performed with the Lipofectamine (Invitrogen) transfection reagent, following the manufacturer’s protocol. Lipofectamine 2000 was used for A549, HepG2/C3A, HT-29, SK-MEL-28, and SK-OV-3 cells. Lipofectamine LTX PLUS was used for MCF7, PC-3, and U2-OS cells. In general, cells attached to the culturing surface were washed with phosphate-buffered saline, and the medium was replaced with 100 μL of Opti-MEM with 2% fetal bovine serum. A total of 100 ng per well in a 96-well plate of the plasmid was then mixed with a 0.3 μL/well of Lipofectamine in Opti-MEM, and 20-min later the mixture was added to the wells. After 6 h of transfection, the cells were then cultured in regular medium for 24 h and subsequently harvested.
Real-time quantitative RT-PCR analysis
Total RNA was extracted from cells with the RNeasy mini kit (Qiagen) and depleted of contaminating DNA with RNase-free DNase (Qiagen). Equal amounts of total RNA (1 μg) were reverse-transcribed using the qScript cDNA Synthesis kit (Quanta Biosciences). The first-strand cDNA was used as a template. Real-time PCR was carried out using SYBR green fluorescence. Two microliters of RT were used in a 25-μL reaction. Each sample was assayed in three independent RT reactions and triplicate reactions were performed and normalized to the GAPDH expression levels. Negative controls included the absence of enzyme in the RT reaction and the absence of template during PCR. Relative quantification of gene expression was performed with the comparative CT method. Primers used for quantitative RT-PCR analyses were synthesized by Sigma-Aldrich.
High-throughput quantitative RT-PCR analysis
The first-strand cDNA, synthesized using the qScript cDNA Synthesis kit, was first amplified for specific target amplification (STA). Briefly, a 12-cycle preamplification reaction was performed for each sample in 5 µL by pooling all primer pairs (final concentration, 50 nM), 1.25 µL cDNA, and 2.5 µL 2× PreAmp Master Mix (Applied Biosystems) following the manufacturer’s protocol. Unincorporated primers were then cleaned up using Exonuclease I (New England Biolabs). Briefly, 2 µL of diluted Exo I at 4 units/µL was added to each 5-µL STA reaction, and then incubated for 30 min at 37°C and 15 min at 80°C. Samples were then diluted five times with low TE buffer. High-throughput qPCRs were performed on the Biomark HD (Fluidigm) in a microfluidic multiplex 48.48 dynamic array chip according to the Fluidigm Advanced Development Protocol with EvaGreen. For each individual assay, 5 µL Assay Mix containing 9 µM forward primer, 9 µM reverse primer, and 1× Assay Loading Reagent was loaded into the Assay Inlets on the chip. For each sample, 5 µL Sample Mix containing 2.25 µL diluted sample in 1× DNA binding Dye Sample Loading Reagent and 1× SsoFast EvaGreen Supermix (Bio-Rad) was loaded into the sample inlets. The Biomark’s default Fast thermo cycling program with a melting step was used for the real time PCR reactions and fluorescence detection.
miRNA library screen by RPPA
The miRNA library was designed and synthesized by Dharmacon. MDA-MB-231 cells were seeded (3750 cells/well) and transfected with 50 nM miRNA mimics. After 48 h, cells were lysed and RPPA analysis was carried out as previously described (Zhang et al. 2009; Hennessy et al. 2010; Lu et al. 2011); mimics tested and antibodies used are given in Supplemental Table S4. Cells were washed with PBS, then lysed in 1% Triton X-100, 50 mM HEPES (pH 7.4), 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM Na pyrophosphate, 1 mM Na3VO4, 10% glycerol, containing freshly added protease and phosphatase inhibitors. Cellular proteins were denatured by 1% SDS (with β-mercaptoethanol) and diluted in six twofold serial dilutions in dilution buffer (lysis buffer containing 1% SDS). Serial diluted lysates were arrayed on nitrocellulose-coated FAST slides (Whatman Inc.). Each slide was probed with a validated primary antibody plus a biotinconjugated secondary antibody.
The signal was amplified using a DakoCytomation-catalyzed system (Dako) and visualized by DAB colorimetric reaction. The slides were scanned, analyzed, and quantified using a customized-software Microvigene (VigeneTech Inc.) to generate spot intensity. Each dilution curve was fitted with the logistic model (“Supercurve Fitting” developed by the Department of Bioinfomatics and Computational Biology at the MD Anderson Cancer Center, “http://bioinformatics.mdanderson.org/OOMPA”). The program fits a single curve using all the samples (in the dilution series) on a slide with the signal intensity as the response variable and the dilution steps as the independent variable. Protein expression for each well and each antibody was normalized as SEM relative to mock transfections, with P-values for individual miRNA mimics calculated using a single-sample Student’s t-test against mock transfections. Mimics for RPPA experiments (159 miRNA mimics in total) were chosen from a preliminary test of 879 mimics, identifying transfections that lead to highest total fold change across profiling antibodies.
Statistical analysis
All experiments were performed at least in triplicate and representative results are shown. All data are shown as the mean ± SE. Student’s t-tests were used to evaluate statistical significances between different treatment groups.
Data access
The RPPA data from this study, including 163 RPPA experiments following miRNA transfections, where each experiment has readouts from 158 antibodies as given in Supplemental Table S4, are available from The Cancer Proteome Atlas (TCPA; http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html) under accession number TCPA00000001. Cupid source code is available for download from SourceForge project CupidTool at http://cupidtool.sourceforge.net/.
Supplementary Material
Acknowledgments
We acknowledge the generous funding provided by the NIH under the following grant awards: (1) Roadmap grant for a Center for the Multiscale Analysis of Genetic Networks (MAGNet) (U54CA121852); (2) Genetic Network Inference with Combinational Phenotypes (R01CA109755); (3) In Silico Research Centers of Excellence NCI-caBIG 29XS192 and 12ST1103; and (4) LINCS grants 1U01HL111566-01 and 5U01CA164184-02. RPPA was supported by an NCI CCSG grant (P30 CA016672). G.B.M. and P.R. were supported by U54 CA112970. The results published here are in part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI as of January 2011. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/. The dbGaP accession number for the data analyzed in this work is phs000178.v4.p4, dated January 24, 2011. P.R., G.B.M., P.S., and A.C. conceived and supervised the project and participated in its computational and experimental design. H.S.C. and P.S. designed and implemented the computational methods; H.S.C., P.R., A.A.I., H.R.K., and P.S. analyzed data; P.S. and P.R. designed the experimental assays. D.L.N., X.Y., and A.I. performed the experiments; H.S.C., P.S., and A.C. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.178194.114.
References
- Betel D, Koppal A, Agius P, Sander C, Leslie C. 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. 2009. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem 284: 1971–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer A, Hausser J. 2014. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays 36: 617–626. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AP, Tooney PA, Cairns MJ. 2013. Context-specific microRNA function in developmental complexity. J Mol Cell Biol 5: 73–84. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. 2011. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2: 27:1–27:27. [Google Scholar]
- Darbellay G, Vajda I. 1999. Estimation of the information by an adaptive partitioning of the observation space. IEEE Trans Inf Theory 45: 1315–1321. [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. 2011. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44: 839–847. [DOI] [PubMed] [Google Scholar]
- Elefant N, Altuvia Y, Margalit H. 2011. A wide repertoire of miRNA binding sites: prediction and functional implications. Bioinformatics 27: 3093–3101. [DOI] [PubMed] [Google Scholar]
- Erhard F, Haas J, Lieber D, Malterer G, Jaskiewicz L, Zavolan M, Dolken L, Zimmer R. 2014. Widespread context-dependency of microRNA-mediated regulation. Genome Res 24: 906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Wen J, Lees M, Hoyer-Hansen M, Farkas T, Krogh A, Jaattela M, Lund AH. 2011. microRNA-101 is a potent inhibitor of autophagy. EMBO J 30: 4628–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. 2007. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics 8: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. 2009. MicroRNAs in cancer. Annu Rev Med 60: 167–179. [DOI] [PubMed] [Google Scholar]
- Gotte M, Mohr C, Koo CY, Stock C, Vaske AK, Viola M, Ibrahim SA, Peddibhotla S, Teng YH, Low JY, et al. 2010. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene 29: 6569–6580. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, Davies MA, Liu W, Coombes K, Meric-Bernstam F, et al. 2010. A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clin Proteomics 6: 129–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. 2011. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 39: D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human microRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CJ, Martiniez A, Shi XB, Yang J, Evans CP, Dobi A, deVere White RW, Kung HJ. 2014. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 33: 2495–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284. [DOI] [PubMed] [Google Scholar]
- Leivonen SK, Makela R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellstrom K, Sahlberg N, et al. 2009. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene 28: 3926–3936. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838. [DOI] [PubMed] [Google Scholar]
- Lu Y, Muller M, Smith D, Dutta B, Komurov K, Iadevaia S, Ruths D, Tseng JT, Yu S, Yu Q, et al. 2011. Kinome siRNA-phosphoproteomic screen identifies networks regulating AKT signaling. Oncogene 30: 4567–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217. [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. 2011. MicroRNAs can generate thresholds in target gene expression. Nat Genet 43: 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. 2009. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res 37: D155–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. 2012. Functional microRNA targets in protein coding sequences. Bioinformatics 28: 771–776. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm M, Hackenberg M, Langenberger D, Frishman D. 2010. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics 11: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. 2011. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147: 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejnar CE, Zdobnov EM. 2012. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res 40: 11673–11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. 2009. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 37: D105–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT, Xu LD, Wang YY, Du L, Zhang YP, et al. 2011. MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res 39: 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wei Q, Mao L, Liu W, Mills GB, Coombes K. 2009. Serial dilution curve: a new method for analysis of reverse phase protein array data. Bioinformatics 25: 650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.