Summary
Induced pluripotent stem (iPS) cells hold great promise for personalized regenerative medicine. However, recent studies show iPS cell lines carry genetic abnormalities, suggesting reprogramming may be mutagenic. Here we show that ectopic expression of the reprogramming factors increases the levels of phosphorylated histone H2AX, one of the earliest cellular responses to DNA double strand breaks (DSBs). Further mechanistic studies uncover a direct role of the homologous recombination (HR) pathway, a pathway essential for error-free repair of DNA DSBs, in reprogramming. This role is independent of the use of integrative or non-integrative methods to introduce reprogramming factors, despite the latter being considered a safer approach that circumvents genetic modifications. Finally, deletion of the tumor suppressor p53 rescues the reprogramming phenotype in HR-deficient cells primarily through restoration of reprogramming-dependent defects in cell proliferation and apoptosis. These novel mechanistic insights have important implications for the design of safer approaches to create iPS cells.
Introduction
Pioneering work by Yamanaka and colleagues has identified key transcription factors that enable reprogramming of somatic cells to a pluripotent state (Takahashi and Yamanaka, 2006). This technology has been used to generate human iPS cells, which closely resemble embryonic stem (ES) cells in differentiation potential, self-renewal capacity, transcriptional profile, and epigenetic state (Hochedlinger and Plath, 2009; Okita and Yamanaka, 2011). Like ES cells, iPS cells can be differentiated into a wide range of cell types, allowing the generation of patient-specific cells suitable for cell replacement therapy and disease modeling.
Despite this great promise, a number of studies suggest that reprogramming and subsequent expansion of iPS cells in culture leads to accumulation of diverse genetic abnormalities at chromosomal, subchromosomal and nucleotide levels (Gore et al., 2011; Hussein et al., 2011; Laurent et al., 2011; Mayshar et al., 2010). The source of these genetic lesions remains under debate. Some reports attribute it primarily to clonal capture of variant cells within the donor cell population (Cheng et al., 2012; Young et al., 2012), yet another study suggests that approximately half of the mutations arise de novo during reprogramming (Gore et al., 2011). This has prompted us to examine whether reprogramming is a novel trigger of DNA damage, and the roles of the homologous recombination (HR) DNA repair pathway in reprogramming.
We used a drug-inducible system to discriminate the effects of reprogramming from viral integration, since the latter is known to cause DNA Double Strand Breaks (DSBs). The results show that ectopic expression of the reprogramming factors is sufficient to induce DNA DSBs, providing a plausible molecular mechanism for genetic abnormalities observed in iPS cell lines. Furthermore, efficient reprogramming requires key HR genes, including Brca1, Brca2 and Rad51, independent of the methods used to introduce the reprogramming factors. Finally, deletion of the tumor suppressor p53 largely restores normal reprogramming in HR-deficient mouse embryonic fibroblasts (MEFs), accompanied by a correction of reprogramming-dependent defects in cell proliferation and apoptosis. These findings provide novel mechanistic insights into reprogramming and have important implications for designing rational approaches to generate lesion-free iPS cells suitable for clinical applications.
Results
Reprogramming induces DSBs
DNA DSBs can be triggered by a number of DNA damaging agents such as γ-irradiation and oxidative stress. Excessive accumulation of DSBs in a cell leads to growth arrest, apoptosis, or mutations in the genome. Ectopic expression of Oct4, Sox2, Klf4 and c-Myc or Oct4, Sox2 andKlf4, hereafter referred to as 4F or 3F respectively, allows reprogramming of mouse embryonic fibroblasts (MEFs) to a pluripotent state (Hochedlinger and Plath, 2009; Okita and Yamanaka, 2011). Transduction of 4F or 3F using constitutive retroviral expression vectors has been shown to increase the number of cells with phosphorylated histone H2AX (γH2AX) nuclear foci, one of the earliest cellular responses to DSBs (Kawamura et al., 2009; Muller et al., 2012). However, it is unclear whether the DSBs are caused by reprogramming or viral transgene integration, as the latter is known to cause DSBs (Skalka and Katz, 2005).
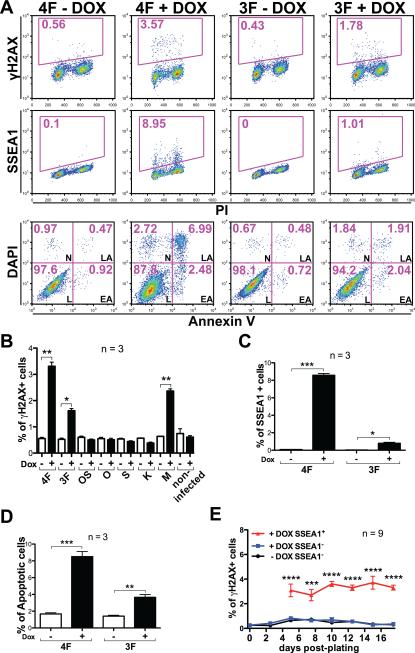
To determine whether there is a direct link between epigenetic reprogramming and increased DNA DSBs, we used doxycycline-inducible lentiviral vectors (FUW-tetO) to express reprogramming factors in wild-type MEFs, and assessed γH2AX through flow cytometry (Huang and Darzynkiewicz, 2006). The effects of reprogramming genes were determined by comparing the same pool of infected cells with or without doxycycline treatment. We found that 4F- and 3F-infected MEFs showed a ~6- and a 3-fold increase respectively of γH2AX+ cells after 5 days of doxycycline treatment compared to infected-but-untreated or non-infected MEFs, whereas doxycycline treatment alone on non-infected MEFs had no effects (Figure 1A, 1B). This correlated with the acquisition of an early reprogramming marker (SSEA1), and a marked increase in the percentage of cells undergoing apoptosis identified by Annexin V staining (Figure 1A, 1C, 1D). Expressing c-Myc alone also had an effect, consistent with a previous report (Karlsson et al., 2003); whereas expressing other reprogramming factors individually or in combination (Oct4 and Sox2) had no significant effect (Figure 1B).
Figure 1. Reprogramming induces DSBs and apoptosis.
(A) Representative Fluorescence-activated cell sorting (FACS) plots of 4F- and 3F-infected wild-type MEFs stained for γH2AX, SSEA1 and Annexin V after cells were cultured with or without doxycycline (DOX) for 5 days. Numbers indicate percentages of positive cells. PI: Propidium Iodide; DAPI: 4',6-diamidino-2-phenylindole; L: alive; EA: early apoptotic; LA: late apoptotic; N: necrotic. (B) Quantification of the percentage of γH2AX+ cells in wild-type MEFs infected with reprograming genes in combination or individually. OS: Oct4-Sox2; O: Oct4; S: Sox2; K: Klf4; M: c-Myc. (C, D) Quantification of the percentage of SSEA1+ (C) and Annexin V+ (D) cells in wild-type MEFs transduced with 4F and 3F. Apoptotic cells are the sum of EA and LA cells. (E) Time-lapse, flow cytometric quantification of γH2AX+ cells present in reprogrammable MEFs with or without DOX treatment; cells were separated based on the expression of SSEA1. In all column graphs of this study, error bars indicate SEM, and p values by two-tailed student t-test <0.05, 0.01, 0.001 and 0.0001 are indicated by one, two, three or four asterisks, respectively.
Because non-integrative methods are thought to generate safer iPS cells for clinical use, we measured γH2AX+ cells during reprogramming using a non-integrative approach based on the use of “reprogrammable”-MEF (Carey et al., 2010; Stadtfeld et al., 2010). We generated reprogrammable MEFs by combining an allele constitutively expressing the reverse tetracycline-controlled transactivator (rtTA) from the Rosa26 locus with a doxycycline-inducible polycistronic reprogramming cassette (OKSM) targeted to the Col1A1 locus (Stadtfeld et al., 2010). This system allows homogeneous expression of the reprogramming factors ideal for studies of reprogramming. Using flow cytometry, we analyzed the percentage of γH2AX+ cells at different time points after doxycycline treatment. Additionally, we used the pluripotency cell surface marker SSEA1 to identify early reprogramming cells in doxycycline-treated conditions (Brambrink et al., 2008). We observed the same low levels of γH2AX expression in both untreated MEFs and SSEA1− cells in the doxycycline-treated condition (Figure 1E). In contrast, there was a significant increase in the percentage of γH2AX+ cells in the SSEA1+ population in doxycycline-treated cells. This increase occurred early, and remained constant during the reprogramming process. These results demonstrate that reprogramming, rather than viral integration, is directly responsible for the accumulation of γH2AX in cells.
Reprogramming is impaired in Brca1 and Brca2 mutant MEFs
In mammalian cells, three pathways have been described for repair of DSBs: HR, nonhomologous end joining (NHEJ), and single-strand annealing (SSA) (Moynahan and Jasin, 2010). HR is responsible for accurate repair of DNA damage using the sister chromatid as a template. In contrast, repairs by NHEJ and SSA are intrinsically error-prone, and can lead to deletions and other types of mutations. Previous studies have shown that fibroblasts defective for the Fanconi anemia (FA) complementation group are resistant to reprogramming using classic viral infection-based methods (Muller et al., 2012; Raya et al., 2009). These studies suggest a potential link between HR and reprogramming, as several FA pathway components have been shown to promote HR (Nakanishi et al., 2005). However, a direct role of HR in reprogramming has not been established, because FA proteins also have distinct functions independent of HR.
We examined the role of Brca1 and Brca2, two genes essential for homology-directed DNA repair, during reprogramming using homozygous MEFs generated from three hypomorphic mutant alleles. Brca1Tr carries an insertion within exon 11, leading to a truncated Brca1 protein with 924 amino acids (Ludwig et al., 2001). The second Brca1 allele, Brca1S1598F, contains a point mutation in the Brca1 C-terminal (BRCT) domain, which disrupts the interaction of Brca1 with the phosphorylated isoforms of several repair proteins including Abraxas/CCDC98, BACH1/FancJ, and CtIP (Shakya et al., 2011). The Brca2Δ27 allele harbors a deletion of exon 27, generating a truncated protein lacking 187 C-terminal amino acids (McAllister et al., 2002). All three mutations impair homology-directed DNA repair. Adult mice homozygous for each of these mutations are identified from crosses of heterozygous animals, suggesting these mutations do not significantly affect cell growth or survival in vivo.
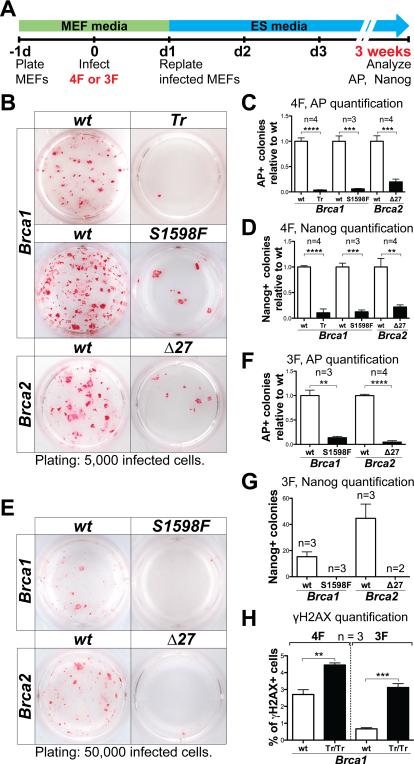
In wild-type MEFs, we typically detected ~300 alkaline phosphatase (AP)+ colonies, and ~100 Nanog+ colonies 3 weeks after plating of 50,000 4F-infected cells using the constitutive retroviral expression vector pMXs (Figure 2A, 2B). In contrast, the numbers of AP+ and Nanog+ colonies were significantly reduced (up to ~20 fold) in Brca1 and Brca2 homozygous mutant MEFs when compared to wild-type control MEFs (Figure 2B-D). By picking colonies with iPS-like morphology, we were able to establish Brca2 mutant iPS cell lines with comparable efficiency (~40%) to wild-type controls (Figure S1A). Brca2 mutant iPS cells were indistinguishable from control wild-type iPS cells in expression of Nanog and other pluripotency markers by real-time quantitative RT-PCR (qRT-PCR), immunohistochemical analysis (Figure S1B, S1D). The rates of proliferation and apoptosis were not significantly different between Brca2 mutant iPS cell lines compared to control wild-type lines (Figure S1E-H). Therefore, the reprogramming phenotypes observed in Brca2 mutant MEFs are not due to impaired proliferation and/or increased apoptosis of HR-deficient iPS cells formed during reprogramming. However, we were not able to establish bona fide iPS cell line from Brca1 mutant MEFs out of 10 colonies picked (Figure S1A). The best Brca1 mutant lines appeared partially reprogrammed, exhibiting only occasional Nanog staining by immunohistochemical analysis. Compared to Brca2 mutant and wild-type iPS cell lines, Brca1 mutant lines exhibited limited up-regulation of the pluripotency gene Nanog, accompanied by incomplete silencing of the fibroblast marker Col6a1 and the reprogramming transgenes (Figure S1B-S1D). These data show that both Brca1 and Brca2 are required for efficient reprogramming, and Brca1 may also be required for iPS cell line establishment.
Figure 2. Reprogramming is impaired in Brca1 and Brca2 mutant MEFs.
(A) Schematics of all virus-mediated reprogramming experiments in this study. MEFs were infected with 4F or 3F 1-day after plating, and replated in 12-well dishes the next day on irradiated MEFs at densities specified (indicated bellow the AP-staining pictures for all figures in this study). AP and Nanog staining was performed after 3 weeks (unless otherwise noted). (B-D) Representative AP staining (B), and quantification of AP+ (C) and Nanog+ (D) colonies generated with 4F-reprogramming from Brca1Tr/Tr, Brca1S1598F/S1598F and Brca2Δ27/Δ27 MEFs, compared with wild-type (wt) MEFs from littermate controls. (E-G) Representative AP staining (E), and quantification of AP+ (F) and Nanog+ (G) colonies generated with 3F-reprogramming. (H) Quantification of the percentage of γH2AX+ cells in 4F- and 3F-infected, Brca1Tr/Tr mutant and control wild-type MEFs after 5 days of DOX treatment. See also Figure S1.
Next, we examined whether mutations in Brca1 and Brca2 affect 3F-reprogramming without c-Myc, the overexpression of which alone increases DNA DBSs. Using doxycyclineinducible lentiviral expression vectors (FUW-tetO) to express the 3F, we detected ~150 AP+ colonies, and ~40 Nanog+ colonies 3 weeks after plating of 50,000 infected, doxycycline-treated wild-type MEFs. As in 4F-reprogramming, both Brca1 and Brca2 homozygous mutant MEFs showed an up-to 20-fold reduction in the number of AP+ colonies (Figure 2E, 2F). Moreover, no Nanog+ colonies were detected (Figure 2G) from mutant MEFs. These results support a critical role of Brca1 and Brca2 in both 3F- and 4F-reprogramming independent of the infection method used to introduce the reprogramming factors.
Finally, to establish a direct link between reprogramming and HR-mediated DNA repair, we compared the percentage of γH2AX+ cells in Brca1Tr/Tr versus wild-type MEFs during reprogramming (Figure 2H). We detected a significant increase in the percentage of γH2AX+ cells in both 4F- and 3F-expressing mutant cells compared to wild-type controls. These data, together with the established roles of Brca1 and Brca2 in HR, strongly suggest a direct involvement of HR-mediated DNA DSB repair in reprogramming.
HR genes play a direct role during reprogramming
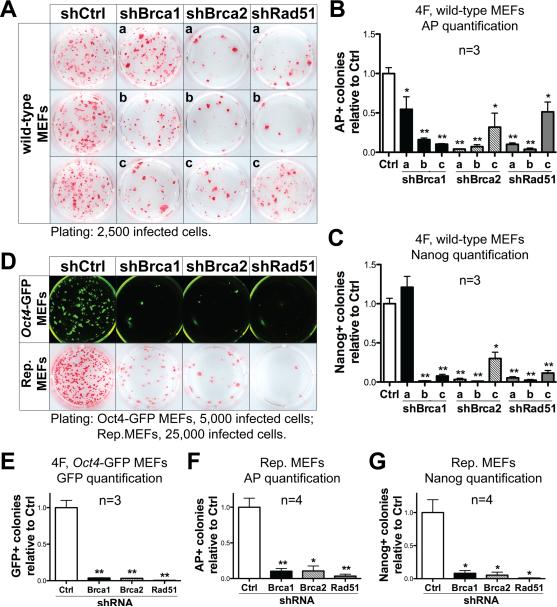
Brca1 and Brca2 mutant MEFs may have accumulated genetic or cellular alterations during their culture before reprogramming, which could prevent the formation of iPS colonies. Additionally, mutant MEFs show a small but significant decrease (<3% decrease for FUW-tetO vectors) in gene transduction efficiency compared to wild-type controls (Figure S2A, S2B), raising the possibility that mutant MEFs may reprogram less efficiently due to a requirement of HR genes for viral integration and transgene expression. To determine whether HR genes are directly required for reprogramming, we introduced 4F while simultaneously expressing short hairpin RNAs (shRNAs) against individual HR genes (Brca1, Brca2 and Rad51) in wild-type MEFs. As expected, knockdown of HR genes had no significant effect on the transduction of a GFP reporter or the reprogramming transgenes (Figure S2C, S2D). Significantly, a decrease in reprogramming efficiency was observed with all shRNAs, except for one due to insufficient knockdown of the transcript (Figure 3A-C and Figure S3D). The most efficient shRNA (shRad51-b) reduced the number of AP+ colonies by ~60 fold. Likewise, we performed shRNA-mediated knockdown of Brca1, Brca2 and Rad51 in 3F-reprogramming experiments, and observed a marked decrease of AP+ and Nanog+ colonies (Figure S3A-C, S3E). Similar results were obtained using an additional pluripotency marker gene Oct4, by conducting reprogramming experiments on MEFs carrying one copy of the Oct4-GFP transgenic reporter allele (Szabo et al., 2002) (Figure 3D, 3E and Figure S3F, S3G).
Figure 3. HR genes are directly required during reprogramming.
(A-C) Representative AP staining (A), and quantification of AP+ (B) and Nanog+ (C) colonies generated with 4F-reprogramming and a panel of shRNAs targeting Brca1 (shBrca1-a, b, c), Brca2 (shBrca2-a, b, c) and Rad51 (shRad51-a, b, c) compared to the shRNA control vector (shCtrl). Lower-case letters refer to individual shRNAs targeting each HR gene. shBrca1-c, shBrca2-b and shRad51-b were used for further experiments. (D) Upper panel: Representative fluorescence images of Oct4-GFP+ colonies generated with 4F and shRNAs targeting HR genes. Lower panel: Representative AP staining images from reprogrammable (Rep.) MEFs infected with shRNAs against HR genes. (E) Quantification of Oct4-GFP+ colonies from experiments using 4F-infected Oct4-GFP MEFs and acute HR-gene knockdown. (F, G) Quantification of AP+ (F) and Nanog+ (G) colonies from experiments using reprogrammable MEFs and acute HR-gene knockdown. See also Figures S2 and S3.
The requirement of HR genes is independent of viral integration
Experiments described above introduced reprogramming genes using classic viral infection-based methods commonly used in reprogramming studies. However, viral integration triggers DNA DSBs (Skalka and Katz, 2005), which may necessitate HR-mediated DNA repair. Therefore we proceeded to determine the requirement of HR genes in the absence of viral infection using reprogrammable MEFs. We infected reprogrammable MEFs with shRNAs targeting HR genes, and added doxycycline to initiate reprogramming. Using a control shRNA, we detected on average ~600 alkaline AP+ colonies (Figure 3D, 3F), and ~500 Nanog+ colonies (Figure 3G) from 50,000 reprogrammable MEFs after ~3 weeks of doxycycline treatment. shRNAs against Brca1, Brca2 and Rad51 all lead to a marked decrease in the number of both AP+ and Nanog+ colonies (Figure 3D, 3F, 3G and Figure S3J). These results demonstrate that DNA damage increases during reprogramming independent of viral integration, and that the HR pathway is also required for efficient reprogramming using non-integrative methods.
p53 deletion rescues reprogramming defects of HR-deficient MEFs
Because cells with excessive DNA damage are typically eliminated through p53-dependent apoptosis or growth arrest, we hypothesized that deletion of p53 would rescue the reprogramming defects in HR-deficient MEFs. This would be consistent with an established role of the p53 pathway in limiting the rate of reprogramming (Spike and Wahl, 2011).
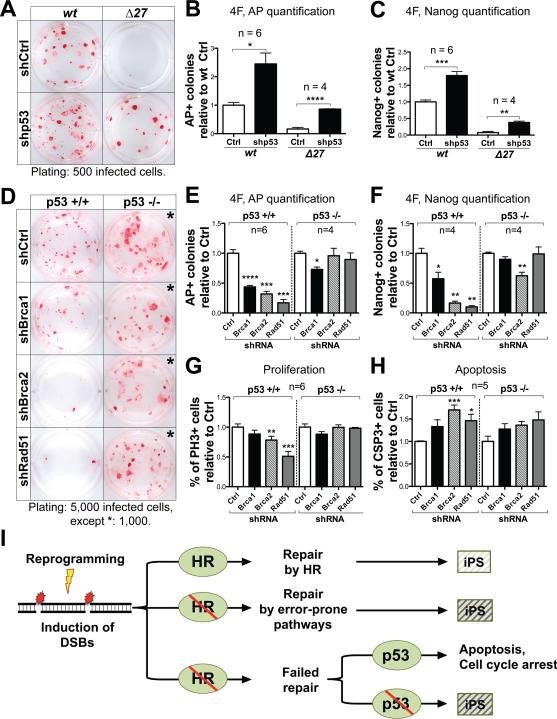
We performed 4F-reprogramming on MEFs derived from Brca2 homozygous mutant and wild-type embryos, and used a well-characterized shRNA to simultaneously suppress p53 (Hemann et al., 2003). Down-regulating p53 significantly increased the reprogramming efficiency in both mutant and wild-type MEFs, though the reprogramming efficiency of mutant MEFs was not rescued to wild-type levels (Figure 4A-C). The partial rescue may be due to incomplete inactivation of p53 using the knockdown approach (Figure S4A). To further investigate the role of p53 in HR-deficient MEFs, we generated p53 null mutant MEFs (Jacks et al., 1994) and performed 4F-reprogramming experiments while using shRNAs against Brca1, Brca2 and Rad51. In wild-type control MEFs, knockdown of HR genes caused a significant reduction in the number of AP+ and Nanog+ colonies (Figure 4D-F and Figure S4B). A ~20-fold increase in the numbers of AP+ and Nanog+ colonies was observed in p53 null mutant MEFs compared to wild-type control MEFs, consistent with previous reports. However, knockdown of HR genes generally had no significant effects on reprogramming of p53 null mutant MEFs (Figure 4D-F).
Figure 4. Down-regulating p53 rescues the reprogramming phenotype of HR-defective MEFs.
(A-C) Representative AP staining (A) and quantification of AP+ (B) and Nanog+ (C) colonies generated with 4F-reprogramming from Brca2Δ27 homozygous mutant and wild-type MEFs infected with an shRNA targeting p53 (shp53) or vector control (shCtrl). (D-F) Representative AP staining (D) and quantification of AP+ (E) and Nanog+ (F) colonies generated with 4F-reprogramming from p53 null and wild-type MEFs under acute HR-gene knockdown. All staining were performed 16 days after replating of infected cells. (G, H) Quantification of the percentage of Phospho-Histone H3+ (PH3+) (G) and Cleaved Caspase-3+ (CSP3+) (H) cells, 6 days post-infection of 4F and HR-gene knockdown in p53 null mutant and wild-type MEFs. (I) Our results support a critical role of the HR pathway for efficient reprogramming. We propose a model in which reprogramming increases the level of DNA damage, which is responsible for the genetic aberrations observed in iPS cell lines (indicated by a light shaded box). A defective HR pathway may lead to increased genetic aberration (indicated by dark shaded boxes), or the elimination of abnormal cells through p53-mediated cell cycle arrest or apoptosis. See also Figure S4.
To further investigate the cellular mechanisms, we analyzed cell proliferation and apoptosis during reprogramming by immunostaining for the mitotic marker Phospho-Histone H3 and the apoptotic marker cleaved Caspase-3. During 4F-reprogramming of wild-type control MEFs, HR deficiency caused a significant decrease in the percentage of proliferating cells, and an increase of apoptotic cells (Figure 4G, 4H and Figure S4C, S4D). In contrast, during 4F-reprogramming of p53 null MEFs, HR deficiency failed to cause any significant defects in cell proliferation or apoptosis (some increase was observed in apoptosis, though not statistically significant) (Figure 4G, 4H and Figure S4C, S4D). These results suggest that a defective HR pathway leads to an increased number of cells accumulating DNA damage during reprogramming. p53-mediated growth arrest and apoptosis is responsible for the elimination of these cells, and consequently a significant decrease in reprogramming efficiency. Although down-regulating p53 rescues the reprogramming phenotype in HR-deficient MEFs, it may also allow the generation of iPS cells with genetic aberrations (Figure 4I).
Discussion
Current reprogramming strategies rely on ectopic expression of defined sets of pluripotency-associated transcription factors (Hochedlinger and Plath, 2009; Okita and Yamanaka, 2011). The recent development of non-integrative methods to introduce reprogramming genes theoretically circumvent undesirable genetic modifications in iPS cells caused by transgene insertions in classic reprogramming approaches (Gonzalez et al., 2011). However, surveys of iPS cells generated using both integrative and non-integrative methods reveal significant genetic abnormalities (Gore et al., 2011; Hussein et al., 2011; Laurent et al., 2011; Mayshar et al., 2010). We show here that ectopic expression of the reprogramming factors increases the levels of the DNA DSB marker γH2AX independent of viral integration. This effect may be linked to oncogenic activities of the reprogramming factors (Daley, 2008). On the other hand, epigenetic remodeling, including global DNA demethylation may also contribute to DNA damage during reprogramming. Although the mechanisms by which 5-methylcytosine is converted into cytosine in CpG islands are not yet well understood, prevailing models suggest that this conversion involves potentially mutagenic DNA modifications that need to be processed through DNA repair mechanisms (Teperek-Tkacz et al., 2011).
Our results show that an intact HR pathway is required to achieve efficient reprogramming, even in the absence of potential genome modifying agents such as the oncogene c-Myc or viral-integration. Complete loss-of-function of HR genes during reprogramming may lead to even more profound effects. HR genes may also have functions in addition to DNA repair during reprogramming. For example, Brca1 is implicated in basal transcriptional regulation (Mullan et al., 2006) and transcriptional activation of several genes including Sox2 (Kondo and Raff, 2004). These additional roles may explain the stronger reprogramming phenotype observed in Brca1 mutant MEFs compared to Brca2 mutant cells. However, differences in genetic background and/or severity of the hypomorphic alleles used in this work may also contribute to the phenotypic differences. Recent evidence that core components of the nucleotide excision repair pathway act as ES cell-specific transcriptional co-activators regulating the expression of Nanog (Fong et al., 2011) raises the interesting possibility that additional DNA repair pathway components may also be coopted in ES cells to maintain pluripotency.
Finally, a better understanding of the role of DNA repair pathways during reprogramming will contribute to the identification of safer approaches to create iPS cells. The generation of desired cell types for regenerative medicine can also be achieved using more direct approaches, such as lineage reprogramming. Compared to pluripotency reprogramming, lineage reprogramming may involve less extensive epigenetic remodeling, and it does not typically rely on ectopic expression of classic oncogenes. For regenerative medicine, it will be crucial to determine whether lineage reprogramming induces similar levels of DNA damage as pluripotency reprogramming, and assess its mutagenic impact.
Experimental Procedures
Reprogramming and generation of iPS cell lines
For reprogramming experiments, passage 2 MEFs were seeded at 2×105 cells per well of a 6-well dish. MEFs were infected twice on the next day with fresh viral supernatants. The day after infection, MEFs were replated at different densities as specified densities on irradiated MEF feeder layers and cultured in mouse ES cell media (Knockout DMEM supplemented with 15% Hyclone FBS, L-glutamine, penicillin/streptomycin, nonessential amino acids, β-mercaptoethanol, and 1000 U/ml LIF). See extended experimental procedures for a detailed description of the reprogramming experiments performed in this article.
Knockdown of gene expression using shRNAs
To knockdown expression of Brca1, Brca2 and Rad51 genes, pLKO.1-puro lentiviral vectors expressing three different shRNAs per gene were obtained from Sigma (MISSION shRNA constructs). In all experiments, knockdown efficiency was assessed by qRT-PCR analyses 6 days after infection by comparing with the expression of the corresponding gene in cells infected with an empty pLKO.1-puro control virus (Sigma, SHC001) (Figure S3D, S3E, S3G, S3J, S4B). We used a well-characterized shRNA (MLS-shp53)(Hemann et al., 2003) to knockdown p53 expression, and used the empty vector (MLS-empty) as a control (Figure S4A).
Alkaline phosphatase and immunofluorescence staining
AP staining was performed using Vector Red Alkaline Phosphatase Substrate Kit following manufacturer's guidelines (Vector Laboratories, SK-5100). For nuclear immunostaining, cells were fixed with 4% paraformaldehyde in PBS for ~10 minutes followed by standard immunofluorescence staining procedures. The following primary antibodies were used: Nanog (Cosmobio Japan, REC-RCAB0002P-F), Oct4 (Santa Cruz Biotechnology, sc-5279), Klf4 (Santa Cruz Biotechnology, sc-20691), Sox2 (Santa Cruz Biotechnology, sc-17320). For SSEA1 surface marker expression analysis, live cells were directly stained for 30 minutes with an SSEA1 antibody conjugated with Alexa 488 (Santa Cruz Biotechnology, sc-21702 AF488) in PBS with 0.2% BSA.
Proliferation and apoptosis immunofluorescence analysis
For proliferation and apoptosis analyses, cells (infected on the previous day, or not infected) were plated at 104 cells per well of a 48-well dish. Five days after plating, cells were fixed with 4% paraformaldehyde in PBS for ~10 minutes. Immunofluorescence staining using either a Phospho-Histone H3 (Ser10) antibody (PH3) (Cell Signaling Technology, 9701S) or a Cleaved Caspase-3 (Asp175) antibody (CSP3) (Cell Signaling Technology, 9661S) was performed following standard procedures. In both cases, detection was achieved using a donkey anti-rabbit Alexa 488 secondary antibody (Life Technologies, A21206) combined with DAPI nuclear staining. Plates were imaged in multiple fluorescence channels using a Thermo Scientific Cellomics ArrayScan HCS Reader (PH3:Objective 10X, channel 1 dye XF53_386_23, channel 2 dye XF53_485_20; CSP3:Objective 10X, channel 1 dye BGRFR_386_23, channel 2 dye XF53_485_20) (Supplementary Fig. 7a, 7b). Automated image analysis of PH3+ cells (nuclear staining) was performed using Target Activation.V4 BioApplication, whereas quantification of CSP3+ cells (cytoplasmic staining) was performed using Compartmental Analysis.V4 (Figure S4C, S4D).
Flow cytometric analysis of γH2AX and SSEA1
Cells were first incubated with the SSEA1 antibody conjugated with Alexa 488 (described above) for 30 minutes. After washing steps, cells were fixed in 70% ice-cold ethanol, and stored at −20°C for up to 2 weeks. Next, cells were incubated with an Anti-phospho-Histone H2A.X (Ser139) antibody (Millipore, 05-636) followed by the Alexa 647 Goat Anti-Mouse IgG1 secondary antibody (Life Technologies, A21240) for γH2AX detection. Finally, cells were stained with PI solution (PBS containing 5μg/ml PI and 100μg/ml RNAse A) prior to flow cytometric analysis using Becton-Dickinson FACSCalibur.
Flow cytometric analysis of Annexin V and SSEA1
For apoptosis assays, flow cytometry was performed on cells stained with Annexin V–FITC (BD Pharmingen, 556547) and DAPI. In some experiments, cells were also stained with SSEA1-APC (R&D systems FAB2155A). Briefly, cells were washed twice with PBS, and stained with 0.5 ul of Annexin V–FITC (or with 0.5 ul of Annexin V–FITC and 4 ul SSEA1-APC) in 100 ul binding buffer (10 mM HEPES, pH 7.4, 140 mM NaOH, 2.5 mM CaCl2) for 30 minutes at room temperature in the dark. Next, cells were washed twice with the binding buffer, and then resupended in binding buffer containing 1μg/ml DAPI. Apoptotic cells were detected using a Beckman Coulter CyAn ADP Analyzer. Both early apoptotic (Annexin V+, DAPI−) and late apoptotic (Annexin V+, DAPI+) cells were included in cell death quantifications.
Statistical analysis
All values are shown as mean ± SEM. p values were calculated using two-tailed student's t-test; p<0.05 (*) was considered significant.
Supplementary Material
Acknowledgments
We thank Shinya Yamanaka, Rudolf Jaenisch, Didier Trono and Bob Weinberg for providing viral expression and packaging vectors through Addgene, Wenjun Guo for the FUW-tetO vector, Hans-Guido Wendel and Elisa Oricchio for p53 shRNA constructs, Kathryn Anderson, Hisham Bazzi, and Raymond Wang for providing mutant mice for MEF isolation, Zengrong Zhu, Katia Manova-Todorova and Sanghoon Oh for assisting with image acquisition and analysis, Diane Domingo and Jennifer Wilshire for assistance with flow cytometry, members of the Huangfu and Jasin laboratories, Mark Tomishima and Wenjun Guo for critical reading of the manuscript. This study was funded in part by the Louis V. Gerstner Jr. Young Investigators Award and MSKCC Society Special Projects Committee. F.G. was supported by the NYSTEM fellowship from the Center for Stem Cell Biology of Sloan-Kettering Institute.
References
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat Methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hansen NF, Zhao L, Du Y, Zou C, Donovan FX, Chou BK, Zhou G, Li S, Dowey SN, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harbor symposia on quantitative biology. 2008;73:171–174. doi: 10.1101/sqb.2008.73.041. [DOI] [PubMed] [Google Scholar]
- Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147:120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Boue S, Izpisua Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming a la carte. Nat Rev Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Darzynkiewicz Z. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol Biol. 2006;314:73–80. doi: 10.1385/1-59259-973-7:073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narva E, Ng S, Sourour M, Hamalainen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci U S A. 2003;100:9974–9979. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T, Fisher P, Ganesan S, Efstratiadis A. Tumorigenesis in mice carrying a truncating Brca1 mutation. Genes Dev. 2001;15:1188–1193. doi: 10.1101/gad.879201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- McAllister KA, Bennett LM, Houle CD, Ward T, Malphurs J, Collins NK, Cachafeiro C, Haseman J, Goulding EH, Bunch D, et al. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 2002;62:990–994. [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- Muller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K, Moreau LA, Schambach A, Park IH, London WB, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366:2198–2207. doi: 10.1098/rstb.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M, Rio P, Sleep E, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R, Reid LJ, Reczek CR, Cole F, Egli D, Lin CS, deRooij DG, Hirsch S, Ravi K, Hicks JB, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334:525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ 12 Suppl. 2005;1:971–978. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- Spike BT, Wahl GM. p53, Stem Cells, and Reprogramming: Tumor Suppression beyond Guarding the Genome. Genes & cancer. 2011;2:404–419. doi: 10.1177/1947601911410224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, Hubner K, Scholer H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Teperek-Tkacz M, Pasque V, Gentsch G, Ferguson-Smith AC. Epigenetic reprogramming: is deamination key to active DNA demethylation? Reproduction. 2011;142:621–632. doi: 10.1530/REP-11-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MA, Larson DE, Sun CW, George DR, Ding L, Miller CA, Lin L, Pawlik KM, Chen K, Fan X, et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.