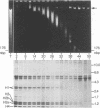
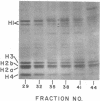
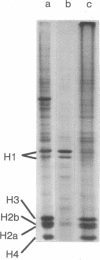
Abstract
Component α DNA is a homogeneous, highly repetitive fraction that comprises nearly a quarter of the African green monkey (Cercopithecus aethiops) genome. By restriction enzyme analysis, it has a repeat periodicity of 176 ± 4 nucleotide base pairs, corresponding closely with the length of DNA contained within a nucleosome. The sequence is organized into large blocks of constitutive heterochromatin. A method is described here for the isolation of intact polynucleosomal arrays containing only component α sequences. Isolated monkey nuclei are treated with EcoRI, which releases only component α nucleosomal arrays; the arrays are then fractionated and purified by sedimentation in sucrose gradients. The method permits a compositional analysis of the proteins associated with a constitutively repressed, heterochromatic sequence.
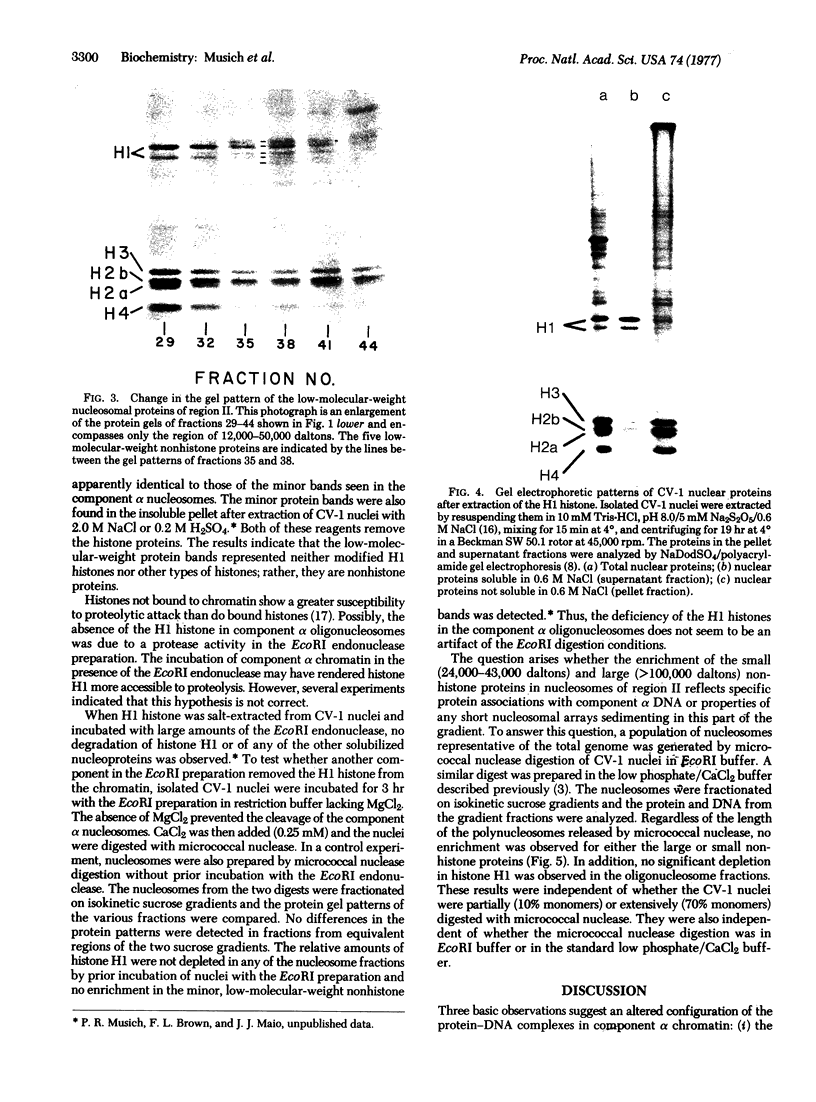
The major differences in the proteins associated with component α nucleosomes that distinguish them from the bulk DNA nucleosomes are a decrease in the content of the H1 histones in the component α nucleosomes and a concomitant increase in the amount of certain nonhistone proteins. The specific observations are: (i) In the component α nucleosomes, 65-70% of the proteins were nonhistone proteins; this contrasts with the value, 40%, for nonhistone proteins associated with nucleosomes containing bulk DNA. (ii) The amount of H1 histone in chromatin containing predominantly bulk DNA was about 13.7%. However, the H1 histone was depleted and possibly absent in component α oligonucleosomes. (iii) Coincident with the decrease in the H1 histones and in the same molecular weight range (24,000-43,000), there appeared five minor nonhistone proteins. The minor, low-molecular-weight, nonhistone proteins were not detected in chromatin containing bulk DNA but they represented nearly 12% of the protein in component α nucleosomes. The resistance to salt extraction (0.6-2.0 M NaCl) indicates that the low-molecular-weight nonhistone proteins are tenaciously bound to the component α nucleosomes. In addition, a class of high-molecular-weight (>100,000) nonhistone proteins was enriched 5- or 6-fold in component α oligonucleosomes. The relative amounts of the nucleosome core histones were not changed.
Keywords: EcoRI restriction endonuclease, nucleosome fractionation, mammalian repetitive DNA sequences, histone and nonhistone proteins
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berezney R., Coffey D. S. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Bostock C. J., Christie S., Hatch F. T. Accessibility of DNA in condensed chromatin to nuclease digestion. Nature. 1976 Aug 5;262(5568):516–519. doi: 10.1038/262516a0. [DOI] [PubMed] [Google Scholar]
- Doenecke D., McCarthy B. J. Movement of histones in chromatin induced by shearing. Eur J Biochem. 1976 May 1;64(2):405–409. doi: 10.1111/j.1432-1033.1976.tb10316.x. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Noll M., Kornberg R. D. Electron microscopy of defined lengths of chromatin. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman L., Huang R. C. Reconstitution of chromatin. The sequential binding of histones to DNA in the presence of salt and urea. J Mol Biol. 1972 Feb 28;64(1):1–8. doi: 10.1016/0022-2836(72)90317-8. [DOI] [PubMed] [Google Scholar]
- Lipchitz L., Axel R. Restriction endonuclease cleavage of satellite DNA in intact bovine nuclei. Cell. 1976 Oct;9(2):355–364. doi: 10.1016/0092-8674(76)90125-2. [DOI] [PubMed] [Google Scholar]
- Maio J. J. DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J Mol Biol. 1971 Mar 28;56(3):579–595. doi: 10.1016/0022-2836(71)90403-7. [DOI] [PubMed] [Google Scholar]
- Maio J. J., Schildkraut C. L. Isolated mammalian metaphase chromosomes. II. Fractionated chromosomes of mouse and Chinese hamster cells. J Mol Biol. 1969 Mar 14;40(2):203–216. doi: 10.1016/0022-2836(69)90469-0. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Senior M. B., Olins A. L., Olins D. E. Chromatin fragments resembling v bodies. Science. 1975 Jan 17;187(4172):173–175. doi: 10.1126/science.1111096. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]