Abstract
MicroRNAs control gene expression by inhibiting translation or promoting degradation of their target mRNAs. Since the discovery of the first microRNAs, lin-4 and let-7, in C. elegans, hundreds of microRNAs have been identified as key regulators of cell fate determination, lifespan, and cancer in species ranging from plants to humans. However, while microRNAs have been shown to be particularly abundant in the brain, their role in the development and activity of the nervous system is still largely unknown. In this review, we describe recent advances in our understanding of microRNA function at synapses, the specialized structures required for communication between neurons and their targets. We also propose how these advances might inform the molecular model of memory.
Keywords: Memory, microRNA, miRNA, Plasticity, RISC, Synapse
INTRODUCTION
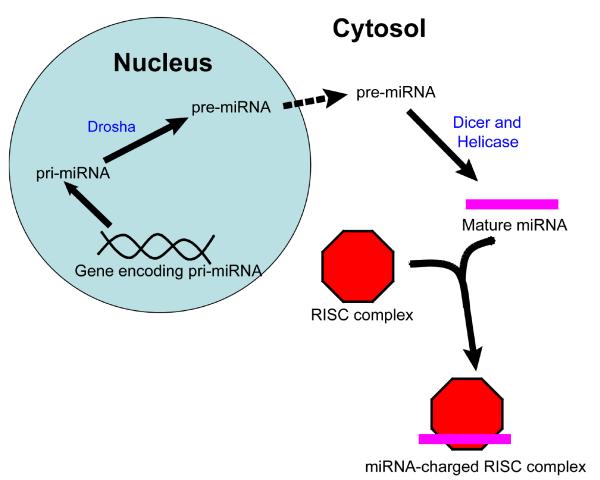
MicroRNAs (miRNAs) were first identified in the nematode C. elegans as key regulators of developmental transitions, and since their discovery have been identified in species ranging from plants to humans (1-3). These short non-coding RNAs are initially transcribed as long primary transcripts (pri-miRNAs) varying in size from hundreds to thousands of nucleotides. They are then cleaved by the RNase III enzyme Drosha to ~70-100nt stem-loop precursor miRNAs (pre-miRNAs), which are exported to the cytoplasm by Exportin 5 (3). Finally, pre-miRNAs are processed by a second RNase III enzyme, Dicer, to a duplex of 21-22nt, which is incorporated into the RNA-induced silencing complex (RISC) (Fig. 1; (3)). MiRNA complementary sites are generally located in the 3′UTR of mRNA, with the 3′ end of the sites (the ‘seed region’) thought to be the most important in determining target specificity (4). Upon miRNA binding, mRNAs are then localized to the processing bodies (P-bodies), where they are either deadenylated and degraded or translationally inhibited (5). The extent of complementarity between miRNAs and their targets may influence whether transcripts are degraded (3).
Fig. 1.
A simplified diagram of miRNA biogenesis, beginning with miRNA transcription in the nucleus and concluding with the loading of the mature miRNA into RISC in the cytoplasm.
MiRNA expression has been detected at various levels in a number of cell types, and miRNAs are particularly abundant in the nervous system, where they have been shown to be key regulators during development as well as in mature neurons (6, 7). A particularly interesting area of research is the role of miRNAs in the development and function of synapses, the structures that mediate communication between nerve cells. Both precursor and mature miRNAs have been identified in synaptic fractions of the mouse brain (8), and a number of recent studies have implicated miRNAs or proteins known to be required for miRNA processing, such as Dicer, in synapse formation or activity. In this review, we will describe the advances stemming from these studies, and propose how they might contribute to a revised molecular model of memory formation and storage.
MiRNAs and mRNA localization
As stated above, miRNAs are first cleaved to pri-miRNAs in the nucleus by Drosha, and then exported to the cytoplasm for further processing by Dicer and incorporation into the RISC complex. It is becoming increasingly clear that the cellular compartment(s) where pre-miRNAs are processed to their mature form could have important consequences for gene regulation. In large part, mRNAs do not freely diffuse throughout the cell, but are instead targeted to specific locations by ribonucleoprotein (RNP) complexes that are often part of larger RNA transport granules (for an excellent recent review of mRNA localization, see (9)). As a consequence, miRNA-mediated regulation must not only depend on its sequence complementarity with target mRNAs, but also on whether they are localized to the same cellular space at the same time. If mature miRNAs are present in the cell body, for instance, they are likely to regulate a distinct (and likely larger) pool of transcripts than if they were found at an individual synapse. Moreover, since miRNAs often function combinatorially like transcription factors, the range of mature miRNAs in a particular site could affect which transcripts are targeted and to what degree they are down-regulated (10).
Multiple mechanisms likely control whether specific mRNAs and miRNAs are co-localized within a neuron. Not only could mature miRNAs bind to mRNAs prior to their recognition by RNA binding proteins (RBPs) for transport. Since both miRNAs and RBPs are known to recognize mRNA transcripts primarily through cis-localization elements in their 3′UTRs, it is also conceivable that the binding of a miRNA/RISC complex to one site could sterically hinder the recognition of a neighboring regulatory site by RBPs, leading to the retention of both the mRNA and mature miRNA in a particular cellular compartment. Alternatively, cis-acting localization tags could be present in either pre-miRNA or mature miRNA sequences, and lead to independent transport of miRNAs to synapses. The possibility that pre-miRNAs could be cleaved at the synapse to form mature miRNAs is supported by the finding that miRNA processing machinery is not localized exclusively to the soma. Immunostaining has revealed that both Dicer and the Argonaute protein eiF2c, a core RISC component, are found in dendritic spines (11), the location of the majority of excitatory synapses in the mature mammalian nervous system (reviewed in (12)). Moreover, Dicer and Argonaute appear to be enriched in post-synaptic densities (PSDs), the thick and electron-dense regions of the post-synaptic membrane (11).
MiRNA processing and the fragile x mental retardation protein
In addition to targeting transcripts to specific regions, including synapses, RBPs also regulate translation of bound mRNAs both during and after transport (reviewed in (13) and (14)). Given the importance of miRNAs in post-transcriptional regulation, an attractive model is that miRNAs function together with RBPs to control the local translation of synaptic RNAs. Indeed, recent studies of the fragile X mental retardation protein (FMRP), which is known to bind RNA lend support to the possibility that local translational inhibition is coordinated by miRNA processing machinery and RBPs.
Fragile X syndrome (FXS), the most common heritable cause of mental retardation, appears to be primarily caused by transcriptional silencing of the FMR1 gene (15, 16). This gene encodes FMRP, which negatively regulates dendritic protein synthesis. Ultimately, the release of FMRP-mediated translational inhibition in FXS is thought to interfere with the normal morphological changes required for memory formation, and while the cognitive deficits observed in FXS patients range in severity, individuals with this disorder generally display deficits in working memory (15).
Several recent studies have linked FMRP function to miRNAs. The Drosophila homolog of FMRP (dFMR1/dFXR) was shown to associate with RISC (17) and to interact with Dicer and the miR-2b miRNA in S2 cells (16, 17) and in vivo, the activity of dFMR1 during growth of larval synapses is controlled at least in part by the RISC protein Argonaute 1 (18). Moreover, in a study of olfactory classical conditioning in Drosophila, protein synthesis-dependent long-term memory (LTM) required the presence of FMRP and its interaction with Argonaute 1, while excess protein synthesis led to memory defects (19). These results suggest that FMRP and miRNAs likely function together to inhibit translation in dendrites. One attractive model is that miRNAs help to identify the proper mRNA transcripts for suppression by FMRP (18). However, few miRNAs that associate with FMRP have been identified, and a complete molecular understanding of how FMRP/Argonaute 1-mediated translational inhibition differs from previous models of FMRP regulation has yet to be elucidated.
MiRNAs and synapse development and function
Although a number of miRNAs have been shown to be important in neuronal development and differentiation (6, 7), little is known about the ways in which miRNAs might direct synapse initiation or maturation. In 2008, evidence emerged that let-7, one of the first miRNAs to be discovered (1), may also play a role in development of Drosophila neuromuscular junctions (NMJs), an important model system for synapse studies (20, 21). In the absence of let-7 as well as the lin-4 homolog miR-125, abdominal NMJ maturation is abnormal, and this can be explained by a failure to down-regulate the abrupt (ab) gene in muscle cells (21). When let-7 and miR-125 single mutants were compared, miR-125 mutants possessed normal NMJs while let-7 mutants continued to display striking defects in NMJ structure, demonstrating that let-7 is responsible for this phenotype (20). Interestingly, members of the murine let-7 family are expressed in neurons and muscles (22, 23), leading to the possibility that they might play a conserved role in synapse development in mammals.
Notably, a second miRNA, miR-1, has been identified for its role after development in maintaining homeostasis in a C. elegans neuromuscular junction (24). In wild-type animals, body wall muscles contract upon binding of acetylcholine (ACh) to nicotinic acetylcholine receptors (nAChRs) at neuromuscular junctions. miR-1 is capable of modulating the muscles’ responsiveness to Ach by targeting two nAChR receptor subunits, unc-29 and unc-63. miR-1 also targets MEF-2, a transcription factor found in C. elegans muscle that modulates RAB-3-dependent ACh secretion through an unknown retrograde signaling mechanism. Ultimately, the authors propose that miR-1 provides a rapid means for altering ACh sensitivity and release in order to maintain balanced synaptic function. In future studies, it will be interesting not only to determine how miR-1 senses changes in ACh responses and transduces the signal to unc-29 and unc-63, but also to identify how MEF-2 signals to the presynaptic neuron to restrict ACh release through RAB-3.
Within the central neuron, the shape, size and number of postsynaptic structures must also be tightly regulated during development and throughout the life of a neuron. miR-134 is a brain-specific miRNA found near synapses in mammalian dendrites that controls dendritic spine size. Schratt et al. showed that over-expression of miR-134 leads to decreased spine width and volume due to the down-regulation of the Lim-domain-containing protein kinase 1 (Limk1), and that BDNF stimulation leads to a release in inhibition by miR-134 and enhanced Limk1 translation (25). The authors suggest that miR-134 might be important for restricting Limk1 translation during its transport to synapses. Surprisingly, miR-134 associates with polysome fractions following BDNF treatment, suggesting that miR-134 continues to bind Limk1 during protein synthesis (25).
In addition to its effects on miR-134 regulation, BDNF has also been shown to trigger the activation of miR-132 transcription by the calcium response element binding protein (CREB), a transcription factor that controls many cellular a variety of adaptive responses in the nervous system (26). miR-132 is required to inhibit translation of P250GAP, a known Rho family GTPase-activating protein, and induce dendritic growth. It is likely that miR-132 targets P250GAP directly, since transfection of a mutant form of P250GAP containing a modified miR-132 target site leads to a decrease in activity-dependent dendritic morphogenesis. The finding that activity leads to the inhibition of one miRNA and activation of another suggests that regulation of RISC and other proteins necessary for miRNA biogenesis and function might be influenced by the specific RNAs to which they are bound. This would allow for the differential expression of distinct populations of miRNA targets prior to and during neuronal stimulation.
Taken together, these studies of Drosophila, C. elegans, and mammalian synapses have illustrated several essential roles that miRNAs play in synapse structure and function. In the future, it will be interesting to characterize functions of many of the other miRNAs known to be expressed in muscles or the nervous system. These studies will require overcoming known obstacles in miRNA research, including the presence of multiple copies or related families of miRNA genes as well as the absence of marked phenotypes - or the presence of lethality - in genetic mutants (4). Moreover, they may mandate identifying mutants for genes known to interact with miRNAs. Several of the studies described in this review (19, 26, 27) have inhibited suspected miRNA targets utilizing a method of gene knockdown termed RNA interference (RNAi), which also relies on RISC for function (28, 29). Potentially oversaturating RNA silencing pathways through RNAi could result in phenotypes that are difficult to interpret or that confound further analysis, as was observed in (30).
MiRNAs and the molecular model of memory
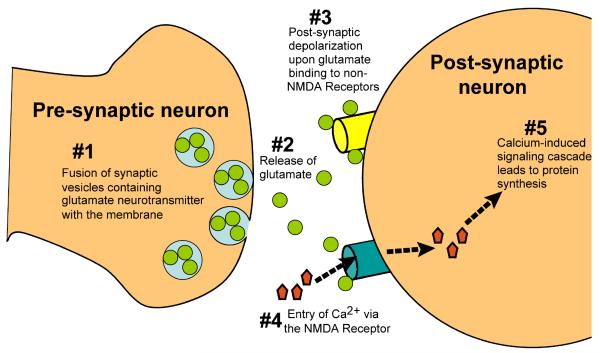
It has long been recognized that new protein synthesis is required for formation of long-term memories (LTMs) (31). Fig. 2 shows a simplified view of the current molecular model of memory, a process reviewed thoroughly by Hernandez and Abel (32). In brief, the long-term strengthening of the synaptic connections between cells which underlies LTM formation, also known as long-term potentiation (LTP), stems from the transduction of electrical stimulation at the postsynaptic membrane into chemical signals, including a rise in intracellular calcium levels and the activation of Ca2+/calmodulin-dependent protein kinase II (CaMKII). These cellular changes ultimately lead to enhanced gene transcription and translation, including synaptic protein synthesis.
Fig. 2.
Long-term memory formation consists of several sequential steps. Following depolarization of the presynaptic neuron, vesicles containing the glutamate neurotransmitter fuse with the presynaptic membrane and glutamate is released (steps 1 and 2). Binding of glutamate to the non-NMDA receptor opens the receptor channel, leading to postsynaptic Na+ influx and membrane depolarization (step 3). Depolarization results in removal of an Mg2+ block in the NMDA receptors, permitting Ca2+ entry into the cell (step 4), CaMKII activation, and ultimately protein synthesis (step 5).
One of the factors known to be involved in local translational control at synapses is cytoplasmic polyadenylation element binding protein (CPEB), which binds to CPE elements in 3′UTRs of target mRNAs and regulates polyadenylation. When CPEB is phosphorylated during neuronal activity, the cleavage and polyadenylation specificity factor (CPSF) is recruited to the target mRNA and translational inhibition is released (33). Strikingly, deletion of CPEB or removal of its phosphorylation site results in disruption of local translational control and defects in memory formation and/or synaptic changes associated with LTP. For a more detailed discussion of CPEB regulation as well as other known local translational control mechanisms, see (34).
Interestingly, the 3′UTR of CaMKII contains binding sites for CPEB, CPSF, and miRNAs in close proximity (35), introducing the possibility that CPEB phosphorylation and release of miRNA inhibition could function redundantly to ensure CaMKII up-regulation following neuronal stimulation. Findings from a 2006 study in Drosophila are consistent with this hypothesis. The authors observed that upon activity, miRNA processing was impaired through the selective degradation of the RBP and RISC component Armitage, leading to enhanced synaptic targeting and translation of CaMKII (36). Moreover, Schratt et al. showed that Limk1 repression by an individual miRNA, miR-134, was also abrogated upon activation of rat synaptoneurosomes by BDNF (25). However, data also suggest that miRNA processing and regulation may not always be inhibited during LTP. miR-132 transcription is increased following BDNF treatment (26), and Lugli et al. described evidence that a rise in intracellular Ca2+ upon postsynaptic stimulation activates the calpain protease, which in turn leads to release of activated Dicer and eIF2c, a known RISC component, from the PSD (11).
If miRNA processing machinery can be degraded in some cases and activated in others, the overlapping miRNA, CPEB, and CPF sites in the CaMKII 3′UTR could provide a means of independently regulating CaMKII with respect to other CPE-containing mRNAs. To investigate this possibility, it will not only be important to validate the physiological relevance of these cis-regulatory elements, but also to further dissect local changes in the translation of individual mRNAs both during and after activity. With the identification of additional synaptic miRNAs and their targets, it will be possible to determine whether they function together with CPEB or play an opposing role during memory formation.
While performing these experiments, it will be important to distinguish whether phenotypes in Dicer and known RISC components are due exclusively to their roles in miRNA biogenesis, or whether the proteins might also regulate independent processes during synaptic plasticity. A recent study by Berdnik et al. lends support to the latter possibility. Through a forward genetic screen in Drosophila, the authors observed that both pasha and Dicer-1 were required for normal olfactory projection neuron morphogenesis (37). Interestingly, mutations in the core RISC components AGO-1 and AGO-2 did not lead to detectable defects. While this could have been due to the persistence of protein from parental cells or the possibility that the AGO1 allele was hypomorphic rather than a true null, an alternative explanation is that pasha and Dicer-1 play pivotal roles in neural development that are unrelated to RISC function.
CONCLUSIONS
In this review, we have presented pioneering work that reveals a new role for miRNAs in synaptic development and function. These studies not only demonstrate that miRNA-mediated inhibition is important for maintaining homeostasis at the neuromuscular junction, but they also show that release of this inhibition can be an important part of synaptic plasticity in the central nervous system. Protein synthesis has long been identified as essential for the formation of long-term memories, and it is therefore exciting, but not entirely surprising, that miRNAs are involved in this process. This is a field that has developed quickly, and with the identification of numerous brain-specific miRNAs in recent years (23), it promises to continue to yield exciting discoveries in the near future.
Acknowledgements
We thank members of the Slack lab for helpful discussions. FJS is supported by grants from the NIH and McDonnell Foundation.
REFERENCES
- 1.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosik KS. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 8.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin KC, Ephrussi A. mRNA Localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem. Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 12.Hering H, Sheng M. Dendritic spines: structure, dynamics and regulation. Nat. Rev. Neurosci. 2001;2:880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 13.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 14.Kiebler MA, DesGroseillers L. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 15.Garber KB, Visootsak J, Warren ST. Fragile X syndrome. Eur. J. Hum. Genet. 2008;16:666–672. doi: 10.1038/ejhg.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 19.Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat. Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 23.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 24.Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 26.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wayman GA, Davare M, ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 29.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 30.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 31.Gold PE. Protein synthesis and memory. Introduction. Neurobiol. Learn. Mem. 2008;89:199–200. doi: 10.1016/j.nlm.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol. Learn. Mem. 2008;89:293–311. doi: 10.1016/j.nlm.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 34.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashraf SI, Kunes S. A trace of silence: memory and microRNA at the synapse. Curr. Opin. Neurobiol. 2006;16:535–539. doi: 10.1016/j.conb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Berdnik D, Fan AP, Potter CJ, Luo L. MicroRNA processing pathway regulates olfactory neuron morphogenesis. Curr. Biol. 2008;18:1754–1759. doi: 10.1016/j.cub.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]