Abstract
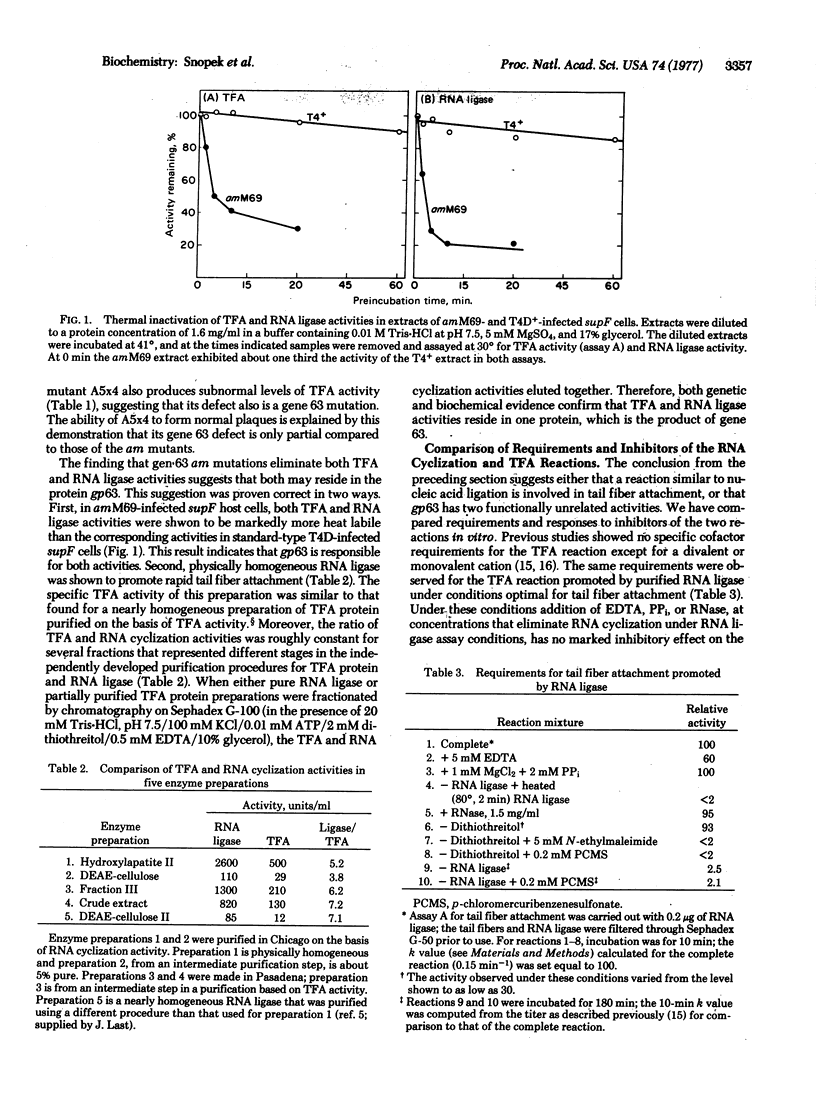
RNA ligase and tail fiber attachment activities, normally induced following bacteriophage T4 infection of Escherichia coli, are not induced when gene 63 amber mutants of T4 infect nonpermissive host cells. Both activities are induced when these mutants infect permissive hosts, or when revertants of these mutants infect nonpermissive hosts. When one of these mutants infects a host that carries supF, both activities are more than normally heat labile. RNA ligase, purified to homogeneity, promotes the tail fiber attachment reaction in vitro with a specific activity similar to that of the most highly purified preparations of gene 63 product isolated on the basis of tail fiber attachment activity. We conclude that T4 RNA ligase is gene 63 product. The RNA ligase and tail fiber attachment reactions differ in requirements and in response to some inhibitors, suggesting that the two activities of the gene 63 product may be mechanistically unrelated.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cranston J. W., Silber R., Malathi V. G., Hurwitz J. Studies on ribonucleic acid ligase. Characterization of an adenosine triphosphate-inorganic pyrophosphate exchange reaction and demonstration of an enzyme-adenylate complex with T4 bacteriophage-induced enzyme. J Biol Chem. 1974 Dec 10;249(23):7447–7456. [PubMed] [Google Scholar]
- Depew R. E., Snopek T. J., Cozzarelli N. R. Characterization of a new class of deletions of the D region of the bacteriophage T4 genome. Virology. 1975 Mar;64(1):144–145. doi: 10.1016/0042-6822(75)90086-0. [DOI] [PubMed] [Google Scholar]
- Dickson R. C. Assembly of bacteriophage T4 tail fibers. IV. Subunit composition of tail fibers and fiber precursors. J Mol Biol. 1973 Oct 5;79(4):633–647. doi: 10.1016/0022-2836(73)90068-5. [DOI] [PubMed] [Google Scholar]
- Edgar R. S., Wood W. B. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci U S A. 1966 Mar;55(3):498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M., Lucas Z. J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXV. Purification and properties of deoxyribonucleic acid polymerase induced by infection with phage T4. J Biol Chem. 1968 Feb 10;243(3):627–638. [PubMed] [Google Scholar]
- Kaufmann G., Kallenbach N. R. Determination of recognition sites of T4 RNA ligase on the 3'-OH and 5' -P termini of polyribonucleotide chains. Nature. 1975 Apr 3;254(5499):452–454. doi: 10.1038/254452a0. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Littauer U. Z. Covalent joining of phenylalanine transfer ribonucleic acid half-molecules by T4 RNA ligase. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3741–3745. doi: 10.1073/pnas.71.9.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last J. A., Anderson W. F. Purification and properties of bacteriophage T4-induced RNA ligase. Arch Biochem Biophys. 1976 May;174(1):167–176. doi: 10.1016/0003-9861(76)90335-0. [DOI] [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Lewis A. S. Purine nucleoside phosphorylase of rabbit liver. Mechanism of catalysis. J Biol Chem. 1977 Jan 25;252(2):732–738. [PubMed] [Google Scholar]
- Ohtsuka E., Nishikawa S., Sugiura M., Ikehara M. Joining of ribooligonucleotides with T4 RNA ligase and identification of the oligonucleotide-adenylate intermediate. Nucleic Acids Res. 1976 Jun;3(6):1613–1623. doi: 10.1093/nar/3.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber R., Malathi V. G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sninsky J. J., Last J. A., Gilham P. T. The use of terminal blocking groups for the specific joining of oligonucleotides in RNA ligase reactions containing equimolar concentrations of acceptor and donor molecules. Nucleic Acids Res. 1976 Nov;3(11):3157–3166. doi: 10.1093/nar/3.11.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snopek T. J., Sugino A., Agarwal K. L., Cozzarelli N. R. Catalysis of DNA joining by bacteriophage T4 RNA ligase. Biochem Biophys Res Commun. 1976 Jan 26;68(2):417–424. doi: 10.1016/0006-291x(76)91161-x. [DOI] [PubMed] [Google Scholar]
- Sugino A., Goodman H. M., Heyneker H. L., Shine J., Boyer H. W., Cozzarelli N. R. Interaction of bacteriophage T4 RNA and DNA ligases in joining of duplex DNA at base-paired ends. J Biol Chem. 1977 Jun 10;252(11):3987–3994. [PubMed] [Google Scholar]
- Tessman I. Mutagenic treatment of double- and single-stranded DNA phages T4 ans S13 with hydroxylamine. Virology. 1968 Jun;35(2):330–333. doi: 10.1016/0042-6822(68)90275-4. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Cameron V. Equimolar addition of oligoribonucleotides with T4 RNA ligase. Nucleic Acids Res. 1977 Jan;4(1):85–98. doi: 10.1093/nar/4.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderslice R. W., Yegian C. D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology. 1974 Jul;60(1):265–275. doi: 10.1016/0042-6822(74)90384-5. [DOI] [PubMed] [Google Scholar]
- Walker G. C., Uhlenbeck O. C., Bedows E., Gumport R. I. T4-induced RNA ligase joins single-stranded oligoribonucleotides. Proc Natl Acad Sci U S A. 1975 Jan;72(1):122–126. doi: 10.1073/pnas.72.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Luftig R. B., Wilson J. H., Eddleman H., Lyle H., Wood W. B. Assembly of bacteriophage T4 tail fibers. II. Isolation and characterization of tail fiber precursors. J Mol Biol. 1970 Nov 28;54(1):15–31. doi: 10.1016/0022-2836(70)90443-2. [DOI] [PubMed] [Google Scholar]
- Wilson J. H. Function of the bacteriophage T4 transfer RNA's. J Mol Biol. 1973 Mar 15;74(4):753–757. doi: 10.1016/0022-2836(73)90065-x. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Henninger M. Attachment of tail fibers in bacteriophage T4 assembly: some properties of the reaction in vitro and its genetic control. J Mol Biol. 1969 Feb 14;39(3):603–618. doi: 10.1016/0022-2836(69)90148-x. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]


