Abstract
The post-translational modification of proteins with ubiquitin can take on many forms, including the decoration of substrates with polymeric ubiquitin chains. These chains are linked through one of the seven lysine residues in ubiquitin, with the potential to form a panoply of linkage combinations as the chain length increases. The ensuing structural diversity of modifications serves a variety of signaling functions. Still, some linkages are present at a much higher level than others in cellulo. Although ubiquitination is an enzyme-catalyzed process, the large disparity of abundancies led us to the hypothesis that some linkages might be intrinsically faster to form than others, perhaps directing the course of enzyme evolution. Herein, we assess the kinetics of ubiquitin dimer formation in an enzyme-free system by measuring the rate constants for thiol–disulfide interchange between appropriate ubiquitin variants. Remarkably, we find that the kinetically expedient linkages correlate with those that are most abundant in cellulo. As the abundant linkages also appear to function more broadly in cellulo, this correlation suggests that the more accessible chains were selected for global roles.
Keywords: mixed disulfide, protein–protein interaction, thiol-disulfide interchange, ubiquitin
Introduction
Ubiquitin is a small, globular, and structurally robust protein that is highly conserved among eukaryotes. The structure, referred to as a β-grasp, consists of a five-stranded β-sheet and an α-helix, with a flexible tail of four residues protruding at the C terminus [Fig. 1(A)].1 The addition of ubiquitin chains to proteins serves as an important post-translational modification in eukaryotic cells.2–6 Ubiquitin is attached to a target protein through an isopeptide bond between the C-terminal glycine residue of ubiquitin and the ε-nitrogen of a lysine residue on the target protein or another ubiquitin. In cellulo, the formation of the isopeptide bond occurs through the concerted actions of three enzymes, known generally as the ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin–protein ligases (E3).
Figure 1.
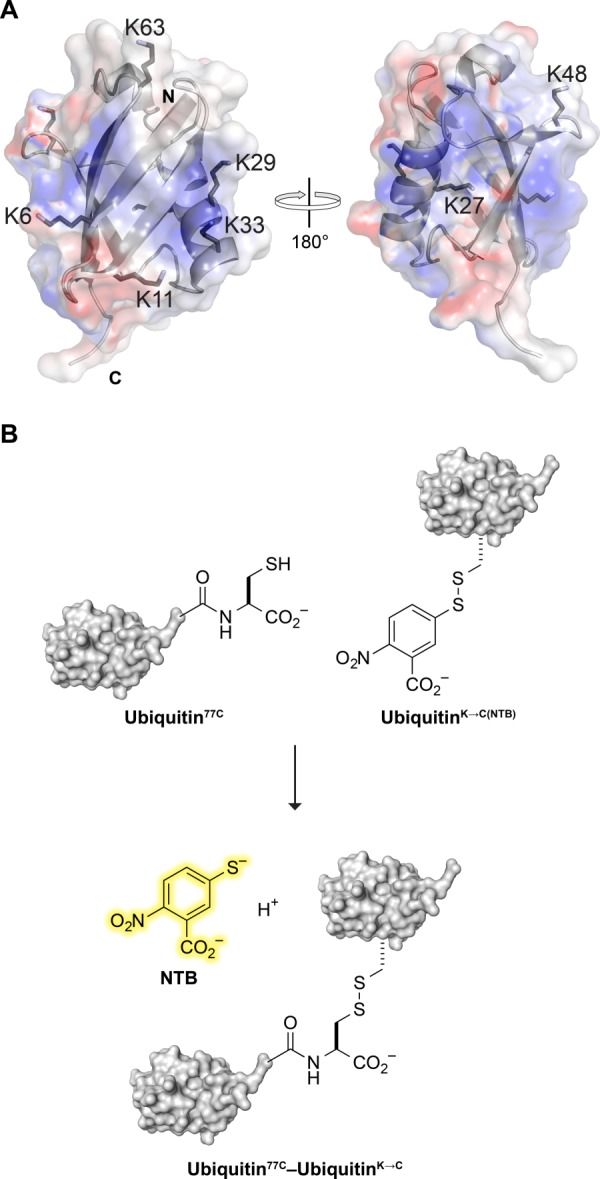
Ubiquitin dimer formation. (A) Ribbon diagram of ubiquitin showing the location of its 7 lysine residues and N-and C-termini. The translucent surface depicts electrostatic potential (blue: positive; red: negative). The image was created with the program PyMOL and PDB entry 1ubq.45 (B) Scheme for a chromogenic assay of ubiquitin dimer formation. Thiol–disulfide interchange between a ubiquitin variant with a C-terminal cysteine residue and an NTB mixed disulfide of a K→C ubiquitin variant releases NTB.
Ubiquitin contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) that can serve as sites of attachment for additional ubiquitin molecules [Fig. 1(A)].7,8 Recent studies of ubiquitin polymers have found a diverse set of ubiquitin linkages present in cellulo, including all seven homopolymeric ubiquitin chains.7 A comprehensive analysis revealed that the various linkages exist in vastly different abundances, with K48 (29%), K11 (28%), and K63 (16%) being the most plentiful linkages, and K6 (11%), K27 (9%) K33 (4%), and K29 (3%) being more rare.8
The consequences for a ubiquitin-modified target depend on the architecture of the attachment.9–13 Perhaps the most well-studied modification, K48-linked chains, are known to serve as potent signals for proteasomal degradation.14 In contrast, K63-linked chains are not related to proteasomal degradation, but instead serve roles in DNA repair and cytokine signaling.15,16 K11-linked chains are believed to play an important role in marking substrates for degradation in endoplasmic reticulum-associated degradation (ERAD), and in cell-cycle regulation.8,17–19 Less is known about the functions of the rare linkages through K6, K27, K29, and K33. Formation of the K6 linkage is known to be catalyzed by the BRCA1, providing a potential function in DNA damage response.20,21 K27 has been linked tentatively to the protein kinase C signaling pathway.22 Relatedly, K29-and K33-linked chains have been associated with the regulation of AMP-activated protein kinase (AMPK)-related kinases.23
The different isopeptide linkages impart different quaternary structure on the polyubiquitin chains. Known three-dimensional structures indicate that K63-linked chains take on an “extended” conformation, whereas K6-, K11-, and K48-linked chains adopt “compact” conformations in which the ubiquitin moieties packed together tightly.24–27 The different orientations of the ubiquitin chains display different surface patches to proteins containing ubiquitin-binding domains,11,28 and presumably enable linkages to play distinct roles.
The variations in abundance and function associated with the isopeptide linkages raises the question: Why are particular linkages used for particular functions? One possibility is that certain linkages are inherently accessible, and that cellular machinery evolved to use these linkages for functions of special importance. The process of ubiquitination is dictated by the concerted actions of the E1, E2, and E3 enzymes. Of these enzymes, E2 and E3 have the potential to impart linkage specificity. These enzymes do so by positioning the ubiquitins to favor the attachment of the donor ubiquitin to a particular lysine of the acceptor ubiquitin. Although some of these enzymes are known to impart only particular linkages, others are promiscuous. For example, E2 Ubc5 is known to catalyze the formation of all seven possible linkages.29
We hypothesized that particular ubiquitin–ubiquitin linkages are inherently faster to form than others, and that these linkages are used preferentially by the cell to signal critical events. To test this hypothesis, we developed an enzyme-free system to model dimer formation. As disulfide bonds had been used previously to mimic the covalent attachment of ubiquitin,30–33 we used disulfide bonds as a proxy for isopeptide linkages. Here, we measure the rate constants for the formation of all seven ubiquitin dimers. The resulting data support our hypothesis, and have implications for the evolution of ubiquitination.
Results
Rate constants of dimerization
All seven lysine-to-cysteine ubiquitin variants were produced in E. coli and purified in a yield of 3–30 mg L−1, with UbiquitinK27C providing the lowest yield. Purified variants were treated with DTNB and characterized by MALDI–TOF (Supporting Information Fig. S1). We could not produce ubiquitin with an additional C-terminal cysteine residue in E. coli, even with the aid of a protease inhibitor cocktail (data not shown), suggesting an inherent instability of this variant. To overcome this problem, the C-terminal glycine residues were replaced with two alanine and a cysteine residue. The resulting protein, Ubiquitin77C, was purified from E. coli in a yield of 50 mg L−1. Only the thiolate form of a cysteine residue is nucleophilic in water.34 Accordingly, we choose Ubiquitin77C as the nucleophile and UbiquitinK→C(NTB) as the electrophile (rather than the inverse) to keep the nucleophile constant and thereby eliminate the effect of nucleophile pKa on the reaction rate. The AlaAlaCys peptide was synthesized to mimic the C terminus of Ubiquitin77C, but control for issues of either favorable or unfavorable interactions that might arise with a globular protein.
Plotting vo versus [Ubiquitin77C] (or [AlaAlaCys]) as in Eq. 2 generated statistically significant slopes for all UbiquitinK→C(NTB) variants, with R2 > 0.85 in all cases [Fig. 2(A)]. The y-intercepts did not differ significantly from zero, indicating a good fit to Eq. 1. The high quality of the fit to a second-order rate equation precludes a first-order reaction from the noncovalent association of ubiquitin monomers under the experimental conditions. After the kinetic assays, the formation of each Ubiquitin77C–UbiquitinK→C dimer was verified by MALDI–TOF mass spectrometry.
Figure 2.
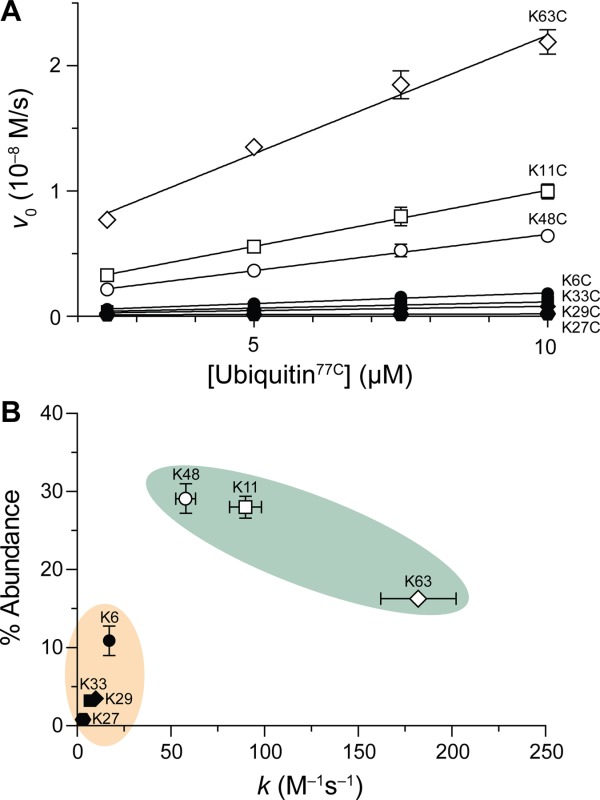
Intrinsic rate of ubiquitin dimer formation. (A) Graph of the rates of formation of ubiquitin dimers between Ubiquitin77C (0–10 µM) and varying concentrations of a UbiquitinK→C(NTB) variant (10 µM). Data are for the release of 5-thio-2-nitrobenzoate during the reaction shown in Figure 1B. (B) Plot of the natural abundance of linkages8 versus dimerization rate constants. Green shading indicates linkages that form quickly and are common in cellulo; orange shading indicates linkages that form slowly and are rare in cellulo.
The K63C variant showed the fastest rate of dimer formation of the seven [Fig. 2(A)]. K11C and K48C were also fast to dimerize. The K6C, K29C, K27C, and K33C variants were all appreciably slower to form dimers. Ubiquitin77C was faster to dimerize with each of the seven UbiquitinK→C(NTB) variants than was AlaAlaCys (Supporting Information Table SI).
Comparing dimerization rate to abundance in cellulo
Given the observation from the kinetics data that some linkages were significantly slower to form than others, we compared these rates to the percent abundance of these linkages in cellulo as determined by mass spectrometry. For this comparison, we used abundancies in yeast,8 as we used yeast ubiquitin in our experiments. Although linkage abundancies can vary between cell type and treatment conditions, other studies corroborate the data in yeast.7,35
We observed that the linkages fall into two categories: fast/abundant or slow/rare [Fig. 2(B)]. Our definition of “abundant” is based upon whether the proportion of each linkage in cellulo was above 14%, which is the occurrence that would be observed if the seven linkages were distributed evenly. The statistical partitioning of these linkages into these two groups was confirmed by a cluster analysis partitioning around medoids.36,37
Solvent-accessible surface area
To determine if the reactivity we observed was related to the accessibility of the residues, we calculated the solvent-accessible surface area (SASA) with of the native lysine residues as well as the substituted cysteine residues [Fig. 3(A)]. For both types of residue, position 63 was calculated to be very highly exposed, which correlates with our observation that this residue was the most reactive. Positions 11 and 48 were likewise highly exposed and reactive. In contrast, although position 33 was calculated to be highly exposed, it was among the slowest to react. Positions 6 and 29 have an intermediate level of exposure, but showed reactivity similar to that of buried position 27. Interestingly, positions 27, 29, and 33 are located on the α-helix of ubiquitin [Fig. 1(A)], and show similar reactivity despite having varying levels of solvent accessibility.
Figure 3.
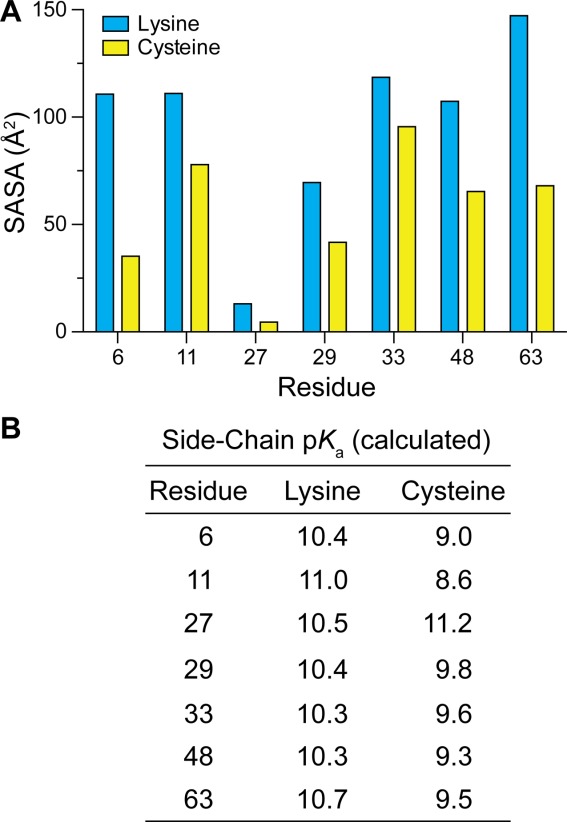
Parameters relevant for the intrinsic rate of ubiquitin dimer formation. (A) Calculated solvent-accessible surface area (SASA) values for lysine side chains in wild-type ubiquitin and cysteine side chains in K→C variants. (B) Calculated values of pKa for lysine side chains in wild-type ubiquitin and cysteine side chains in K→C variants.
Side-chain pKa values
Values of pKa for the ammonium group of lysine residues and thiol group of cysteine residues were calculated with the program PROPKA.38 There was little variability in the calculated pKa values, aside from that of buried residue Cys27.
Discussion
Using disulfide bond formation to mimic ubiquitin dimerization, we find that certain linkages are inherently faster to form. This differential reaction rate could be attributed to several factors. Because ubiquitin is a globular protein, we suspected that accessibility of the seven positions would play an important role in the rate of linkage formation. SASA calculations on the crystalline protein do provide a partial explanation for the relative reaction rates. The nucleophile in the fastest (K63) and slowest (K27) reacting variants had the most and least solvent exposure in the solid state. Other linkages, however, adhered less well to this constraint.
The seven UbiquitinK→C(NTB) variants reacted more quickly with Ubiquitin77C than with the AlaAlaCys peptide (Supporting Information Table SI). This finding is also consistent with unfavorable steric interactions playing a minor role in dimer formation. Rather, the faster rate of each protein–protein reaction is consistent with favorable interactions between the two ubiquitin monomers. Indeed, a noncovalent ubiquitin·ubiquitin complex has been detected in solution, and shown to have an equilibrium dissociation constant of Kd = 5 mM.39 Our experiments were performed at a ubiquitin concentration of 10 µM, which is the endogenous concentration of ubiquitin in human cells.40 Under our conditions, the rate of dimer formation did not deviate significantly from a second-order rate equation [Eq. 1], and we could not detect the formation of a noncovalent complex despite the intrinsic affinity.39
The amino group of a lysine residue is the relevant nucleophile in cellulo. The relative rates of lysine Nε acetylation are known to be K6 ≈ K48 ≈ K63 > K33 > K11 > K27 ≈ K29.41 Likewise, the K27C and K29C variants were slow to dimerize in our system [Fig. 2(A)]. K6 was acetylated more readily than anticipated from our data, and K11 was less so. Calculated pKa values of both lysine and cysteine residues suggests little differentiation, aside from the high pKa for the unreactive K27C variant [Fig. 3(B)]. Although neither solvent exposure nor pKa provides a complete explanation for the reactivity observed in our study, each likely contributes to that reactivity.
The covalent attachment of ubiquitin or ubiquitin-like proteins is the only known post-translational modification of a protein with another protein.42 Thus, the formation of a ubiquitin dimer is constrained by unique steric demands. In hydrogen–deuterium exchange experiments, the main-chain amides of K11, K48, and K63 have been found to be more susceptible to exchange than those of the other four lysine residues.43,44 This finding is consistent with K11, K48, and K63 being in surface loops, whose flexibility could relieve steric constraints during a reaction with another ubiquitin monomer. In contrast, the other lysine residues reside in highly ordered elements of secondary structure (i.e., α-helices and β-sheets).45 Accordingly, the kinetic data [Fig. 2(A)] correlate with the accessibility of each lysine residue to solvent and thus its availability for a chemical reaction.
We found another correlation, one between the rate of formation of a dimer and the abundance of its linkage in cellulo. Most notably, the linkages were clearly divisible into two groups: fast/abundant, or slow/rare [Fig. 2(B)]. Interestingly, these groupings relate to the types of functions that these linkages serve. For example, fast/abundant linkages serve global functions in the cell, with K11 and K48 playing large roles in directing substrates for proteasomal degradation, and K63 for repair pathways.46,47 The K11R variant results in hypersensitivity to ER stress, reflecting the important role K11-linked chains play in ERAD.8 The K48R substitution is known to be lethal in budding yeast, evidencing the critical nature of this connectivity.48 The phenotype induced by the K63R variant is likewise prominent—a hypersensitivity to DNA damage.49 In comparison, the linkages categorized as slow/rare appear to have more specific functions. To date, linkages to K6, K27, K29, and K33 have been found in only a few substrates, and the biological function of these linkages appears to be less consequential than those to K11, K48, and K63.46,47
The site-selectivity of the cellular ubiquitination machinery also supports our hypothesis. The human genome encodes ∼35 E2 and >600 E3 enzymes. E2s are abundant in human cells (>4 × 104 molecules/cell)50 and take on broad roles in comparison to E3s. Those E2s that act in a linkage-specific manner build K11, K48, and K63 linkages. In comparison, E3s tend to be less abundant, and known linkage-specific E3s forge rare isopeptide bonds. For example, the E3s BRCA1,20,21 Parkin,51 KIAA10,52 and Cul3-KLHL2053 generate K6-, K27-, K29-, and K33-linked chains, respectively.
Conclusions
We provide the first data on the intrinsic ability of a ubiquitin dimer to form without the imposition of an enzyme. Nonetheless, we do not ignore that ubiquitination is an enzyme-catalyzed process in eukaryotic cells. In that light, we propose that the complex cellular ubiquitination machinery, comprising hundreds of enzymes, evolved to enhance differential native rates of dimerization. Further, the correlation between the rate at which a dimer forms and the natural abundance of its analogous isopeptide suggests that the more readily formed linkages were recruited for the most broadly important cellular functions, while other linkages were destined for more narrow roles.
Methods
Chemicals
Ultrapure water with a resistivity of ≥18 MΩ cm−1 was generated with an Arium Pro water purification system from Sartorius (Bohemia, NY). The 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) and HPLC-grade solvents were from Sigma–Aldrich (St. Louis, MO). cDNA encoding Saccharomyces cerevisiae ubiquitin was codon-optimized for expression in Escherichia coli and synthesized by Bio Basic (Toronto, Canada). The vector pTXB, and NdeI and SapI restriction enzymes were from New England BioLabs (Ipswich, MA).
Instrumentation
The mass of each ubiquitin variant and dimer was confirmed by matrix-assisted laser desorption/ionization time-of-flight (MALDI–TOF) mass spectrometry with a Voyager-DE-PRO Biospectrometry Workstation from Applied Biosystems (Foster City, CA). Absorbance measurements were made with an infinite M1000 plate reader from Tecan (Männedorf, Switzerland). Rate constants were calculated with Prism 6 software from GraphPad (La Jolla, CA).
Protein production
cDNA encoding S. cerevisiae ubiquitin was inserted into the pTXB expression vector between the NdeI and SapI sites. Site-directed mutagenesis with the QuickChange kit from Agilent Technologies (Santa Clara, CA) was used to generate Ubiquitin77C, in which Gly75 and Gly76 are replaced with alanine residues and a cysteine residue is added to the C terminus. Site-directed mutagenesis was also used to create all seven individual lysine-to-cysteine variants, UbiquitinK→C. Constructs were verified by Sanger sequencing at the University of Wisconsin–Madison Biotechnology Center Sequencing Facility. For protein production, plasmids were transformed into competent BL21(DE3) cells and grown for 6 h at 37°C and then 23°C for 12 h in an autoinduction medium containing ampicillin (100 µg mL−1).54
Protein purification
Proteins were purified by methods similar to those described previously.55 Briefly, E. coli cell pellets were resuspended in lysis buffer (which was 50 mM Tris–HCl buffer, pH 7.6, containing 1 mM TCEP) and lysed at 22 kPSI in a T Series Cell Disrupter 2.2 kW from Constant Systems Limited (Northants, UK). The debris was cleared by centrifugation at 15,000g for 45 min at 4°C. Precipitation with 0.5% v/v perchloric acid removed the majority of other proteins. The slurry was clarified by centrifugation at 8000g for 20 min at 4°C, and the supernatant was dialyzed against 50 mM sodium acetate buffer, pH 5.0, overnight at 4°C. Ubiquitin variants were purified by cation-exchange chromatography using a HiTrap SP HP column and an ÄKTA system from GE Healthcare (Piscataway, NJ) with a linear gradient of NaCl (0.00–1.00 M) in 50 mM sodium acetate buffer, pH 5.0. Ubiquitin variants were characterized by SDS–PAGE and MALDI–TOF mass spectrometry, and were stored under Ar(g) at 4°C until their use.
Semisynthesis of UbiquitinK→C(NTB) variants
The UbiquitinK→C variants contain a single cysteine residue, which was reacted with DTNB to form a mixed disulfide with 2-nitro-5-thiobenzoate (NTB). Purified UbiquitinK→C variants were incubated overnight at 4°C with 1.25 mM DTNB in 125 mM HEPES–NaOH buffer, pH 8.0, containing EDTA (12.5 mM) before dialysis against 50 mM sodium acetate buffer, pH 5.0. The dialyzed solution of UbiquitinK→C(NTB) variants was purified via strong cation-exchange (vide supra) and stored under argon at 4°C.
Peptide synthesis
The tripeptide AlaAlaCys was synthesized by segment condensation of the corresponding Fmoc-protected amino acids on a solid phase using a Prelude peptide synthesizer from Protein Technologies (Tuscon, AZ) at the University of Wisconsin–Madison Biotechnology Center. The resin was preloaded Cys(Trt)−2-chlorotrityl resin (0.47 mmol g−1). Fmoc-deprotection was achieved by treatment with a solution of piperidine (20% v/v) in N,N-dimethylformamide. The added amino acid (4 equiv) was converted to an activated ester with HCTU and N-methylmorpholine. Each residue was double-coupled between Fmoc-deprotection steps. Peptide was cleaved from the resin with 5 mL of 85.5:5.0:5.0:5.0:2.5 trifluoroacetic acid/phenol/water/thioanisole/1,2-ethanedithiol, precipitated from ethyl ether at 0°C, and isolated by centrifugation. Semi-preparative HPLC was used to purify the peptide (gradient: 10–50% B over 60 min, where buffer A was H2O containing trifluoroacetic acid (0.1% v/v) and buffer B was acetonitrile containing trifluoroacetic acid (0.1% v/v). A 100-µM scale synthesis yielded 0.013 g (49.3%) of purified peptide. LC/MS [M+H]+: calc'd 264.31, found 264.05. 1H NMR (600 MHz, Methanol-d4) δ 8.29 (d, J = 7.9 Hz, 1H), 4.66–4.61 (m, 1H), 4.48 (q, J = 7.1 Hz, 1H), 3.96 (q, J = 7.0 Hz, 1H), 3.01 (dd, J = 14.0, 4.4 Hz, 1H), 2.92 (dd, J = 14.0, 6.3 Hz, 1H), 1.55 (d, J = 7.1 Hz, 3H), 1.43 (d, J = 7.2 Hz, 3H). 13C NMR (126 MHz, Methanol-d4) δ 175.83, 174.01, 172.04, 57.12, 51.75, 51.33, 27.95, 19.14, 18.88.
Kinetics assays of dimerization
We developed a chromogenic assay for the formation of ubiquitin dimers [Fig. 1(B)]. This assay uses two types of ubiquitin variants. The nucleophilic variant (Ubiquitin77C) has a C-terminal cysteine residue. The electrophilic variants (UbiquitinK→C(NTB)) were the seven individual lysine-to-cysteine variants as mixed disulfides with NTB. Attack of the nucleophilic thiolate forms a ubiquitin dimer in which the disulfide linker is only one atom longer than the isopeptide linker of native dimers. The simultaneous production of NTB enables monitoring by absorbance at 412 nm (ε = 14,150 M−1 cm−1),56 providing an expedient measurement of the rate of ubiquitin dimerization.
Kinetic assays were performed at 25°C in 50 mM HEPES–NaOH buffer, pH 8.0, containing EDTA (0.10 mM) and NaCl (100 mM). A UbiquitinK→C(NTB) variant (10 µM) was reacted with Ubiquitin77C (0–10 µM) or AlaAlaCys (0–10 µM) for 10 min, while the absorbance at 412 nm was recorded continuously. Assays were performed in duplicate, and data were averaged across three trials.
Data were assumed to fit to a second-order rate equation, as in Eq. 1
| 1 |
Equation 2 was used to determine the value of kobs as the slope of the line of the initial velocity (v0) for varying concentrations of Ubiquitin77C.
| 2 |
Finally, Eq. 3 was used to determine the value of the second-order rate constant k from those of kobs and [UbiquitinK→C(NTB)] = 10 µM.
| 3 |
For the analysis of AlaAlaCys reactivity, [AlaAlaCys] replaced [Ubiquitin77C] in Eqs. 1 and 2.
Solvent-accessible surface area in ubiquitin
The solvent-accessible surface area (SASA) of lysine residues in wild-type ubiquitin and cysteine residues (with a conserved χ1 angle) in UbiquitinK→C variants was calculated with the program PyMOL from Schrödinger (New York, NY) using PDB entry 1ubq45 and a probe size of 1.4 Å2.
Acknowledgments
The authors thank A. J. Ellison for synthesis and characterization of the AlaAlaCys peptide, Prof. E. R. Strieter, J. D. Vasta, and R. W. Newberry for contributive discussions, and C. Cook for assistance with statistical analyses, all from the University of Wisconsin–Madison.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J Biol Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- Rose I. Ubiquitin at Fox Chase (Nobel lecture) Angew Chem Int Ed. 2005;44:5926–5931. doi: 10.1002/anie.200500995. [DOI] [PubMed] [Google Scholar]
- Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture) Angew Chem Int Ed. 2005;44:5932–5943. doi: 10.1002/anie.200501724. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin–proteasome system, and onto human diseases and drug targeting. Angew Chem Int Ed. 2005;44:5944–5967. doi: 10.1002/anie.200501428. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Spasser L, Brik A. Chemistry and biology of the ubiquitin signal. Angew Chem Int Ed. 2012;51:6840–6862. doi: 10.1002/anie.201200020. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Ye Y, Blaser G, Horrocks MH, Ruedas-Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492:266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S, Dikic I. Selectivity of the ubiquitin-binding modules. FEBS Lett. 2012;586:2705–2710. doi: 10.1016/j.febslet.2012.04.053. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Chem Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. K11-Linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Bremm A, Komander D. Emerging roles for Lys11-linked polyubiquitin in cellular regulation. Trends Biochem Sci. 2011;36:355–363. doi: 10.1016/j.tibs.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Morris JR, Solomon E. BRCA1:BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet. 2004;13:807–817. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Ooka S, Sato K, Arima K, Okamoto J, Klevit RE, Fukuda M, Ohta T. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1–BARD1 ubiquitin ligase. J Biol Chem. 2004;279:3916–3924. doi: 10.1074/jbc.M308540200. [DOI] [PubMed] [Google Scholar]
- Okumura F, Hatakeyama S, Matsumoto M, Kamura T, Nakayama KI. Functional regulation of FEZ1 by the U-box-type ubiquitin ligase E4B contributes to neuritogenesis. J Biol Chem. 2004;279:53533–53543. doi: 10.1074/jbc.M402916200. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- Bremm A, Freund SMV, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat Chem Biol. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Abeywardana T, Dhall A, Marotta NP, Varkey J, Langen R, Chatterjee C, Pratt MR. Semisynthetic, site-specific ubiquitin modification of α-synuclein reveals differential effects on aggregation. J Am Chem Soc. 2012;134:5468–5471. doi: 10.1021/ja300094r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar RA. Reactivity and pH dependence of thiol conjugation to N-ethylmaleimide: detection of a conformational change in chalcone isomer. Biochemistry. 1990;29:3684–3690. doi: 10.1021/bi00467a014. [DOI] [PubMed] [Google Scholar]
- Phu L, Izrael-Tomasevic A, Matsumoto ML, Bustos D, Dynek JN, Fedorova AV, Bakalarski CE, Arnott D, Deshayes K, Dixit VM, Kelley RF, Vucic D, Kirkpatrick DS. Improved quantitative mass spectrometry methods for characterizating complex ubiquitin signals. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.003756. .003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2011. ) R: a language and environment for statistical computing. The R Foundation for Statistical Computing, Vienna, Austria. ISBN: 3–900051-07-0. Available at: http://www.R-project.org/. Accessed: November 6, 2014.
- Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. 2014. ) Cluster: cluster analysis basics and extensions. R package version 1.15.3.
- Rostkowski M, Olsson MHM, Søndergaard CR, Jensen JH. Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct Biol. 2011;11:6. doi: 10.1186/1472-6807-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang W-P, Xing Q, Ren X, Liu M, Tang C. Noncovalent dimerization of ubiquitin. Angew Chem Int Ed. 2003;51:469–472. doi: 10.1002/anie.201106190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielak GJ, Li C, Miklos AC, Schlesinger AP, Slade KM, Wang GF, Zigoneanu IG. Protein nuclear magnetic resonance under physiological conditions. Biochemistry. 2009;48:226–234. doi: 10.1021/bi8018948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Kruppa GH, Young MM, Schoeniger J. A top-down method for the determination of residue-specific solvent accessibility in proteins. J Mass Spectrom. 2004;39:322–328. doi: 10.1002/jms.587. [DOI] [PubMed] [Google Scholar]
- Walsh CT. Posttranslational modification of proteins: expanding nature's inventory. Greenwood Village, CO: Roberts and Company; 2006. [Google Scholar]
- Sterling HJ, Williams ER. Real-time hydrogen/deuterium exchange kinetics via supercharged electrospray ionization tandem mass spectrometry. Anal Chem. 2010;82:9050–9057. doi: 10.1021/ac101957x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Abzalimov RR, Bobst CE, Kaltashov IA. Conformer-specific characterization of nonnative protein states using hydrogen exchange and top-down mass spectrometry. Proc Natl Acad Sci USA. 2013;50:20087–20092. doi: 10.1073/pnas.1315029110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Iwai K. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life. 2004;56:193–201. doi: 10.1080/1521654042000223616. [DOI] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitin-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claasen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol Systems Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsa N, Norkett R, Wauer T, Mevissen TET, Wu H-C, Foltynie T, Bhatia K, Hirst WD, Komander D, Plun-Favreau H, Kittler JT. Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem. 2014;289:14569–14582. doi: 10.1074/jbc.M114.563031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W-C, Lee Y-R, Lin S-Y, Chang L-Y, Tan YP, Hung C-C, Kuo J-C, Liu C-H, Lin M-Y, Xu M, Chen ZJ, Chen R-H. K33-Linked polyubiquitination of coronin 7 by Cul3–KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol Cell. 2014;54:586–600. doi: 10.1016/j.molcel.2014.03.035. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Express Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


