Abstract
Despite of the common usage of glucocorticoids (GCs), a significant portion of asthma patients exhibit GC insensitivity. This could be mediated by diverse mechanisms, including genomics. Recent work has suggested that measuring changes in gene expression may provide more predictive information about GC insensitivity than baseline gene expression alone, and that expression changes in peripheral blood may be reflective of that in the airway. Through gene expression omnibus (GEO) analysis, we identified IRF1 whose expression is affected by GC treatment in airway smooth muscle (ASM) cells, normal human bronchial epithelial (NHBE) cells, and lymphoblastoid cell lines (LCLs). Significant IRF1 down-regulation post GC exposure was confirmed in two cultured airway epithelial cell lines and primary NHBE cells (p<0.05). We observed large inter-individual variation for GC induced IRF1 expression changes among primary NHBE cells tested. Significant down-regulation of IRF1 was also observed in six randomly selected LCLs (p<0.05) with variable degree of down-regulation among different samples. In peripheral blood mononuclear cells (PBMCs) obtained from healthy volunteers, variable down-regulation of IRF1 by GC was also shown. NFKB1, a gene whose expression is known to be down-regulated by GC and the degree of down-regulation reflective of GC response, was used as a control in our study. IRF1 shows more consistent down-regulation across tissue types when compared to NFKB1. Our results suggest GC induced IRF1 gene expression changes in peripheral blood could be used as a marker to reflect GC response in the airway.
Keywords: glucocorticoid, gene expression, airway, blood
INTRODUCTION
Asthma is a chronic condition that affects approximately 300 million people globally and kills 250,000 people annually [1]. Inhaled glucocorticoids (GCs) are the most effective medication for the control of persistent asthma, and oral GCs are the first line treatment for acute asthma exacerbations [2]. Despite the broad utility of these agents and the proven efficacy in the general population, individual patient’s responses to GC treatment vary significantly. For example, estimates of asthma cases exhibiting GC insensitivity range from 10% (in all patients with asthma) to 25% (in adolescents with severe asthma) [3, 4], making GC insensitivity a significant cause of asthma morbidity and mortality. Further, large inter-individual differences have been observed for the time/dosage needed to achieve disease exacerbation control. In addition to the negative effects on patient health, it is estimated that 2/3 of the $14 billion spent on asthma in the U.S. annually is for the sickest 15% of patients in emergency room and hospital settings. Many of these visits could be prevented by early identification of GC resistant patients and the use of an alternative therapy [5].
Like asthma, GC insensitivity is a complex human trait mediated by diverse mechanisms, including genetics [4, 6–14]. Considerable efforts have been dedicated to identify genes whose expression can be used as a marker of GC sensitivity. For example, one recent study used a 6-gene expression signature from sputum samples to predict inhaled corticosteroid sensitivity [15]. While it is ideal to evaluate gene expression at the site of GC action, e.g., airway tissues, direct airway sampling via sputum induction or bronchoscopy is technically challenging, uncomfortable for patients and often impractical in clinical practice [16]. Human peripheral blood on the other hand is readily accessible. A recent study in patients with idiopathic pulmonary fibrosis found that four gene expression levels quantified in patients’ peripheral blood mononuclear cells (PBMCs) were predictive of poor outcome from the disease [17]. Asthma researchers are beginning to use PBMCs as a surrogate model system for GC response marker discovery [5, 18, 19]. Indeed, Hakonarson et al. reported that baseline expression of 11 genes in PBMCs correctly predicted GC response phenotype in 84% of patients in an independent sample, with baseline expression of NFKB1 alone making an accurate prediction in 81.25% of patients [5].
Despite the easy access of PBMCs from patients, tissue specific gene expression remains a major concern when using this model for drug response prediction in the airway. In this study, we proposed to study gene expression changes in response to GC treatment in PBMCs. Our hypothesis is that certain gene expression changes are drug specific and tissue independent; while baseline gene expression levels often differ among tissue types, GC-induced expression changes can be shared among different tissues for certain genes/pathways. This hypothesis is supported by a previous report that change in NFKB1 expression following ex vivo dexamethasone treatment was significantly correlated with lymphocyte GC sensitivity, and that individuals with greater NFKB1 down-regulation showed a greater lymphocyte GC sensitivity [19].
In this study, we evaluated gene expression changes in multiple tissue types using existing genome level high throughput datasets to identify candidate markers. Furthermore, we evaluated these markers in airway derived primary cells and cell lines as well as peripheral blood derived material to confirm that expression changes in peripheral blood were reflective of expression changes in the airway and to elucidate the potential predictive value of these markers.
METHODS
Gene target selection
We examined publically available high throughput data sets (from gene expression omnibus (GEO)) that assayed gene expression in control and dexamethasone treated samples using microarray. Three data sets were evaluated. They are airway smooth muscle (ASM) cells (treated with 1μM dexamethasone for 4 and 24 hours; GSE34313), normal human bronchial epithelial (NHBE) cells (treated with 1μM dexamethasone for 8 and 24 hours; GSE1815), and lymphoblastoid cell lines (LCLs) (treated with 1μM dexamethasone for 8 hours; GSE29342). A two-tailed Student t-test was performed between control and dexamethasone treated samples at each time point in the datasets. Genes that show potentially differential expression between dexamethasone treated and control sample (p<0.05) were then compared between the three GEO data sets to identify genes whose expression are affected by dexamethasone treatment at all time points and in all cell types. Note that the use of p<0.05 as a threshold is for filtering purpose only. In addition, we selected NFKB1, a gene that has rich literature support for its role in GC sensitivity, as a reference.
Cell lines, primary cells and drugs
Human airway epithelial cell lines (1HAEo- and 16HBE14o-) were obtained from Dr. Gruenert at University of California San Francisco. These cell lines were cultured in plates coated with LHC basal medium, (Cat# 12677-019;Invitrogen, CA), bovine serum albumin (Cat# A9647; Sigma, MO), Vitrogen 100 (Cat# 354231; BD Biosciences, NJ), human fibronectin (Cat# 354008; BD Biosciences, NJ) and maintained using MEM with Earle’s salts supplemented with 10% heat inactivated fetal bovine serum (FBS), 1% L-glutamine and 1% pencillin/streptomycin. Primary airway epithelial cells were obtained from lungs donated for research use by the Regional Organ Bank of Illinois. Lungs were collected from subjects for purposes of transplantation but rejected for this use. The use of these lungs was considered to be exempt by the University of Chicago Biological Sciences Division IRB. We have previously described their collection and culture [20, 21]. HapMap LCLs were purchased from the Coriell Institute of Medicine (Camden, NJ). Peripheral blood mononuclear cells (PBMCs) were obtained from two healthy donors at three independent time points. Collection and use of these samples was performed under the approval of the University of Chicago Biological Sciences Division IRB (#14911). From each of the donors, 10 mL whole blood was collected at each of the three time points. PBMCs were isolated through density gradient separation method using ACCUSPIN™ System-Histopaque®-1077 (Cat# A7054; Sigma, MO). Dexamethasone, a synthetic GC, was purchased from Sigma (Cat# A4902; St. Louis, MO) and dissolved in 100% filtered ethanol and RPMI media to 100 μM stock solution.
Dexamethasone treatment effect on selected genes in primary human airway epithelial cells and cell lines
1HAEo- and 16HBE14o- cells were plated at a concentration of 0.5 million and 0.7 million cells per well respectively, in a 6-well plate and were treated with 1 μM dexamethasone solution or control solution (RPMI containing 0.04% ethanol v/v). Cells were harvested for RNA isolation using QIAzol lysis reagent (Cat# 79306; Qiagen, MD) at baseline, 6 hours, or 24 hours after treatment. Primary NHBE cells were treated with 10 μM dexamethasone solution for 24 hours prior to RNA isolation.
Dexamethasone treatment effect on gene expression in LCLs and PBMCs
Six randomly selected HapMap cell lines (GM07029, GM11823, GM19200, GM19210, GM18521, GM19221) were cultured in RPMI media with 15% FBS and 1% L-glutamine. Each LCL was plated at a concentration of 1×106 cells/mL. After overnight incubation, cells were pelleted for the zero hour control, or treated with control or dexamethasone solutions, such that final concentration of 1 μM dexamethasone was achieved. Six and 24 hours after addition of dexamethasone/control, cells were pelleted for subsequent RNA isolation, cDNA synthesis, and gene expression quantification. Following PBMCs isolation, cells were treated with control or 1 μM dexamethasone for 6 hours.
Total RNA was isolated from cells according to the manufaturer’s protocol using the RNeasy Mini Kit (Cat# 74104; Qiagen, CA). Reverse transcription was performed using a high capacity cDNA reverse transcription kit (Cat# 4368819; Applied biosystems, CA). Gene expression was quantified through quantitative PCR (qPCR) using TaqMan Fast Advanced Master Mix (Cat# 4444557; Applied Biosystems, CA). Primers specific to IRF1 (Hs00971960_m1), NFKB1 (Hs00765730_m1), B2M (4326319E) and ACTB (Hs99999903m1) were purchased from Applied Biosystems and were used in the appropriate reactions. We selected ACTB for the housekeeping gene for experiments in the 1HAEo- cell line and B2M for all other experiments. All PCR reactions were performed in triplicate per sample using ViiA7 PCR cycler and detection system (4453536, Applied Biosystems).
Data Analysis
Quantity of each gene expressed was normalized to ACTB for experiments in the 1HAEo- cell line and to B2M in all other cell lines. Normalized expression of control and dexamethasone treatment groups were compared using a two-tailed Student’s t-test with α=0.05.
RESULTS
Identification of gene targets
Upon examining three independent GEO datasets for dexamethasone induced gene expression changes in ASM [22] (GSE34313) and NHBE [23] (GSE1815) and LCLs [24] (GSE29342), we found 8 genes whose expression was changed in all 3 tissues: KLF9, IRF1, FKBP5, HBEGF, PPAT, ARHGEF2, MT1E and PLAU (Table 1). We chose to follow up on IRF1 gene, given the significant effects of dexamethasone treatment on this gene at all time points in all 3 tissue types (ASM, NHBE and LCLs) (Table 1). NFKB1, the reference gene with extensive literature support in the GC sensitivity, did not show significant down-regulation with dexamethasone treatment at all time points. However, it was significantly down-regulated at 4 hours and at 24 hours (in 4 of the 5 probe sets evaluated) in ASM cells; but only 1 of the 3 probe sets showed significant down-regulation at 8 hours in NHBE cells.
Table 1.
GC induced differential gene expression in three independent GEO datasets derived from three tissue types.
| NHBE (GEO1815) | ASM (GEO34313) | LCL (GEO29342) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| 8-hour | 24-hour | 4-hour | 24-hour | 8-hour | ||||||
|
|
||||||||||
| Gene name | log2FC | p value | log2FC | p value | log2FC | p value | log2FC | p value | Log2FC | p value |
| KLF9 | 1.02 | 2.10E-03 | 0.47 | 1.04E-01 | 1.37 | 2.53E-06 | 1.92 | 6.97E-04 | 0.81 | 1.96E-47 |
| IRF1 | −1.49 | 7.77E-04 | −4.69 | 2.93E-05 | −1.64 | 2.37E-06 | 0.62 | 3.16E-02 | −0.43 | 1.06E-21 |
| FKBP5 | 2.39 | 2.91E-05 | 2.39 | 5.62E-04 | 3.73 | 9.39E-06 | 3.71 | 1.38E-04 | 2.44 | 2.50E-54 |
| HBEGF | −0.62 | 1.36E-03 | −0.94 | 6.61E-02 | −3.01 | 3.78E-05 | −5.76 | 2.14E-05 | −0.14 | 8.44E-13 |
| PPAT | 0.60 | 9.53E-03 | 0.56 | 5.43E-02 | −0.24 | 7.62E-06 | −0.69 | 4.34E-03 | −0.19 | 6.04E-17 |
| ARHGEF2 | −0.77 | 1.03E-02 | −0.36 | 4.51E-02 | −0.77 | 6.16E-05 | −0.48 | 9.15E-03 | −0.10 | 1.47E-05 |
| MT1E | 0.45 | 3.97E-03 | 0.73 | 3.61E-04 | 1.31 | 2.74E-06 | 1.30 | 6.68E-03 | 0.48 | 5.95E-05 |
| PLAU | −0.95 | 8.30E-05 | −0.54 | 1.35E-04 | −0.91 | 3.85E-05 | −0.75 | 6.53E-03 | −0.10 | 1.33E-03 |
NHBE, normal human bronchial epithelial cells. ASM, airway smooth muscle cells. LCL, lymphoblastoid cell lines. FC, fold change. All cells were treated with 1 μM dexamethasone.
Evaluating expression changes in airway epithelial cell lines and primary NHBE cells
In the 1HAEo- cell line (Figure 1, Supplemental Figure 1A, 1B), both NFKB1 (p=0.009, 0.039) and IRF1 (p=0.0001; 0.0050) showed significant down-regulation at 6 and 24 hours, respectively, with 1 μM dexamethasone treatment. In the 16HBE14o- line (Figure 2, Supplemental Figure 1C, 1D), NFKB1 was significantly down-regulated at 6 hours (p=0.024), but was significantly up-regulated at 24 hours (p=0.008) by dexamethasone treatment. IRF1 was similarly significantly down-regulated at 6 hours (p<0.0001), but showed no difference at 24 hours (p=0.9014) with dexamethasone treatment. In primary NHBE cells, we observed a clear inter-individual variability in gene expression response to dexamethasone treatment (10 μM 24h, Figure 3). Specifically, IRF1 was significantly down-regulated in subjects 198 (p<0.0001) and 266 (p=0.008), but it was significantly up-regulated with dexamethasone treatment in subjects 286 (p=0.008) and 242 (p=0.0004). NFKB1 was down-regulated with dexamethasone treatment in NHBE cells collected from subjects 198 (p=0.005) and 266 (p=0.035); it underwent no change in subject 286 (p=0.247) and was up-regulated in subject 242 (p<0.0001).
Figure 1.
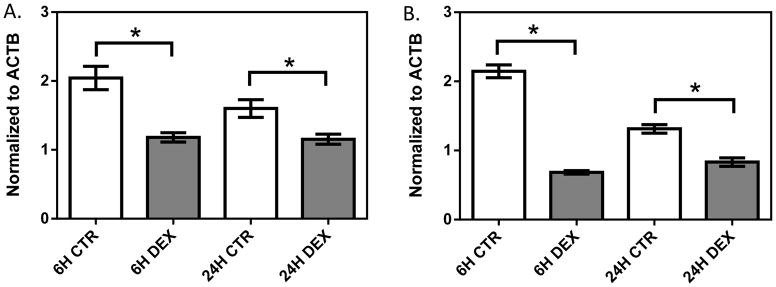
The effect of dexamethasone treatment on NFKB1 (A) and IRF1 (B) in the 1HAEo- cell line. 1HAEo- cells were treated with 1 μM dexamethasone for 6 and 24 hours. Gene expression was normalized to ACTB and each experiment was performed in triplicate. * represent p<0.05 a two-tailed unpaired Student’s t-test.
Figure 2.
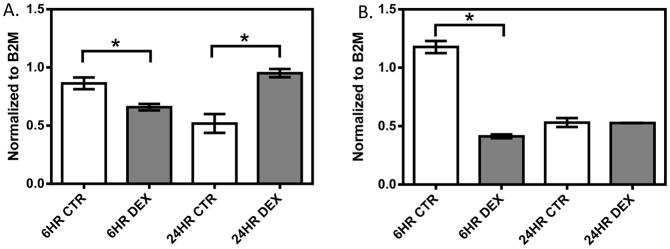
The effect of dexamethasone treatment on NFKB1 (A) and IRF1 (B) in the 16HBE14o- cell line. 16HBE14o- cells were treated with 1 μM dexamethasone for 6 and 24 hours. Gene expression was normalized to B2M and each experiment was performed in triplicate. * represent p<0.05 a two-tailed unpaired Student’s t-test.
Figure 3.
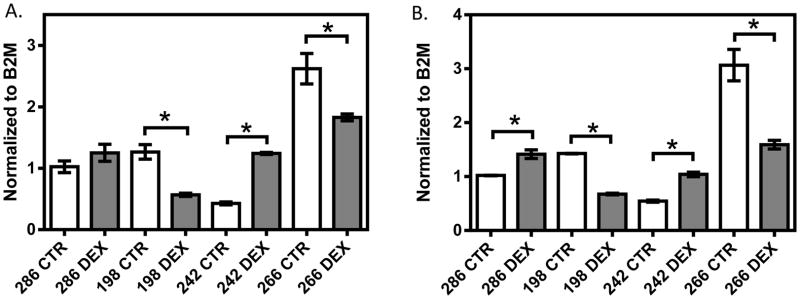
The effect of dexamethasone treatment on NFKB1 (A) and IRF1 (B) in primary NHBE cells from four donors. Normal primary NHBE cells were treated with 10 μM dexamethasone for 24 hours. Gene expression was normalized to B2M and each experiment was performed in triplicate. * represent p<0.05 a two-tailed unpaired Student’s t-test.
Gene expression changes in LCLs and PBMCs
In six randomly selected LCLs, we observed significant decreases in IRF1 expression in the dexamethasone treatment group at 6 hours (p=0.015) and at 24 hours (p=0.008) when compared to those of control group (Figure 4). Significant decrease in NFKB1 expression was observed after 6 hours of dexamethasone treatment (p=0.03), however, there is no significant difference in NFKB1 expression at 24 hours in the same six cell lines (p>0.05). The degree of IRF1 and NFKB1 expression change varied among samples.
Figure 4.
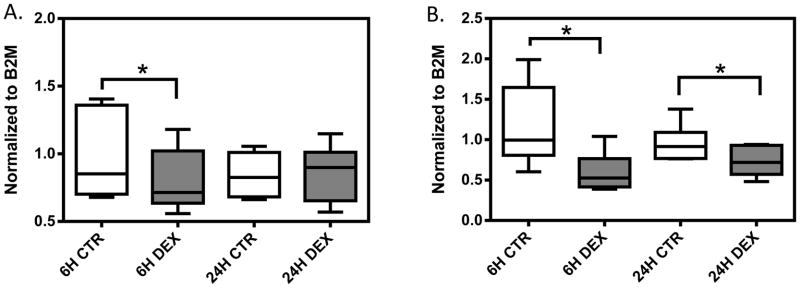
Gene expression in response to dexamethasone treatment in six randomly selected LCLs. A box represents the median and interquartile range of normalized gene expression for: A) NFKB1, and B) IRF1. Whiskers extend to the minimum and maximum values of normalized gene expression for each treatment group. Gene expression level was measured at 6 and 24 hours after 1μM dexamethasone treatment. Expression was normalized to housekeeping gene B2M. * represent p<0.05 a two-tailed unpaired Student’s t-test.
PBMCs were obtained from three independent blood draws from 2 healthy volunteers. Followed by dexamethasone (1 μM) or control treatment for 6 hours, qPCR was performed to quantify NFKB1 and IRF1 expression (Figure 5, Supplemental Figure 2). In both individuals, no significant difference was observed between the control group and dexamethasone treatment group in NFKB1 expression (p>0.05 for both healthy donors). On the other hand, expression of IRF1 was significantly reduced after dexamethasone treatment when compared to the control (p = 5.4×10−5 and p=7.5×10−5 for subject 14911-06 and 14911-18, respectively). Furthermore, the dexamethasone induced IRF1 down-regulation is fairly consistent among independent experiments for a given donor’s PBMCs (Supplemental Figure 2); while the degree of IRF1 down-regulation varies between the two donors.
Figure 5.
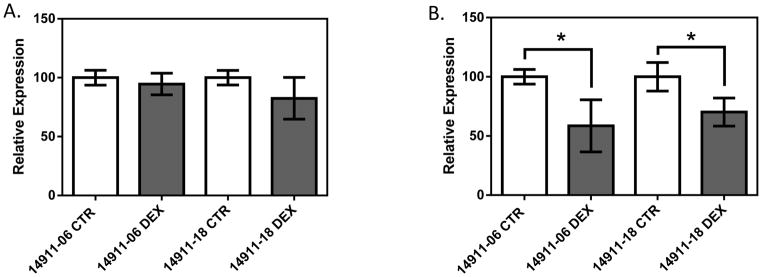
Gene expression in response to dexamethasone treatment in PBMCs from two apparently healthy volunteers for A) NFKB1 and B) IRF1. The two subjects’ IDs are 14911-06 and 14911-18. Independent blood collections were performed at three time points from the same donors, followed by dexamethasone treatment (1 μM, 6 hours) and qPCR. Each bar represents 9 data points collected in triplicate from each experiment at three separate time points. CTR stands for control; while DEX stands for dexamethasone treatment. Gene expression is normalized to B2M. * represent p<0.05 a two-tailed unpaired Student’s t-test.
DISCUSSION
As GC insensitivity may affect up to 10% of the world’s 300 million asthmatics, the ability to predict which patients will be resistant to GC and initiate alternative therapies would benefit both patients and society as a whole [1, 4]. Previous studies estimate that ~2/3 of the $14 billion spent annually on asthma treatment in the U.S. annually goes to the 15% of patients with the most severe asthma for care in emergency and hospital settings [5]. Accordingly, significant effort has been dedicated to the identification of biomarkers that predict inter-individual variability in gene response. Previous work has focused on predicting GC response using baseline gene expression [5], but we tested whether we could use changes in drug stimulated gene expression as a predictor of response. Our rationale were that 1) baseline expression varies among tissues and is affected by collection conditions; and 2) smaller expression variability associated with baseline gene expression limits its predictive power, since it has been reported that there is an average 2-fold expression difference among baseline expression of all genes in humans [25]; while the drug induced gene expression changes could vary to a much larger degree(e.g., 92-fold MDR1 transcript expression induction by digoxin in human intestine epithelial cell line [26]). With prediction in mind, it is important to develop a model system to quantify expression changes that can be evaluated prior to administering the drug to patients. Based on prior research which suggests that gene expression and expression changes in peripheral blood may be reflective of airway tissues, [5, 18, 19], we chose to evaluate expression changes in peripheral blood-derived cells.
Our work started by establishing links between drug-targeted tissues, in this case ASM and NHBE cells, and peripheral blood derived materials. We used three independent GEO data sets [22–24] to select GC responsive genes. We identified 8 such genes whose expression levels were significantly affected by dexamethasone treatment in all three tissue types. Among them, genetic variants in PLAU have been reported to associate with asthma and with airway hyperresponsiveness [27]. Further, multiple studies have indicated the role of IRF1, interferon regulator factor 1, in asthma risk in several ethnicities [28–32]. As a proof of concept, our work followed up on IRF1, however, all other genes could be further examined as peripheral gene expression changes markers for GC sensitivity.
In airway epithelial cell lines, we confirmed that NFKB1 and IRF1 were consistently down-regulated by dexamethasone treatment at 6 hours. We observed large inter-individual variability in response to GC treatment showed by IRF1 or NFKB1 expression changes in NHBE cells obtained from normal subjects. Furthermore, we demonstrated that although down-regulation is the general trend of IRF1 and NFKB1 expression after dexamethasone treatment in LCLs and PBMCs, the degree of expression change also varies among individuals.
NFKB1 is strongly expressed as a subunit of NF-κB in all cell types of interest and has a well-documented association with asthma and GC insensitivity. Baseline expression of NF-κB alone has been shown to correctly predict GC response phenotype in 81.25% of asthmatic cases, [5] and fold change in NFKB1 expression following in vitro dexamethasone treatment was reported to significantly correlate with lymphocyte GC sensitivity [19]. Additionally, it has been well established that a large number of genes are transcribed following NF-κB activation, many of which have been associated with airway inflammation in asthma [5]. While it is predicted that down-regulation of NFKB1 is just one of several important mechanisms influencing down-regulation of inflammatory cytokines with GC treatment, Newton et al. demonstrated that NFKB1 expression was significantly reduced 6 hours post treatment with a 1 μM concentration of dexamethasone in both the A549 alveolar lung adenocarcinoma cell line and in the BEAS-2B lung epithelial cell line [33]. We observed this same down-regulation at 6 hours in LCLs and PBMCs, demonstrating the utility of measuring changes in gene expression in PBMCs for reflecting gene expression changes occurring in the airway, at least for major GC responsive genes such as NFKB1. In addition, we noticed considerable inter-individual variation in the gene expression change in response to dexamethasone treatment in both the airway cells and in PBMCs, which could reflect different clinical responses to GCs in patients.
As noted in our results, we carefully examined IRF1 because the observed down-regulation after in dexamethasone treatment at multiple time points and in all cell types examined from GEO data sets. While not as studied as NFKB1, we observed a larger drug treatment effect on IRF1 expression compared to that of NFKB1. It has been shown that IRF1 expression may be regulated by NFKB1 and a significant association has been observed between NFKB1 genotype and IRF1 expression levels [34]. This suggests that assaying gene expression at a different point in the NF-κB signaling pathway, for example at IRF1, may be a better measure of GC response than changes in NFKB1 expression levels.
Genetic variants in IRF1 have been shown to influence IgE regulation, atopy [30] and are associated with development of asthma in various ethnic groups [28, 29, 31, 32]. A recent study has reported male-specific asthma risk associations in European Americans at the IRF1 locus [35]. It is worth noting that although genetic variants at IRF1 locus have been linked to asthma risk, the discovery that IRF1 expression changes may be an indicator of GC sensitivity and that the change of IRF1 expression in response to GC treatment can be seen in ASM, NHBE and peripheral blood cells are unique findings in our study. Tliba et al elegantly demonstrated an interaction between IRF1, the glucocorticoid receptor (GR) and fluticasone (an inhaled GC) in ASM cells [36]. In addition, IRF1 is up-regulated in GC resistant cells [37] and acts to reduce GC signaling by sequestering GRIP-1, a co-activator protein necessary for the transactivation activity of the glucocorticoid receptor [37]. Taken together, these studies support a role of IRF1 expression in GC sensitivity.
The strength of our work lies in the incorporation of multiple modalities (in silico independent datasets integration, candidate genes replication in blood cell lines/primary cells as well as airway epithelial cells). However, one limitation is that the numbers of different biological samples evaluated were small. Furthermore, the NHBE samples were derived from healthy donors rather than asthma patients. Therefore, larger studies in asthma patients are warrantied and will provide additional evidence whether markers identified from this study can be of use in the clinic.
In summary, our systematic evaluation of GEO data sets for ASM cells, NHBE cells and LCLs identified IRF1 as a candidate GC responsive gene whose expression change could be predictive of GC response. Our findings support future research into the development of an ex vivo assay of gene expression changes in PBMCs in response to GC treatment for the prediction of clinical GC response.
Supplementary Material
Supplemental Figure 1: The effect of dexamethasone treatment on NFKB1 (A, C) and IRF1 (B, D) in the 1HAEo- cell line (A, B) and in the 16HBEo- cell line (C, D). Gene expression was normalized to ACTB in 1HAEo- cell line; while expression was normalized to B2M in the 16HBEo- cell line. Data presented represent 3 independent experiments performed in triplicate.
Supplemental Figure 2: Gene expression in response to dexamethasone treatment in PBMCs from two apparently healthy volunteers for NFKB1 (A, B, C) and IRF1 (D, E, F). The two subjects’ IDs are 14911-06 and 14911-18. Independent blood collections were performed at three time points from the same donors, followed by dexamethasone treatment (1 μM, 6 hours) and qPCR. Each experiment is labeled as Exp1, Exp2 and Exp3. CTR stands for control; while DEX stands for dexamethasone treatment. Gene expression is normalized to B2M. Each experiment was performed in triplicate. * represent p<0.05 a two-tailed unpaired Student’s t-test.
Acknowledgments
This study was supported by the NIH/NIGMS grant K08GM089941, NIH/NCATS [UL1RR024999], NIH/NIGMS grant U01GM61393, NIH/NIDDK grant 2T35DK062719-26 and by Natural Science Foundation of Ningbo City (2014A610235), China. Human tissue used for this research project was provided by Gift of Hope Organ & Tissue Donor Network through the generous gift of donor families. The authors thank Ms. Randi Stern for her assistance in obtaining NHBE cells from the donor lungs. RSH also received support from NIH/NCI grant R21 CA139278, Circle of Service Foundation Early Career Investigator award, University of Chicago Cancer Center Support Grant (#P30 CA14599), Breast Cancer SPORE Career Development Award [CA125183], and a Conquer Cancer Foundation of ASCO Translational Research Professorship award In Memory of Merrill J. Egorin, MD (awarded to Dr. Mark Ratain).
Abbreviations
- ASM
airway smooth muscle
- FBS
fetal bovine serum
- GC
glucocorticoid
- GEO
gene expression omnibus
- LCL
lymphoblastoid cell line
- NHBE
normal human bronchial epithelial
- PBMC
peripheral blood mononuclear cells
Footnotes
Statement of Conflicts: None declared.
References
- 1.Masoli M, Fabian D, Holt S, Beasley R Program GIfA. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59(5):469–78. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 2.Use Inhaled Corticosteroids to Keep Airways Open. Available at: http://www.nhlbi.nih.gov/health/prof/lung/asthma/naci/discover/corticosteriods.htm.
- 3.Chan MTS, Leung DYM, Szefler SJ, Spahn JD. Difficult-to-control asthma: Clinical characteristics of steroid-insensitive asthma. Journal of Allergy and Clinical Immunology. 1998;101(5):594–601. doi: 10.1016/S0091-6749(98)70165-4. [DOI] [PubMed] [Google Scholar]
- 4.Durham A, Adcock IM, Tliba O. Steroid resistance in severe asthma: current mechanisms and future treatment. Current pharmaceutical design. 2011;17(7):674–84. doi: 10.2174/138161211795428984. [DOI] [PubMed] [Google Scholar]
- 5.Hakonarson H, Bjornsdottir US, Halapi E, Bradfield J, Zink F, Mouy M, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(41):14789–94. doi: 10.1073/pnas.0409904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes PJ, Ito K, Adcock IM. Corticosteroid resistance in chronic obstructive pulmonary disease: inactivation of histone deacetylase. Lancet. 2004;363(9410):731–3. doi: 10.1016/S0140-6736(04)15650-X. [DOI] [PubMed] [Google Scholar]
- 7.Bouazza B, Krytska K, Debba-Pavard M, Amrani Y, Honkanen RE, Tran J, et al. Cytokines Alter Glucocorticoid Receptor Phosphorylation in Airway Cells. American Journal of Respiratory Cell and Molecular Biology. 2012;47(4):464–73. doi: 10.1165/rcmb.2011-0364OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi Y, Mercado N, Barnes PJ, Ito K. Defects of protein phosphatase 2A causes corticosteroid insensitivity in severe asthma. PLoS One. 2011;6(12):e27627. doi: 10.1371/journal.pone.0027627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes PJ. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. Journal of Allergy and Clinical Immunology. 2013;131(3):636–45. doi: 10.1016/j.jaci.2012.12.1564. [DOI] [PubMed] [Google Scholar]
- 10.Ledderose C, Möhnle P, Limbeck E, Schütz S, Weis F, Rink J, et al. Corticosteroid resistance in sepsis is influenced by microRNA-124–induced downregulation of glucocorticoid receptor-α*. Critical Care Medicine. 2012;40(10):2745–53. doi: 10.1097/CCM.0b013e31825b8ebc. [DOI] [PubMed] [Google Scholar]
- 11.Quante T, Ng YC, Ramsay EE, Henness S, Allen JC, Parmentier J, et al. Corticosteroids Reduce IL-6 in ASM Cells via Up-Regulation of MKP-1. American Journal of Respiratory Cell and Molecular Biology. 2008;39(2):208–17. doi: 10.1165/rcmb.2007-0014OC. [DOI] [PubMed] [Google Scholar]
- 12.Hew M, Chung KF. Corticosteroid insensitivity in severe asthma: significance, mechanisms and aetiology. Internal Medicine Journal. 2010;40(5):323–34. doi: 10.1111/j.1445-5994.2010.02192.x. [DOI] [PubMed] [Google Scholar]
- 13.Oakley RH, Jewell CM, Yudt MR, Bofetiado DM, Cidlowski JA. The Dominant Negative Activity of the Human Glucocorticoid Receptor β Isoform SPECIFICITY AND MECHANISMS OF ACTION. Journal of Biological Chemistry. 1999;274(39):27857–66. doi: 10.1074/jbc.274.39.27857. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Li JJ, Foster PS, Hansbro PM, Yang M. Potential therapeutic targets for steroid-resistant asthma. Current Drug Targets. 2010;11(8):957–70. doi: 10.2174/138945010791591412. [DOI] [PubMed] [Google Scholar]
- 15.Baines KJ, Simpson JL, Wood LG, Scott RJ, Fibbens NL, Powell H, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. The Journal of Allergy and Clinical Immunology. 2014;133(4):997–1007. doi: 10.1016/j.jaci.2013.12.1091. [DOI] [PubMed] [Google Scholar]
- 16.Jia G, Erickson RW, Choy DF, Mosesova S, Wu LC, Solberg OD, et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. Journal of Allergy and Clinical Immunology. 2012;130(3):647–54. e10. doi: 10.1016/j.jaci.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herazo-Maya JD, Noth I, Duncan SR, Kim S, Ma S-F, Tseng GC, et al. Peripheral Blood Mononuclear Cell Gene Expression Profiles Predict Poor Outcome in Idiopathic Pulmonary Fibrosis. Science Translational Medicine. 2013;5(205):205ra136–205ra136. doi: 10.1126/scitranslmed.3005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goleva E, Jackson LP, Gleason M, Leung DYM. Usefulness of PBMCs to predict clinical response to corticosteroids in asthmatic patients. Journal of Allergy and Clinical Immunology. 2012;129(3):687–93. e1. doi: 10.1016/j.jaci.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maranville JC, Baxter SS, Torres JM, Di Rienzo A. Inter-ethnic differences in lymphocyte sensitivity to glucocorticoids reflect variation in transcriptional response. The Pharmacogenomics Journal. 2013;13(2):121–9. doi: 10.1038/tpj.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White SR, Fischer BM, Marroquin BA, Stern R. Interleukin-1beta mediates human airway epithelial cell migration via NF-kappaB. American Journal of Physiology Lung Cellular and Molecular Physiology. 2008;295(6):L1018–27. doi: 10.1152/ajplung.00065.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White SR, Martin LD, Abe MK, Marroquin BA, Stern R, Fu X. Insulin receptor substrate-1/2 mediates IL-4-induced migration of human airway epithelial cells. American Journal of Physiology Lung Cellular and Molecular Physiology. 2009;297(1):L164–73. doi: 10.1152/ajplung.90453.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuno K, Haldar SM, Jeyaraj D, Mailloux CM, Huang X, Panettieri RA, et al. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. American Journal of Respiratory Cell and Molecular Biology. 2011;45(3):642–9. doi: 10.1165/rcmb.2010-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawliczak R, Logun C, Madara P, Barb J, Suffredini AF, Munson PJ, et al. Influence of IFN-gamma on gene expression in normal human bronchial epithelial cells: modulation of IFN-gamma effects by dexamethasone. Physiological Genomics. 2005;23(1):28–45. doi: 10.1152/physiolgenomics.00011.2005. [DOI] [PubMed] [Google Scholar]
- 24.Maranville JC, Luca F, Richards AL, Wen X, Witonsky DB, Baxter S, et al. Interactions between Glucocorticoid Treatment and Cis-Regulatory Polymorphisms Contribute to Cellular Response Phenotypes. PLoS Genet. 2011;7(7):e1002162. doi: 10.1371/journal.pgen.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nature Genetics. 2007;39(2):226–31. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haslam IS, Jones K, Coleman T, Simmons NL. Rifampin and digoxin induction of MDR1 expression and function in human intestinal (T84) epithelial cells. British Journal of Pharmacology. 2008;154(1):246–55. doi: 10.1038/bjp.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bégin P, Tremblay K, Daley D, Lemire M, Claveau S, Salesse C, et al. Association of urokinase-type plasminogen activator with asthma and atopy. American Journal of Respiratory and Critical Care Medicine. 2007;175(11):1109–16. doi: 10.1164/rccm.200607-1012OC. [DOI] [PubMed] [Google Scholar]
- 28.Nakao F, Ihara K, Kusuhara K, Sasaki Y, Kinukawa N, Takabayashi A, et al. Association of IFN-gamma and IFN regulatory factor 1 polymorphisms with childhood atopic asthma. The Journal of Allergy and Clinical Immunology. 2001;107(3):499–504. doi: 10.1067/mai.2001.113051. [DOI] [PubMed] [Google Scholar]
- 29.Noguchi E, Shibasaki M, Arinami T, Yamakawa-Kobayashi K, Yokouchi Y, Takeda K, et al. Mutation screening of interferon regulatory factor 1 gene (IRF-1) as a candidate gene for atopy/asthma. Clinical and Experimental Allergy: Journal of the British Society for Allergy and Clinical Immunology. 2000;30(11):1562–7. doi: 10.1046/j.1365-2222.2000.00916.x. [DOI] [PubMed] [Google Scholar]
- 30.Schedel M, Pinto LA, Schaub B, Rosenstiel P, Cherkasov D, Cameron L, et al. IRF-1 gene variations influence IgE regulation and atopy. American Journal of Respiratory and Critical Care Medicine. 2008;177(6):613–21. doi: 10.1164/rccm.200703-373OC. [DOI] [PubMed] [Google Scholar]
- 31.Walley AJ, Wiltshire S, Ellis CM, Cookson WO. Linkage and allelic association of chromosome 5 cytokine cluster genetic markers with atopy and asthma associated traits. Genomics. 2001;72(1):15–20. doi: 10.1006/geno.2000.6435. [DOI] [PubMed] [Google Scholar]
- 32.Wang T-N, Chu Y-T, Chen W-Y, Feng W-W, Shih N-H, Hsiang C-H, et al. Association of interferon-gamma and interferon regulatory factor 1 polymorphisms with asthma in a family-based association study in Taiwan. Clinical and Experimental Allergy: Journal of the British Society for Allergy and Clinical Immunology. 2006;36(9):1147–52. doi: 10.1111/j.1365-2222.2006.02551.x. [DOI] [PubMed] [Google Scholar]
- 33.Newton R, Hart LA, Stevens DA, Bergmann M, Donnelly LE, Adcock IM, et al. Effect of dexamethasone on interleukin-1β-(IL-1β)-induced nuclear factor-κB (NF-κB) and κB-dependent transcription in epithelial cells. European Journal of Biochemistry. 1998;254(1):81–9. doi: 10.1046/j.1432-1327.1998.2540081.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu-Mares W, Sun Z, Bamlet WR, Atkinson EJ, Fridley BL, Slager SL, et al. Analysis of variation in NF-kB genes and expression levels of NF-kB-regulated molecules. BMC Proceedings. 2007;1(Suppl 1):S126. doi: 10.1186/1753-6561-1-s1-s126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers RA, Scott NM, Gauderman WJ, Qiu W, Mathias RA, Romieu I, et al. Genome-wide interaction studies reveal sex-specific asthma risk alleles. Human Molecular Genetics. 2014 doi: 10.1093/hmg/ddu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tliba O, Damera G, Banerjee A, Gu S, Baidouri H, Keslacy S, et al. Cytokines Induce an Early Steroid Resistance in Airway Smooth Muscle Cells. American Journal of Respiratory Cell and Molecular Biology. 2008;38(4):463–72. doi: 10.1165/rcmb.2007-0226OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhandare R, Damera G, Banerjee A, Flammer JR, Keslacy S, Rogatsky I, et al. Glucocorticoid Receptor Interacting Protein-1 Restores Glucocorticoid Responsiveness in Steroid-Resistant Airway Structural Cells. American Journal of Respiratory Cell and Molecular Biology. 2010;42(1):9–15. doi: 10.1165/rcmb.2009-0239RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: The effect of dexamethasone treatment on NFKB1 (A, C) and IRF1 (B, D) in the 1HAEo- cell line (A, B) and in the 16HBEo- cell line (C, D). Gene expression was normalized to ACTB in 1HAEo- cell line; while expression was normalized to B2M in the 16HBEo- cell line. Data presented represent 3 independent experiments performed in triplicate.
Supplemental Figure 2: Gene expression in response to dexamethasone treatment in PBMCs from two apparently healthy volunteers for NFKB1 (A, B, C) and IRF1 (D, E, F). The two subjects’ IDs are 14911-06 and 14911-18. Independent blood collections were performed at three time points from the same donors, followed by dexamethasone treatment (1 μM, 6 hours) and qPCR. Each experiment is labeled as Exp1, Exp2 and Exp3. CTR stands for control; while DEX stands for dexamethasone treatment. Gene expression is normalized to B2M. Each experiment was performed in triplicate. * represent p<0.05 a two-tailed unpaired Student’s t-test.


