Abstract
Global changes in gene expression accompany the development of cancer. Thus, inherited variants in microRNA binding sites are likely candidates for conferring inherited susceptibility. Using an in-silico approach, we compiled a comprehensive list of SNPs predicted to modulate microRNA binding in genes from several key lung cancer pathways. We then investigated whether these SNPs were associated with lung cancer risk in two independent populations. In general, SNPs in microRNA binding sites are rare. However, some allelic variation was observed. We found that rs1126579 in CXCR2 was associated with a reduced risk of lung cancer in both European American (ORTT vs. CC 0.56 [0.37 – 0.88]; P=0.008) and Japanese (ORTT vs. CC 0.62 [0.38 – 1.00]; P=0.049) populations. Further, we found that the SNP disrupted a novel binding site for miR-516a-3p, led to a moderate increase in CXCR2 mRNA and protein expression and increased MAPK signaling. Moreover, analysis of rs1126579 with serum levels of IL-8, its endogenous ligand, supported an interaction whereby rs1126579-T and high serum IL-8 conferred synergistic protection from lung cancer. Our findings demonstrate a function for a 3′UTR SNP in modulating CXCR2 expression, signaling and susceptibility to lung cancer.
Introduction
Cancer is frequently characterized by global changes in gene expression. These changes can be instigated by a multitude of mechanisms, including gene amplification, somatic mutations, gene deletions or inherited susceptibility. The latter includes epigenetic mechanisms, changes in mitochondrial DNA and single nucleotide polymorphisms (SNPs), which are found throughout the genome. When present in transcription factor binding sites within the 5′ promoter region, SNPs can affect protein-DNA interactions and have appreciable effects on both protein transcription and yield (1,2).
MiRNAs, a class of non-coding genes, modulate mRNA translation by primarily binding to the 3′UTR of mRNA transcripts. MiRNAs temper the translation of up to 60% of mRNA transcripts through semi-conservative binding (3–5). The seed region of a miRNA, which corresponds to nucleotides 2–8 from its 5′end, coordinates exact Watson-Crick binding to the target mRNA sequence. Genetic variation within a miRNA binding site has two main consequences on target protein expression, i.e., quantitative and qualitative (6). For example, a SNP in the 3′UTR could alter the thermodynamic interaction between the miRNA and the mRNA sequence, strengthening or weakening binding and therefore modulating output of protein expression. Alternatively, such SNPs could either completely destroy a pre-existing binding site, or create an illegitimate one, leading to a more qualitative effect (6). Moreover, if a miRNA is liberated from a 3′UTR due to the disruption of a binding site, the increased bio-availability of that miRNA could target other mRNAs from protein-coding genes (7).
Having a central epigenetic role in fine-tuning gene expression, miRNA genes and miRNA binding site sequences in protein-coding genes are highly conserved (6,8,9). However, allelic variations in the 3′UTR of miRNA binding sites have been reported (8,9). Due to qualitative and quantitative effects, such as those outlined above, these inherited genetic alterations could have significant functional implications. Indeed, SNPs within miRNA binding sites have been demonstrated as highly penetrant for certain phenotypes. For example, a SNP in the SLITRK1 gene, which is implicated in Tourette’s syndrome, strengthens an existing binding site for miR-184 and augments its downregulation (10). Since this initial report in 2005, several studies have reported associations between SNPs in miRNA binding sites and muscularity in sheep (11), and hypertension (12,13), diabetes (14,15), Crohn’s disease (16) and obesity (15) in humans. Thus, having demonstrated several phenotypic consequences of SNPs in miRNA binding sites, it is highly plausible that such genetic variation could also play a role in carcinogenesis (6,17,18).
Previously, studies have examined the association between SNPs in miRNA binding sites and lung cancer survival (19,20) or risk of small cell lung cancer (21), few have focused on risk of non-small cell lung cancer (18). The requirements for sequence complementarity and stable thermodynamics around the miRNA-3′UTR seed binding site primes sequence variations within these regions as strong candidates for functional SNPs, thus we hypothesized that genetic variation in microRNA binding sites is a contributing factor to lung cancer risk. We analyzed several key biological pathways previously implicated in lung cancer etiology; phase 1/2 metabolism, DNA repair and inflammation. We identified a SNP in the inflammatory gene, CXCR2, that is associated with lung cancer risk. Moreover, we found that it is a functional SNP that alters the binding of miR-516a-3p and expression of CXCR2 mRNA and protein.
Materials and Methods
Study Subjects from National Cancer Institute/Maryland (NCI/MD) Case-Control Study
Patients with histologically confirmed non-small-cell lung cancer (NSCLC) were recruited from seven hospitals in the greater metropolitan area of Baltimore, MD. Population controls were identified from the Department of Motor Vehicles, MD, and frequency matched to cases by age and gender. Written informed consent was obtained from all participants and the study was approved by the Institutional Review Boards of the participating institutions. Inclusion criteria for this on-going case-control study have been previously described (22). Briefly, participants were United States citizens; English-speaking and non-institutionalized. Cases could not have been interviewed previously as a control for the study and population controls could not have a history of malignancy other than skin cancer. Participants took part in a detailed baseline questionnaire that collected extensive information on nutrition, reproductive health, smoking, and alcohol consumption. Never smokers were defined as those who smoked <100 cigarettes during their lifetime. Former smokers were defined as those who reported quitting smoking ≥1 year before the date of interview. Race was self-reported. Blood was obtained by the interviewers and frozen blood components were sent to the Laboratory of Human Carcinogenesis at the National Cancer Institute. Samples were stored at −80°C until use. For this study, genotyping for CXCR2 was carried out on 429 cases and 484 controls (Table 1).
Table 1.
Characteristics of cases and controls
| NCI/MD
|
Japanese
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | P value | Controls | Cases | P value | |||||
| N | % | N | % | N | % | N | % | |||
| 484 | 429 | 384 | 384 | |||||||
| Age | ||||||||||
| (Mean ± SD) | 67.4 ± 8.5 | 66.6 ± 10 | ns | 59 ± 8 | 59 ± 8 | ns | ||||
| Gender | ns | ns | ||||||||
| Male | 236 | 49 | 228 | 53 | 200 | 52 | 200 | 52 | ||
| Female | 248 | 51 | 201 | 47 | 184 | 48 | 184 | 48 | ||
| Smoking Status | <0.0001 | |||||||||
| Never | 193 | 40 | 29 | 7 | ||||||
| Former | 233 | 48 | 213 | 50 | ||||||
| Current | 57 | 12 | 180 | 43 | ||||||
| ns | ||||||||||
| Never | 187 | 49 | 187 | 49 | ||||||
| Ever | 197 | 51 | 197 | 51 | ||||||
| Packyrs | ||||||||||
| (Mean ± SD) | 17.1 ± 24.2 | 45.1 ± 29.3 | <0.0001 | 25.0 ± 29.8 | 38.0 ± 24.7 | 0.0006 | ||||
| Stage | ||||||||||
| I | 149 | 39 | ||||||||
| II | 44 | 11 | ||||||||
| III | 81 | 21 | ||||||||
| IV | 109 | 28 | ||||||||
| 0 | 11 | 3 | ||||||||
| I | 284 | 74 | ||||||||
| II | 89 | 23 | ||||||||
| Histology | ||||||||||
| Adenocarcinoma | 206 | 52 | ||||||||
| Squamous | 105 | 27 | ||||||||
| NSCLC | 73 | 19 | ||||||||
| Other | 9 | 2 | ||||||||
| Invasive Adenocarcinoma | 373 | 97 | ||||||||
| Adenocarcinoma in situ | 11 | 3 | ||||||||
ns denotes not significant
Study Subjects from National Cancer Center/Japan (NCC) Case Control Study
The Japanese case control study consisted of 384 lung adenocarcinoma cases and 384 controls (Table 1). The 384 cases were selected from the National Cancer Center Hospitals (NCCH) cohort (23) enrolled from 1999 to 2008. Informed consent was obtained from each participant. Cases were diagnosed by histological examination of surgical specimens according to WHO classification. The 384 controls were selected by frequency-matching to the 384 cases on age, sex and smoking at a 1:1 (ever/never) ratio from inpatients/outpatients of the NCCH or volunteers enrolled in Keio University used in our previous study (23). The NCCH control subjects were selected from 1999 to 2007 and did not have a history of cancer. The Keio controls had no known malignancy at the time of recruitment and had a voluntary blood draw on the occasion of a health check examination at Keio University in 2002 and 2003. Smoking histories of the subjects were obtained via interview using a questionnaire. Ever smokers were defined as those who had smoked at least one cigarette per day for 12 months or longer at any time in their life, whereas non-smokers were defined as those who had not. For both cohorts, smoking exposure was represented by pack-years, which was defined as the number of cigarettes smoked per day/20 multiplied by years of smoking.
SNP Selection
To identify SNPs in miRNA binding sites that could potentially be associated with lung cancer risk, we pursued SNPs that could have biological relevance. We collated a list of known genes involved in metabolism (phase 1/2), DNA repair, and inflammation; processes previously reported to be involved in lung cancer (Supplementary Table 1). To investigate if these genes had SNPs that fell within the seed region of miRNA binding sites, we used two web-based tools, Patrocles (www.patrocles.org) (24) and PolymiRTS (http://compbio.utmem.edu/miRSNP/) (25). These sites provide access to databases that collate information on 3′UTR sequences, SNPs, and predicted miRNA binding sites, which enabled us to identify SNPs with the potential to create, destroy or modulate a miRNA binding site. The list of SNPs in miRNA binding sites was further filtered by excluding non-polymorphic SNPs (defined as loci where ≤2% of the population were heterozygous) or polymorphic SNPs with a minor allele frequency (MAF) below 10%. These were excluded as our study did not have sufficient power to determine the statistical significance (P<0.05) of low frequency alleles. For some SNPs, there was no minor allele frequency information available, therefore these SNPs were retained and, after a preliminary analysis of 100 control samples, a decision was made whether or not to include them in the final analysis. To increase the likelihood that we would select SNPs with biological function, i.e., SNPs that affect RNA structure and thus alter miRNA-mRNA binding, we used RNAHybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) (26). This program calculates changes in free energy depending on whether the ancestral or derived allele was expressed. This approach identified 90 SNPs that were submitted for genotyping analysis.
Study approval
Experiments were performed in accordance with the Declaration of Helsinki and approved by the National Cancer Institute, MD, USA and the National Cancer Center Hospital, Japan. All subjects gave informed written consent.
Genotyping
NCI/MD
Genomic DNA was isolated from buffy coat samples containing white blood cells from patients in the case-control study using Flexigene DNA Kit (Qiagen), according to the manufacturer’s instructions. One hundred population controls were initially genotyped because the minor allele frequency of many of the selected SNPs was unknown. Case, control and duplicate samples (10%) were randomized and blinded for processing. The genotype concordance for each SNP was at least 99% among duplicates. Genotyping was completed at BioServe (Beltsville, MD) using the iPlexGold assay (Sequenom, San Diego, CA).
NCC/Japan
Genomic DNA was extracted from whole-blood cells using a Blood Maxi Kit (Qiagen, Tokyo, Japan) according to the supplier’s instructions. Genomic DNA for volunteers enrolled in Keio University was extracted from Epstein-Barr virus-transformed B-lymphocytes derived from the collected whole-blood cells. Genotyping for rs1126579 was performed by the Taqman assay (Applied Biosystems, Foster City, CA) according to the supplier’s instructions.
Statistical Analysis
Departures from Hardy-Weinberg equilibrium were evaluated by calculating and then comparing the expected with observed genotype frequencies using χ2 tests. Differences in characteristics between cases and controls were compared by χ2 test for categorical values or Student’s t tests for continuous measures. Calculations in the NCI/MD study were performed using STATA version 12 software (STATA Corp, College Station, TX), or JMP version 8.0.1 software (SAS Institute, Cary, NC) for the Japanese study. All statistical tests were two-sided.
In the NCI/UMD cohort, risk associations between genotypes and lung cancer susceptibility were estimated by odds ratios (OR) using an unconditional model. The base model was adjusted for potential confounding factors; current cigarette smoking status (never/former/current), gender (male/female), age at diagnosis (continuous), and pack-years of cigarette smoking (continuous). In the Japanese cohort, ORs were estimated using a conditional model adjusted for age at diagnosis (<59/≥60), smoking status (ever/never) and gender (male/female). The P-for-trend method that hypothesizes a dose–response effect by the number of variant alleles was conducted by including genotypes in the model as a continuous ordinal variable (Ptrend). If there were 0 individuals with a particular genotype, the exact logistic model was used. Hazard ratios were estimated using a Cox Proportional Hazards model with adjustment for age, gender, stage and histology. Survival times in the NCI/MD study were gathered through a query of the National Death Index (last entry 12/31/2010) and determined as time from diagnosis to last known follow-up; data were available for 405 of the 443 patients. Survival analyses were conducted assessing the risk of lung-cancer specific mortality; non-lung cancer specific deaths were censored. There were 27 non lung cancer-specific deaths in this group. In the Japanese study, the end point of the analysis was overall survival. Data were available on 373 of the 384 participants. Survival analyses were conducted assessing the risk of lung-cancer specific mortality; non-lung cancer specific deaths were censored. There were 8 non lung cancer-specific deaths in this group.
We tested for effect modification (27) by adding a cross-product term to a logistic regression model adjusted for age, gender, smoking status, IL-8 levels and pack-years of smoking. To test whether these strata were significantly different, models with and without the cross-product term were compared using the likelihood ratio test. To determine whether rs1126579 statistically interacted with serum levels of IL-8, we tested for departure from additivity (28). Departure from additivity, or synergistic interaction, refers to a condition where the sum of the risks of rs1126579 and IL-8 is greater than would be expected from each factor alone. IL-8 serum levels were dichotomized using the median of the control distribution as the cut-point. We tested for interaction with IL-8 with reverse coding for the model given the inverse association with lung cancer risk (i.e., CC=1, CT/TT=0) and the need for the exposed group to be at higher risk in such models (29). Four exposure variables were generated for the analysis; A = rs1126579 (CT/TT), serum IL-8 < median; B = rs1126579 (CT/TT), serum IL-8 > median; C = rs1126579 (CC), serum IL-8 < median; D = rs1126579 (CC), serum IL-8 > median. These groups were then compared in a single logistic regression model (adjusted for age, gender, smoking status, and pack-years of smoking). The output of this model was used to estimate two interaction statistics: interaction contrast ratio (ICR) and attributable proportion (AP). When the ICR and AP ≠ 0 there is evidence for departure from additivity (synergistic interaction). ICR is the excess risk due to interaction relative to the risk without either exposure. AP is the proportion of disease attributable to interaction among individuals with both exposures (28).
3′UTR Luciferase assay
Mechanistic interactions between miRNAs with 3′UTRs were functionally tested in vitro using reporter constructs containing fragments of the 3′UTR bearing the C and T alleles of rs1126579 cloned downstream of Renilla Luciferase. These constructs were created through genomic DNA amplification followed by cloning into a psiCHECK-2 vector (Promega, Madison, WI) that has been modified using Gateway technology (Invitrogen, Carlsbad, CA). Constitutive Firefly Luciferase in a separate operon served as a transfection control. The CXCR2 reporter was generated based on Refseq NM_001168298 and encompasses the first 253 bp of the 3′UTR, centered around the SNP of interest, rs1126579, which is in position 127 of the 3′UTR. This construct was verified to bear rs1126579-C and was mutagenized to generate rs1126579-T. The predicted site for miR-516a-3p binding was mutated from AGGAAGC to CAGAGAG (mutated nucleotides underlined) to abolish miRNA binding. All constructs were sequence-verified.
Plasmid DNA and pre-miRNAs were introduced into 293T cells by reverse transfection onto 96-well plates, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Briefly, cells were plated in media without antibiotics several days prior to transfection and were trypsinized and counted on the day of transfection. Plasmid DNA (20 ng/well) and pre-miRNA (0.03 pmol/well) were premixed with Lipofectamine 2000 and then added individually to each well in a volume of 50 μl in sextuplicate. A miRNA that contains a scrambled seed with no known binding targets was used as a control for normalization of miRNA binding activity for each plasmid DNA. Lastly, 45,000 cells/well were added in a volume of 150 μl. Twenty-four hours after transfection, cells were collected in 20 μL of 1X Passive Lysis Buffer (PLB) reagent (Dual-Luciferase® Reporter Assay, Promega, Madison, WI). Ten microliters of cell lysate was removed from each well for relative luciferase activity (RLA) measurement using a plate-reader luminometer. Fifty microliters of LAR II was added to the cell lysates and firefly luciferase activity was measured. The reaction was stopped with the addition of 50 uL Stop & Glo® Reagent, and Renilla luciferase activity was measured. Normalized RLA was calculated by the following formula: RLA = [firefly luciferase]/[renilla luciferase]. The results were further normalized to the negative control pre-miR construct. All experiments were performed in sextuplicate and repeated. The embryonic kidney cell line, 293T, was obtained from American Type Culture Collection and cultured in DMEM, 10% fetal bovine serum, 100 U/ml of penicillin, 0.1 mg/ml of streptomycin and 2 mM L-glutamine. Cells were cultured in a humidified incubator, at 37°C and 5% CO2.
IL-8 Serum Levels
Serum IL-8 levels were measured as previously described (30). Briefly, blood was collected at the time of enrollment into the case control study and allowed to clot. Serum was isolated by centrifugation and stored at −80°C. After thawing, 25 μL of serum were measured for IL-8 concentrations using electrochemiluminescence immunoassay (ECLIA) plates (MesoScale Discovery, Gaithersburg, MD) and analyzed on the MesoScale Discovery 6000 instrument, following the manufacturer’s assay and analysis protocols. Samples were blinded and randomly distributed for order of processing.
mRNA expression analysis by microarray
Gene expression data for pathological stage I-II lung adenocarcinomas from the Japanese case-control study were generated for 226 cases using Affymetrix Human Genome U133 Plus 2.0 Array (31). These data are deposited at NCBI GEO Profiles as GSE31210. Samples were categorized according to rs1125679 genotypes and analyzed for differential gene expression using BRB-Array Tools. Expression differences between tumor and adjacent normal tissues were assessed by 1-way ANOVA.
mRNA expression analysis by RT-PCR
RNA was extracted from cell lines and paired tumor and uninvolved tissue samples using Trizol, according to the manufacturer’s instructions. RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Taqman primers for CXCR2 were used to measure mRNA expression in tumor and uninvolved tissue (Applied Biosystems, Foster City, CA). Expression of 18S was used as a normalization control. Data are expressed as the negative difference of threshold cycle value (Ct of gene of interest − Ct of 18S). Fold changes were calculated using 2^(−ΔΔCt). mRNA expression data were visualized using GraphPad Prism version 6.0 for Windows (GraphPad Software, San Diego, CA, USA). There are three sequence predictions for CXCR2: CXCR2 (uc002vhb.2) at chr2:218126029–218137253 - transcript variant 1, mRNA; CXCR2 (uc002vha.2) at chr2:218126013–218137253 - transcript variant 1, mRNA; CXCR2 (uc002vgz.2) at chr2:218125290–218137253 - transcript variant 2, mRNA. These variants are predicted to form 2 protein-coding gene products: CXCR2 at chr2:218126013–218137253 - (NM_001557) and CXCR2 at chr2:218125290–218137253 - (NM_001168298). The Taqman assay that we used in this experiment, Hs01011557_m1, maps to the exon 1–2 boundary of NM_001557, which would be spliced out of NM_001168298/uc002vgz.2 mRNA. Thus, these data suggest that the main isoform detected by this assay is uc002vha.2.
Immunohistochemistry
Immunohistochemical staining for CXCR2 was performed using serial sections of paraffin-embedded lung tissue from 41 patients in the NCI-MD case-control study. The sections were deparaffinized using xylene, and then rehydrated through graded ethanol. Antigen retrieval was performed via 15 minutes of microwave boiling in Citrate buffer (1X, pH 6.0). After rinsing with PBS, endogenous peroxidase activity was blocked with a 20 minute incubation with Dako peroxidase block (Dako, Carpinteria, CA). Slides were then incubated overnight at 4°C in a 1:50 dilution of primary antibody (rabbit anti-CXCR2, Abcam, UK) in Dako dilution buffer. Slides were developed according to standard Dako protocol, with Hematoxylin as a counterstain. Slides were scored based on both their percentage of CXCR2 positivity and intensity of staining within tumor foci. Percentage positivity was scored as a ‘0’ (0–20%, negative), ‘1’ (20–40%), ‘2’ (40–60%), ‘3’ (60–80%), or ‘4’ (80–100%). Intensity was scored as either a ‘0’ (negative), ‘1’ (weak), ‘2’ (moderate), or ‘3’ (strong). Scores were combined to generate a total score (min, 0; max, 7).
The Cancer Genome Atlas (TCGA) Data Analysis
TCGA is supported by the National Cancer Institute and the National Human Genome Research Institute to chart the molecular landscape of tumor samples for more than 20 types of cancer (https://tcga-data.nci.nih.gov/tcga/). We used level-3 normalized data to infer the effects of rs1126579 on CXCR2 expression (32). We downloaded matched samples by participants’ sequence data, which have been processed by the Illumina GA Sequencer RNA-seq version 2 pipeline (Mapsplice alignment algorithm and the RSEM algorithm) to generate expression values for CXCR2. Genotype calls for rs1126579 were extracted from Affymetrix GenomeWide SNP6.0 platform data in TCGA. These files had previously been processed using Birdseed.
Results
SNPs in miRNA binding sites are highly conserved
Ninety SNPs passed MAF and RNAHybrid selection and were submitted for genotyping. Of these, 15 failed the Sequenom iPlexGold assay design phase (Supplementary Table 2). SNPs with an unknown MAF were initially analyzed in 100 population controls, 32 of these were non-polymorphic (Supplementary Table 3) and not pursued. This latter quotient is reflective of the literature, which describes a low SNP density and rare variants in and around miRNA binding sites (8,9,33). Forty-six SNPs were polymorphic; for 2 of these, the observed genotype frequencies deviated from expected Hardy Weinberg proportions (Supplementary Table 4).
rs1126579 in CXCR2 is associated with a reduced risk, but not survival, in lung cancer
rs1126579, in CXCR2, was associated with a reduced risk of lung cancer in European Americans in a model adjusted for age, gender, smoking status and pack-years of smoking (ORTT vs. CC 0.59 [0.39 – 0.91]; P=0.017) (Table 2). Data for the other SNPs are shown in Supplementary Table 4. As exposure to secondhand smoke can also influence the inflammatory response and other chronic diseases also cause inflammation, we further adjusted our model for presence/absence of asthma, emphysema, chronic bronchitis, asthma asbestosis, environmental tobacco smoke exposure and COPD (ORTT vs. CC 0.56 [0.37 – 0.88]; P=0.008).
Table 2.
Associations between rs1126579 and lung cancer risk
| NCI Maryland Case Control Studya
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Control | Case | Odds Ratio | LCI | UCI | P | ||
| CXCR2 | CC | 121 (25.5%) | 138 (30.9%) | Referent | Referent | |||
| rs1126579 | CT | 238 (50.2%) | 215 (48.5%) | 0.72 | 0.51 | - | 1.03 | 0.071 |
| TT | 115 (24.3) | 90 (20.3% | 0.59 | 0.39 | - | 0.91 | 0.017 | |
| Dominant | 0.68 | 0.49 | - | 0.95 | 0.023 | |||
| Recessive | 0.73 | 0.51 | - | 1.04 | 0.089 | |||
| Ptrend | 0.77 | 0.63 | - | 0.95 | 0.015 | |||
|
Japanese Case Control Studyb
|
||||||||
| CXCR2 | Genotype | Control | Case | Odds Ratio | LCI | UCI | P | |
| rs1126579 | CC | 35 (9%) | 54 (14%) | Referent | Referent | |||
| CT | 170 (44.4%) | 160 (41.7%) | 0.60 | 0.37 | - | 0.97 | 0.035 | |
| TT | 178 (46.5%) | 170 (44.3%) | 0.62 | 0.38 | - | 1.00 | 0.049 | |
| Dominant | 0.61 | 0.39 | - | 0.96 | 0.032 | |||
| Recessive | 0.91 | 0.69 | - | 1.22 | 0.540 | |||
| Ptrend | 0.85 | 0.69 | - | 1.05 | 0.140 | |||
|
Adenocarcinomac
|
||||||||
| CXCR2 | Genotype | Control | Case | Odds Ratio | LCI | UCI | P | |
| rs1126579 | CC | 121 (25.5%) | 72 (32.7%) | Referent | Referent | |||
| CT | 238 (50.2%) | 102 (46.4%) | 0.73 | 0.49 | - | 1.10 | 0.132 | |
| TT | 115 (24.3) | 46 (20.9) | 0.60 | 0.37 | - | 0.98 | 0.040 | |
|
Squamous Cell Carcinomac
|
||||||||
| CXCR2 | Genotype | Control | Case | Odds Ratio | LCI | UCI | P | |
| rs1126579 | CC | 121 (25.5%) | 34 (32.1%) | Referent | Referent | |||
| CT | 238 (50.2%) | 55 (51.9%) | 0.79 | 0.45 | - | 1.42 | 0.437 | |
| TT | 115 (24.3) | 17 (16.0%) | 0.51 | 0.24 | - | 1.08 | 0.078 | |
Adjusted for age, gender (male vs female), smoking status (current/former/never) and pack-years of smoking
Adjusted for age (<59 vs ≥60), gender (male vs female) and smoking status (ever/never)
These analyses were performed on the NCI-MD study only
The allele frequency of rs1126579-T in the European American population was 33%; in Asian populations it is noticeably higher (>60%). We therefore tested cross-population convergence of our findings in a population of Japanese ancestry. Here, although the T allele was the major allele, we again observed an inverse relationship between this SNP and risk of lung cancer (ORTT vs. CC 0.62 [0.38 – 1.00]; P=0.049) (Table 2) (model adjusted for age, gender and smoking status). We tested the allele frequency of rs1126579 in an African American population, but the frequency of the TT allele was close to 0% (CC = 58%, CT=42%, TT=0%) and therefore we did not pursue the relationship between rs1126579 and lung cancer risk in this population.
The Japanese population consisted of lung adenocarcinomas, while the NCI-MD study included both adenocarcinomas and squamous cell carcinomas. We therefore performed a subgroup analysis of the NCI-MD study by tumor histology and asked if the CXCR2 SNP was associated with risk of adenocarcinoma only. Although there was reduced power in the stratified analysis, the analysis suggests that this SNP is associated with risk of both adenocarcinoma and squamous cell carcinoma (Table 2).
We performed subgroup analyses stratified by gender and smoking status, but we did not observe significant differences in either the direction or effect size of the risk estimates (Supplementary Table 5). There was a suggestion in the NCI-MD study that the association between rs1126579-T and lung cancer risk was stronger in females, as compared to males (ORFemale 0.42 vs. ORMale 0.83, respectively), but this pattern was not replicated in the Japanese population (Supplementary Table 5).
We assessed the relationship between rs1126579 and lung cancer specific mortality in both the Japanese and European American cohorts. However, an association between this SNP and lung cancer-specific mortality was not observed (Supplementary Table 6).
rs1126579 disrupts a binding site for miR516a-3p and alters expression of CXCR2
Our original bioinformatics analysis during SNP selection predicted that the T allele of rs1126579 disrupted a binding site for miR-516a-3p (Figure 1A) and that it created a potential binding site for miR-138-1* (also known as miR-138-3p) (Supplementary Figure 1). The interaction between miR-516a-3p with CXCR2, and its potential modulation by the T allele of rs1126579, was evaluated in vitro using a 3′UTR dual luciferase assay. Significantly greater binding between miR-516a-3p and the C allele, as compared to the T allele, was demonstrated by reduced luciferase activity in the presence of the C allele (Figure 1B). However, we did not find evidence that miR-138-1* bound to either the T or C allele (Figure 1B).
Figure 1. rs1126579-T disrupts a binding site for miR-516a-3p and modulates CXCR2 expression.
A) The T allele of rs1126579 is predicted to disrupt a 7-mer binding site for miR-516a-3p. B) 3′ UTR luciferase assay for miR-516a-3p and miR-138-1* binding to the C and T alleles of rs1126579. Relative luciferase levels are decreased in the presence of the T allele and miR-516a-3p, as compared to the C allele and miR-516a-3p, suggesting decreased binding and therefore increased mRNA translation. Mut denotes mutant seed region. C) mRNA levels of CXCR2 are increased in subjects with the CT or TT genotype. D) Protein levels of CXCR2, as determined by immunohiostochemistry, are increased in tumor tissue from individuals with either the CT or TT rs1126579 genotype. Data were analyzed using ANOVA and Tukey’s post-hoc test. E–F) Levels of CXCR2 mRNA isoform expression in lung adenocarcinoma samples stratified by rs1126579 genotype (data from TCGA).
To test whether this differential binding could modulate CXCR2 expression, we measured CXCR2 mRNA (RT-PCR) and protein (IHC) in tumor samples from the NCI-MD study, where tissue was available. Upon stratification by rs1126579 genotype, levels of CXCR2 mRNA (Figure 1C) and protein (Figure 1D) were 1.7 and 2.5 fold increased, respectively, in tissues from individuals carrying the variant T allele (CT vs. CT/TT combined) (Figure 1D). These data however did not reach statistical significance (P=0.07), most likely due to our limited sample availability (Figure 1D). Representative images of CXCR2 protein staining is shown in Supplementary Figure 2. As three isoforms of CXCR2 mRNA exist, we utilized TCGA RNA-seq data to examine whether a specific isoform of CXCR2 is modulated by rs1126579. As shown in Figure 1E–F, expression of CXCR2 – UC002vha.1 was higher in samples with rs1126579-T compared to rs1126579-C, while expression of CXCR2 – UC002VHB.1 showed no difference. Data for CXCR2 – UC002vgz.1 and rs1126579 were not available. Collectively, these data suggest that rs1126579 increases CXCR2 expression in lung tissue by disrupting a binding site for miR-516a-3p.
rs1126579 modulates CXCR2 signaling
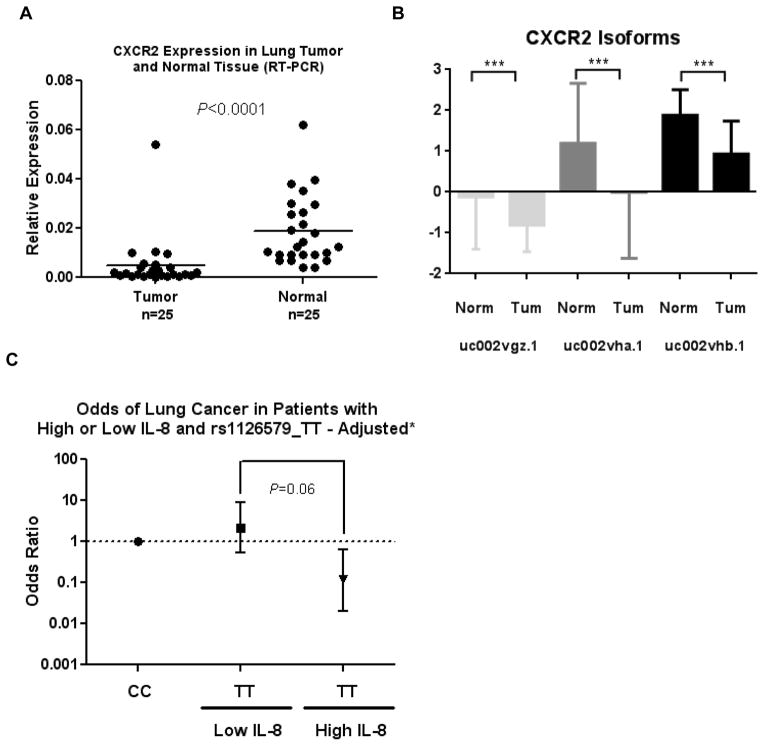
To assess whether the increase in CXCR2 expression resulted in alterations in downstream signaling, we conducted a pathway analysis on gene expression data from 203 tumors that had overlapping CXCR2 SNP data. After stratification by rs1126579 genotype and selection of those genes differentially expressed at P<0.05, a pathway analysis showed that the MAPK pathway was significantly enriched in samples with rs1126579-T (Table 4). We reasoned that if the T allele was associated with reduced risk, loss of miRNA regulation and higher expression of CXCR2, then loss of CXCR2 expression should be observed in tumors. We assessed CXCR2 mRNA expression by RT-PCR in non-involved lung tissue and tumor tissue from the NCI-MD study and observed that CXCR2 expression is decreased in lung cancer (Figure 2A). As mentioned earlier, CXCR2 exists as three isoforms; therefore we asked if the epxression of all isoforms of CXCR2 is decreased in lung cancer. As shown in Figure 2B, expression of all three CXCR2 mRNA isoforms is decreased in lung adenocarcinoma tissue compared to non-involved lung tissue (Figure 2B) (Supplementary Figure 3). Similar patterns and significant downregulation of CXCR2 was observed in lung squamous cell carcinoma (Supplementary Figure 3).
Table 4.
Pathway analysis of genes altered by rs1126579
| Pathway | P-value |
|---|---|
| ERK/MAPK Signaling | 8.22E-06 |
| Molecular Mechanisms of Cancer | 4.92E-05 |
| Renal Cell Carcinoma Signaling | 9.41E-05 |
| Paxillin Signaling | 1.13E-04 |
| NGF Signaling | 1.63E-04 |
Figure 2. CXCR2 expression and signaling is altered in cancer.
A) CXCR2 mRNA levels, as measured by RT-PCR, are lower in lung tumor tissue compared to non-involved lung tissue. Data were analyzed using two-sided paired t-test. B) Expression of all three isoforms of CXCR2 is decreased in lung cancer. Data were analyzed using TCGA RNA-seq data for lung adenocarcinoma. C) Risk of lung cancer in individuals with high and low serum IL-8 levels, dichotomized based on the median of the control population. P value derived from the likelihood ratio test comparing TT against CC in a model adjusted for age, gender, pack-years of smoking and smoking status.
rs1126579 interacts with circulating levels of IL-8
IL-8 is the primary ligand for CXCR2. We previously found that this cytokine is associated with an increased risk of lung cancer (30). As we observed that rs1126579 can modulate CXCR2 expression and is associated with lung cancer risk, we analyzed serum levels of IL-8 on controls (n=147) and cases (n=133) for which we had rs1126579 genotype and serum data. Both the magnitude and direction of the ORs varied in each stratum: The TT genotype of rs1126579 was associated with a reduced risk of lung cancer in subjects with high IL-8 levels (ORTT vs. CC 0.23 [0.07 – 0.86]), while no association was observed in subjects with low IL-8 levels (Table 3) (P for interaction 0.068) (Table 3) suggesting that IL-8 could be a potential effect modifier of the relationship between rs1126589 and lung cancer risk (Figure 2C). To investigate further whether there was any evidence of synergistic interaction, we tested for departure from additivity between serum IL-8 and rs1126579 (28). We found evidence for a synergistic interaction between the T allele of rs1126579 and high levels of IL-8; ICR statistic for CT/TT vs. CC 1.47 (95% C.I. 0.56 – 3.50); AP statistic for CT/TT vs. CC 0.64 (95% C.I. 0.13 – 1.16) (Supplementary Table 7). These statistics imply that individuals with high levels of IL-8 and the CT or TT genotype have a greater reduced risk of lung cancer than would be expected from the independent effects of the TT genotype and high IL-8 alone.
Table 3.
Effect modification of rs1126579 by serum levels of IL-8
| Stratification by serum IL-8 levels
|
|||||
|---|---|---|---|---|---|
| rs1126579 | Control/Case | Low IL-8 | Control/Case | High IL-8 | Pint |
| CC (Reference) | 24/13 | Referent | 18/28 | Referent | |
| CC vs. CT | 39/26 | 1.22 (0.38 – 3.86) P=0.741 | 27/40 | 0.78 (0.28 – 2.15) P=0.625 | 0.068 |
| CC vs. TT | 20/10 | 2.01 (0.52 – 7.78) P=0.312 | 19/16 | 0.23 (0.07 – 0.86) P=0.029 | |
Pint denotes the likelihood ratio test P-value
Discussion
The coalescence of several factors is necessary for a miR-SNP to contribute to a phenotype such as cancer. Firstly, the SNP must alter the thermodynamics of miRNA binding, and secondly, the microRNA designed to bind to the site must be spatially expressed. Thirdly, the gene must play a role in the transformation from a normal to malignant state. Our results highlight the convergence of each of these factors for rs1126579 in CXCR2 and compliment similar findings for a KRAS SNP that modulates let-7 binding and lung cancer risk (18).
Inflammation, which has an intricate role in many malignancies (34), has numerous mediators. We found that a SNP in one of these mediators, CXCR2, is associated with a reduced risk of lung cancer. This association was independently confirmed in a Japanese cohort that was matched on age, gender and smoking status. We further demonstrated that this is a functional SNP and that the T allele of rs1126579 disrupts a binding site for miR-516a-3p (which is endogenously expressed in lung tissue) (35), leading to a moderate increase in CXCR2 mRNA and protein. This increase of mRNA appears to be restricted to a specific isoform of CXCR2, i.e., UC002vha.1. It is not clear why this is the case, but it could be related to the length of mRNA isoform that is expressed in lung tissue, as recent evidence shows that 3′UTR sequence length is tissue specific (36). In addition, Ensembl gene predictions show that just one CXCR2 isoform, UC002vha.1, is long enough to overlap the rs1126579 SNP and thus be modified by it Supplementary Figure 4). This could explain why the SNP primarily affects this transcript, however it is speculative and would need to be further investigation. A pathway analysis highlighted ERK/MAPK signaling, which is downstream of CXCR2, as the top deregulated pathway among rs1126579-T tumors.
Given the role that CXCR2 plays in angiogenesis and inflammation (37–42), and its association with a higher risk of mortality from non-small cell lung carcinoma (43), it was surprising that a SNP which gives rise to increased expression of CXCR2 is associated with a reduced risk of cancer. In lung cancer however, our initial analysis of primary samples from the NCI-MD study indicated that expression of CXCR2 is decreased in lung cancer compared with non-involved lung tissue. We replicated this finding in TCGA and further demonstrated that expression of all three isoforms is decreased in both adenocarcinoma and squamous cell carcinoma. To our knowledge, this is the first such report. Despite its role in pro-inflammatory signaling, CXCR2 is also a primary mediator of p53-dependent senescence in lung (44). Senescence is a potent tumor suppressive mechanism (45–47) and might explain why expression of CXCR2 is decreased in lung cancer. A somatic point mutation in the carboxyl-terminus of CXCR2 (G354W) has been identified in the lung cancer cell line NCI-H1395 by the Catalog of Somatic Mutations in Cancer (COSMIC) (48). The mutation prevents autocrine IL-8-induced receptor internalization and alleviates senescence (44). Decreased expression of CXCR2, as found with the C allele of rs1126579, could be another mechanism to overturn cytokine-driven senescence signaling in lung cancer.
We recently reported that pre-diagnostic levels of IL-8 were associated with an increased risk of lung cancer (30). Here, we found that rs1126579 was associated with reduced risk of lung cancer among individuals with high levels of IL-8. As CXCR2-driven senescence is mediated by IL-8 (49), this finding furthers the possibility that the CXCR2 axis acts as a tumor suppressive mechanism in lung cancer. Thus, the identification of a functional polymorphism in the IL-8 receptor may influence the association between this cytokine in both lung cancer diagnosis and prognosis.
Strengths of our study include the validation of rs1126579 and lung cancer risk associations in two independent populations and functional analysis of the variant. However, there were also some limitations, including restriction of our analysis to binding sites in the 3′UTR of genes. Where possible, future studies should incorporate methods that allow the mining of coding, 5′ and 3′ UTR regions, as miRNAs can also bind to these loci (50). Moreover, studying the existence and contribution of somatic mutations to miRNA binding is likely to have further implications for cancer biology.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Program of the Centre for Cancer Research, National Cancer Institute. This work was also supported in part by Grants-in-Aid from the Ministry of Health, Labor, and Welfare of Japan for the 3rd-term Comprehensive 10-year Strategy for Cancer Control, the National Cancer Center Research and Development Fund (26-A-8), and by the National Cancer Center Research and Development Fund.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have no conflicts of interest to disclose
References
- 1.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119(5):591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 2.De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312(5777):1215–7. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 6.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 8.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(9):3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nature Genetics. 2006;38(12):1452–6. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 10.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310(5746):317–20. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 11.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38(7):813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 12.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81(2):405–13. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanin G, Shenhar-Tsarfaty S, Yayon N, Hoe YY, Bennett ER, Sklan EH, et al. Competing targets of microRNA-608 affect anxiety and hypertension. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv K, Guo Y, Zhang Y, Wang K, Jia Y, Sun S. Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/deletion polymorphism with type 2 diabetes. Biochem Biophys Res Commun. 2008;374(1):101–5. doi: 10.1016/j.bbrc.2008.06.102. [DOI] [PubMed] [Google Scholar]
- 15.Richardson K, Louie-Gao Q, Arnett DK, Parnell LD, Lai CQ, Davalos A, et al. The PLIN4 variant rs8887 modulates obesity related phenotypes in humans through creation of a novel miR-522 seed site. PLoS One. 2011;6(4):e17944. doi: 10.1371/journal.pone.0017944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni S, Qi Y, O’HUigin C, Pereyra F, Ramsuran V, McLaren P, et al. Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease. Proc Natl Acad Sci U S A. 2013;110(51):20705–10. doi: 10.1073/pnas.1312237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29(3):579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 18.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer research. 2008;68(20):8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pu X, Roth JA, Hildebrandt MA, Ye Y, Wei H, Minna JD, et al. MicroRNA-related genetic variants associated with clinical outcomes in early-stage non-small cell lung cancer patients. Cancer Res. 2013;73(6):1867–75. doi: 10.1158/0008-5472.CAN-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zu Y, Ban J, Xia Z, Wang J, Cai Y, Ping W, et al. Genetic variation in a miR-335 binding site in BIRC5 alters susceptibility to lung cancer in Chinese Han populations. Biochem Biophys Res Commun. 2013;430(2):529–34. doi: 10.1016/j.bbrc.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Xiong F, Wu C, Chang J, Yu D, Xu B, Yuan P, et al. Genetic variation in an miRNA-1827 binding site in MYCL1 alters susceptibility to small-cell lung cancer. Cancer Res. 2011;71(15):5175–81. doi: 10.1158/0008-5472.CAN-10-4407. [DOI] [PubMed] [Google Scholar]
- 22.Zheng YL, Loffredo CA, Yu Z, Jones RT, Krasna MJ, Alberg AJ, et al. Bleomycin-induced chromosome breaks as a risk marker for lung cancer: a case-control study with population and hospital controls. Carcinogenesis. 2003;24(2):269–74. doi: 10.1093/carcin/24.2.269. [DOI] [PubMed] [Google Scholar]
- 23.Kohno T, Kunitoh H, Shimada Y, Shiraishi K, Ishii Y, Goto K, et al. Individuals susceptible to lung adenocarcinoma defined by combined HLA-DQA1 and TERT genotypes. Carcinogenesis. 2010;31(5):834–41. doi: 10.1093/carcin/bgq003. [DOI] [PubMed] [Google Scholar]
- 24.Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic acids research. 2010;38(Database issue):D640–51. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, et al. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35(Database issue):D51–4. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–4. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanderWeele TJ. On the distinction between interaction and effect modification. Epidemiology. 2009;20(6):863–71. doi: 10.1097/EDE.0b013e3181ba333c. [DOI] [PubMed] [Google Scholar]
- 28.Skrondal A. Interaction as departure from additivity in case-control studies: a cautionary note. Am J Epidemiol. 2003;158(3):251–8. doi: 10.1093/aje/kwg113. [DOI] [PubMed] [Google Scholar]
- 29.Knol MJ, VanderWeele TJ, Groenwold RH, Klungel OH, Rovers MM, Grobbee DE. Estimating measures of interaction on an additive scale for preventive exposures. Eur J Epidemiol. 2011;26(6):433–8. doi: 10.1007/s10654-011-9554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pine SR, Mechanic LE, Enewold L, Chaturvedi AK, Katki HA, Zheng YL, et al. Increased levels of circulating interleukin 6, interleukin 8, C-reactive protein, and risk of lung cancer. J Natl Cancer Inst. 2011;103(14):1112–22. doi: 10.1093/jnci/djr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–11. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas LF, Saetrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31(1):37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106(29):12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27(21):2380–96. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Araki S, Omori Y, Lyn D, Singh RK, Meinbach DM, Sandman Y, et al. Interleukin-8 is a molecular determinant of androgen independence and progression in prostate cancer. Cancer Res. 2007;67(14):6854–62. doi: 10.1158/0008-5472.CAN-07-1162. [DOI] [PubMed] [Google Scholar]
- 38.Taguchi A, Ohmiya N, Shirai K, Mabuchi N, Itoh A, Hirooka Y, et al. Interleukin-8 promoter polymorphism increases the risk of atrophic gastritis and gastric cancer in Japan. Cancer Epidemiol Biomarkers Prev. 2005;14(11 Pt 1):2487–93. doi: 10.1158/1055-9965.EPI-05-0326. [DOI] [PubMed] [Google Scholar]
- 39.Benoy IH, Salgado R, Van Dam P, Geboers K, Van Marck E, Scharpe S, et al. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res. 2004;10(21):7157–62. doi: 10.1158/1078-0432.CCR-04-0812. [DOI] [PubMed] [Google Scholar]
- 40.Jamieson T, Clarke M, Steele CW, Samuel MS, Neumann J, Jung A, et al. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J Clin Invest. 2012;122(9):3127–44. doi: 10.1172/JCI61067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, DiPietro LA, Elner VM, et al. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science (New York, N Y) 1992;258(5089):1798–801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 42.Addison CL, Daniel TO, Burdick MD, Liu H, Ehlert JE, Xue YY, et al. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J Immunol. 2000;165(9):5269–77. doi: 10.4049/jimmunol.165.9.5269. [DOI] [PubMed] [Google Scholar]
- 43.Saintigny P, Massarelli E, Lin S, Ahn YH, Chen Y, Goswami S, et al. CXCR2 expression in tumor cells is a poor prognostic factor and promotes invasion and metastasis in lung adenocarcinoma. Cancer Res. 2013;73(2):571–82. doi: 10.1158/0008-5472.CAN-12-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133(6):1006–18. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 45.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 46.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 47.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10(1):51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acosta JC, Gil J. A role for CXCR2 in senescence, but what about in cancer? Cancer Res. 2009;69(6):2167–70. doi: 10.1158/0008-5472.CAN-08-3772. [DOI] [PubMed] [Google Scholar]
- 50.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.