Abstract
Previous studies have demonstrated a role for wound healing genes in resolution of cutaneous lesions caused by Leishmania spp. in both mice and humans, including the gene FLI1 encoding Friend leukaemia virus integration 1. Reduction of Fli1 expression in mice has been shown to result in up-regulation of collagen type I alpha 1 (Col1a1) and alpha 2 (Col1a2) genes and, conversely, in down-regulation of the matrix metalloproteinase 1 (Mmp1) gene, suggesting that Fli1 suppression is involved in activation of the profibrotic gene program. Here we examined single nucleotide polymorphisms (SNPs) in these genes as risk factors for cutaneous (CL) and mucosal leishmaniasis (ML), and leishmaniasis per se, caused by L. braziliensis in humans. SNPs were genotyped in 168 nuclear families (250 CL; 87 ML cases) and replicated in 157 families (402 CL; 39 ML cases). Family-based association tests (FBAT) showed the strongest association between SNPs rs1061237 (combined P=0.002) and rs2586488 (combined P=0.027) at COL1A1 and CL disease. This contributes to our further understanding of the role of wound healing in the resolution of CL disease, providing potential for therapies modulating COL1A1 via drugs acting on FLI1.
Keywords: COL1A1, Gene analysis, Cutaneous leishmaniasis, Wound healing
1. Introduction
American tegumentary leishmaniasis (ACL), caused by protozoan parasites of the genus Leishmania, is a major health problem in many low income countries. The infection is associated with a broad spectrum of clinical phenotypes, resulting from both environmental risk factors and a combination of the host’s immune and genetic background. In the last few years we have shown that polymorphisms at genes associated with wound healing and tissue repair are important risk factors for cutaneous leishmaniasis (CL) caused by Leishmania braziliensis (Castellucci et al., 2012; Castellucci et al., 2011). Thus, the FLI1 gene, initially identified and mapped as a gene controlling susceptibility to CL caused by L. major infection in mice (Sakthianandeswaren et al., 2010; Sakthianandeswaren et al., 2005), was also associated with development of CL in humans exposed to L. braziliensis in Brazil. In addition, our data showed that polymorphisms in other wound healing genes related to FLI1 function, in particular genes (TGFB1, TGFBR2, CTGF, SMAD2/3/7) encoding proteins in the transforming growth factor β (TGF-β) signaling pathway, were risk factors for CL. These data support the hypothesis that the normal physiological regulation of the wound healing process is important in the cure of ACL. Of interest, protective alleles for CL at these loci were often acting as risk alleles for mucosal leishmaniasis (ML) caused by L. braziliensis in the same population. This likely reflects the exaggerated pro-inflammatory response associated with ML disease, compared to the measured tumour necrosis factor and interferon-γ responses required to cure CL lesions.
Reduction of Fli1 expression in mice has been shown to result in up-regulation of collagen type I alpha 1 (Col1a1) and alpha 2 (Col1a2) genes and, conversely, in down-regulation of the matrix metalloproteinase 1 (Mmp1) gene, suggesting that Fli1 suppression is involved in activation of the profibrotic gene program (Nakerakanti et al., 2006). Both type I collagens and matrix metalloproteinases play an important role in the normal physiological and pathological conditions of many diseases (Alexakis et al., 2006; Amalinei et al., 2010; Imai et al., 2000; Wynn, 2008). Considering the role of these genes in the wound healing response, together with our previous data showing genetic association of their regulator gene FLI1 with CL in families from Brazil, we extended our analysis of the FLI1 pathway to determine whether polymorphisms at COL1A1, COL1A2 or MMP1 genes could also be involved in the outcome of ACL.
2. Materials and methods
2.1. Study site, diagnosis and sample collection
Our genetic studies are conducted in a region of rural rain forest, Corte de Pedra, Bahia, Brazil, where L. braziliensis is endemic. For host genetic association studies, two family-based cohorts were collected during two study periods, 2000–2004 and 2008–2010, as reported previously along with details of epidemiology and clinical phenotypes of disease (Castellucci et al., 2011; Castellucci et al., 2010; Castellucci et al., 2006). Sample collection for the first cohort was based on ascertainment of index cases of ML from medical records of the Corte de Pedra Public Health Post, and active follow-up to identify and collect all other family members, including those with current or past CL disease. This provided DNA samples (Table 1) from 168 nuclear families that contain 250 CL cases and 87 ML cases. Sample collection for the second cohort was based primarily on incident cases of CL or ML presenting to the health post, with family follow-up to acquire samples from parents and affected siblings, and unaffected siblings if one or both parents were missing. This provided DNA samples (Table 1) from 157 nuclear families that contain 402 CL cases and 39 ML cases. The characteristics of the two cohorts have been described in detail elsewhere (Castellucci et al., 2011), including diagnostic criteria for ML and CL disease.
Table 1.
Characteristics of collections made during the primary (2000–2004) and secondary (2008–2010) sampling periods.
| Primary Sample Period | Secondary Sample Period | |||||
|---|---|---|---|---|---|---|
| CL1 | ML1 | Leishmaniasis1 per se |
CL1 | ML1 | Leishmaniasis1 per se |
|
| No cases | 250 | 87 | 337 | 402 | 39 | 441 |
| Males | 128 | 60 | 188 | 219 | 24 | 243 |
| Females | 122 | 27 | 149 | 183 | 15 | 198 |
| Age at disease | ||||||
| Mean | 19.1 | 30.3 | 22.4 | 21.5 | 26.6 | 21.9 |
| 95% CI | 17.1–21.2 | 25.8–34.7 | 20.3–24.4 | 20.1–22.9 | 20.7–32.4 | 20.6–23.3 |
| No nuclear families | 168 | - | 168 | 157 | - | 157 |
| Total N in families/trios | 767 | - | 767 | 764 | - | 764 |
Unpaired t tests with Welch’s correction for unequal variances show that there are no signficant differences in age range for CL primary vs secondary, ML primary vs secondary, and Leishmaniasis per se primary vs secondary.
2.2. Sample collection and DNA extraction
Blood (8 ml) was taken by venipuncture and collected into dodecyl citrate acid (DCA)-containing vacutainers (Becton Dickinson). Genomic DNA was prepared using the proteinase K and salting-out method (Sambrook et al., 1989).
2.3. Genotyping
Genotyping was performed using pre-designed Taqman® qPCR assays (Life Technologies) for polymorphisms at COL1A1 (rs1061237, rs2586488, rs2075554), COL1A2 (rs388625, rs11770203) and MMP1 (rs5854, rs470747, rs7125062) as presented in Table 2. SNPs were selected pragmatically on the basis of prior use as tagging SNPs in other disease association studies (Erdei et al., 2013; Metlapally et al., 2009), availability of validated predesigned Taqman® qPCR assays, and MAF ≥0.15 for both CEU (Caucasian) and YRI (African) HapMap populations. These two reference populations were selected to mimic as closely as possible ethnic admixture in the population of Bahia. Analysis of linkage disequilibrium using Haploview v4.2 (Barrett et al., 2005) for SNPs with a MAF ≥0.15 for the CEU HapMap populations showed that these SNPs tagged >90% of the COL1A1 gene at D’>0.67 and ~30% cover at r2>0.58, >90% of COL1A2 at D’>0.56 and <20% cover at r2>0.58, and >90% of MMP1 at D’>0.65 and ~30% cover at r2>0.70. All SNPs were in Hardy Weinberg Equilibrium in genetically unrelated founders of the families (data not shown). Missingness (i.e. failure to score on Taqman assays) ranged from 0.80% (8/1008 individuals available for genotyping) to 2.5% (25/1008) across the 8 SNPs. PEDCHECK (O'Connell and Weeks, 1998) was used to determine Mendelian inconsistencies within families and genotypes for these individuals were set to zero for analysis.
Table 2.
Information on the single nucleotide polymorphisms genotyped for COL1A1, COL1A2 and MMP1 in the Brazilian population.
| Gene/Marker | Location | Physical Position (Chr:bp) |
Allelesa | MAFb | Caucasian | African |
|---|---|---|---|---|---|---|
| COL1A2_rs388625 | Intron | 7:94396612 | A/G | 0.48 | 0.48 | 0.42 |
| COL1A2_rs11770203 | Intron | 7:94402145 | G/T | 0.26 | 0.33 | 0.18 |
| MMP1_rs5854 | 3’UTR | 11:102790143 | A/G | 0.25 | 0.35 | 0.19 |
| MMP1_rs470747 | Intron | 11:102790864 | A/G | 0.34 | 0.35 | 0.35 |
| MMP1_rs7125062 | Intron | 11:102792772 | C/T | 0.33 | 0.27 | 0.35 |
| COL1A1_rs1061237 | 3’UTR | 17:50185414 | C/T | 0.30 | 0.31 | 0.42 |
| COL1A1_rs2586488 | Intron | 17:50188065 | A/G | 0.37 | 0.37 | 0.42 |
| COL1A1_rs2075554 | Intron | 17:50196948 | C/T | 0.26 | 0.19 | 0.26 |
Major>minor alleles for this Brazilian population.
Minor allele frequencies (MAF) for this Brazilian population, and for Caucasian (CEU) and African (YRI) populations as indicated.
2.4. Statistical analyses
Linkage disequilibrium (LD) was determined using Haploview v4.2 (Barrett et al., 2005). Family Based Association Tests (FBAT) were carried out under additive, dominant or genotype inheritance models using the –o flag to take both unaffected and affected offspring into account (Laird et al., 2000). ML patients were set to unknown phenotype in the analysis of the CL alone phenotype. Nominal P-values are presented throughout, i.e. without correction for multiple testing. The P-value needed to achieve statistical significance by applying a strict Bonferroni correction to take multiple testing into account was P=0.006 (=0.05/8 SNPs tested). Transmission disequilibrium test power approximations (Knapp, 1999) were used to determine power of the families to detect genetic associations.
3. Results
3.1. Characteristics of the samples and power to detect association
Table 1 provides details of the samples used in this study. Sample collections made during two different time periods, 2000–2004 and 2008–2010, were well-matched geographically (Castellucci et al., 2012) and demographically (Table 1). Table 2 provides details of SNPs genotyped, and demonstrates that all SNPs were at a MAF ≥0.25 in this Brazilian population. The P-value needed to achieve statistical significance taking multiple testing into account is P=0.006 (=0.05/8 SNPs tested). TDT power approximations (Knapp, 1999) showed that the first (250 CL cases) and second (402 CL cases) family datasets had 68% and 89% power, respectively, to detect an odds ratio ≥1.5 at P=0.01 for markers with MAF ≥0.3. The combined 652 trios had 98.7% power to detect association for the same effect size, P-value and MAF. Similarly, for leishmaniasis per se (i.e. CL + ML), power estimations for 337 primary, 441 replication, and 778 combined trios had 91.9%, 91.9%, and 99.6% power, respectively. Power to detect associations with ML disease was low (≤25%) even in the combined dataset.
3.2. Association tests for SNPs at COL1A1, COL1A2 and MMP1 as candidate genes for CL disease
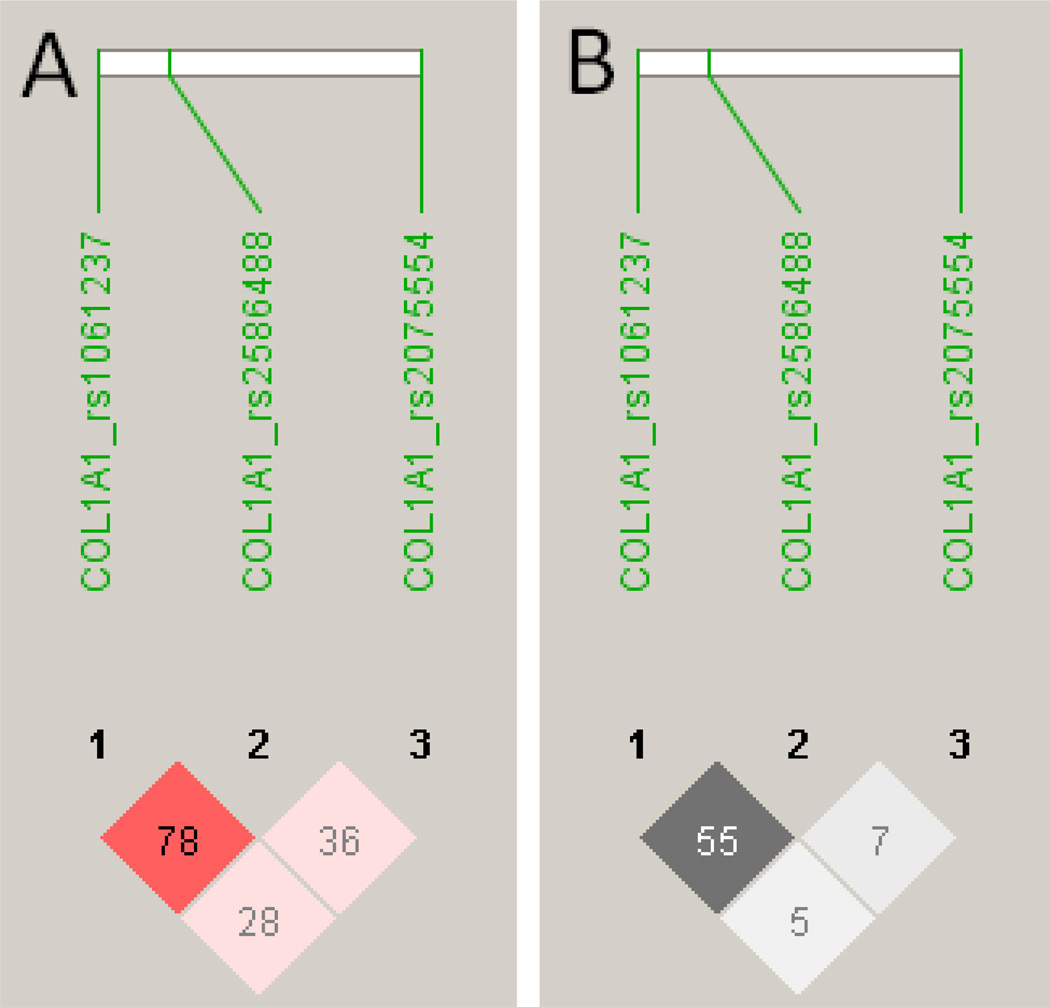
No associations were observed between CL disease and SNPs at COL1A2 or MMP1 (data not shown) used in this study. This does not rule out the possibility that associations could be uncovered given more dense SNP coverage of these two genes. Results of the COL1A1 FBAT analysis for the CL clinical phenotype in the primary (Families 101–168), replication (Families 169–325) and combined data set are presented in Table 3. Analysis under an additive model was sufficient to explain the data. SNP rs1061237 showed a trend for association (P=0.058) in the smaller primary family sample, achieved association at nominal P=0.018 in the replication family sample, and attained a P-value (P=0.002) in the combined analysis that withstands application of a strict correction factor for the 8 SNPs genotyped. SNP rs2586488 also provided evidence (nominal P<0.05) for association in the larger replication sample and the combined analysis. This is consistent with quite strong LD (D’ 0.78; r2 0.55) between these two SNPs (Figure 1). In contrast, there was no evidence for associations between CL disease and SNP rs2075554 and CL disease (Table 3), or of LD between this SNP and the two associated SNPs (Figure 1).
Table 3.
Results of Family-based Association Tests (FBAT) using an additive model of inheritance for associations between tag SNPs at COL1A1 (see Table 2) and CL caused by L. braziliensis in Brazil.
| Gene/Marker | Allele | Allele frequency |
No families |
Var(S) | Z | Pa |
|---|---|---|---|---|---|---|
| Families 101–168 | ||||||
| COL1A1_rs1061237 | C | 0.64 | 64 | 8.07 | 1.90 | 0.058 |
| COL1A1_rs1061237 | T | 0.36 | 64 | 8.07 | −1.90 | 0.058 |
| COL1A1_rs2586488 | G | 0.59 | 64 | 8.70 | 0.74 | 0.462 |
| COL1A1_rs2586488 | A | 0.41 | 64 | 8.70 | −0.74 | 0.462 |
| COL1A1_rs2075554 | C | 0.74 | 56 | 6.14 | 0.87 | 0.382 |
| COL1A1_rs2075554 | T | 0.26 | 56 | 6.14 | −0.87 | 0.382 |
| Families 169–325 | ||||||
| COL1A1_rs1061237 | C | 0.66 | 93 | 6.72 | 2.36 | 0.018 |
| COL1A1_rs1061237 | T | 0.34 | 93 | 6.72 | −2.36 | 0.018 |
| COL1A1_rs2586488 | G | 0.64 | 88 | 5.77 | 2.30 | 0.021 |
| COL1A1_rs2586488 | A | 0.36 | 88 | 5.77 | −2.30 | 0.021 |
| COL1A1_rs2075554 | C | 0.75 | 78 | 6.44 | 0.61 | 0.542 |
| COL1A1_rs2075554 | T | 0.25 | 78 | 6.44 | −0.61 | 0.542 |
| Combined analysis | ||||||
| COL1A1_rs1061237 | C | 0.66 | 157 | 14.84 | 3.07 | 0.002 |
| COL1A1_rs1061237 | T | 0.34 | 157 | 14.84 | −3.07 | 0.002 |
| COL1A1_rs2586488 | G | 0.63 | 152 | 14.66 | 2.21 | 0.027 |
| COL1A1_rs2586488 | A | 0.37 | 152 | 14.66 | −2.21 | 0.027 |
| COL1A1_rs2075554 | C | 0.75 | 134 | 12.61 | 1.08 | 0.279 |
| COL1A1_rs2075554 | T | 0.25 | 134 | 12.61 | −1.08 | 0.279 |
No Families = number of families informative for the FBAT analysis; V(S) is the variance.
A positive Z-score indicates association with disease; a negative Z-score indicates the non-associated or protective allele.
Bold indicates nominal P<0.05
Figure 1.
LD patterns for D’ (A) and r2 (B) were determined in Haploview software v4.2 (Barrett et al., 2005) for the 3 COL1A1 SNPs used in the association analyses. D' values and confidence levels (LOD) are represented as red for D'=1, LOD>2 (none present); shades of pink for varying D’, LOD<0.2; white for D'<1, LOD<2 (none present). r2 values are represented white for r2 = 0, with intermediate values for 0<r2 < 1 indicated by shades of grey. The numbers within the squares represent the D' or r2 scores for pairwise LD.
3.2. COL1A1 SNPs and CL versus ML disease
No associations were observed between ML disease, or leishmaniasis per se, and SNPs at COL1A2 or MMP1 (data not shown). The COL1A1 FBAT analysis for the ML clinical phenotype, and leishmaniasis per se (CL+ML disease) in the primary (Families 101–168), replication (Families 169–325) and combined data set are presented in Supplementary Tables 1 and 2, respectively. Of interest, a trend for association (nominal P=0.079) between rs2586488 and ML disease in the replication families (Supplementary Table 1) showed the minor allele as the risk allele, whereas for CL disease (Table 1) the major allele was the risk allele, reminiscent of earlier findings with other wound healing genes (Castellucci et al., 2012; Castellucci et al., 2011). This contributed to reduction in the strength of associations seen at both rs1061237 and rs2586488 in the analysis of CL+ML as leishmaniasis per se (Supplementary Table 2).
4. Discussion
Here we provide evidence for association between polymorphic variants at COL1A1 and susceptibility to CL disease caused by L. braziliensis in Brazil. This adds to our previous demonstration of roles for wound healing genes in determining the outcome of CL disease, supporting prior murine studies that had highlighted a role for wound healing pathways in the resolution of cutaneous forms of leishmaniasis (Sakthianandeswaren et al., 2005; Sakthianandeswaren et al., 2009).
Collagen type I is a predominant extracellular matrix component of the fibrotic lesion. Extracellular matrix remodeling is a complex and tightly regulated process that occurs during wound repair. In many pathological conditions the balance between extracellular matrix synthesis and degradation is disrupted, leading to abnormal remodeling (Border et al., 1996; Curran and Murray, 1999; Forget et al., 1999; Friedman, 1993; Malemud and Goldberg, 1999; Trojanowska et al., 1998; Vincenti et al., 1994). Leishmaniasis wound repair depends on a balanced immune response, as well as the cooperation of matrix elements and collagens. In addition, previous work suggests that type I collagen remodeling by promastigotes could play an important role during the infection process (Larsen et al., 2006; Petropolis et al., 2014; Stamenkovic, 2003). Even though the importance of the immunological response has been well established in the elimination of Leishmania infection and healing of resulting lesions, the mechanisms involved in skin damage and ulcer resolution needs to be better understood. For example, it needs to be clarified what happens after infection if the promastigotes pass through the dermis extracellular matrix, remaining there until the first contact with macrophages or other potential host cells (Lira et al., 1997; McGwire et al., 2003). Results presented here provide support for a role for polymorphism at COL1A1, which lies within the FLI1 network, in determining the outcome of L. braziliensis infection. This is consistent with our previous demonstration (Castellucci et al., 2011) of association between SNPs at FLI1 that are in strong linkage disequilibrium with functional elements known to influence FLI1 expression through epigenetic (CpG motifs) and enhancer activities. Epigenetic repression of the FLI1 gene is associated with enhanced COL1A1 expression (Wang et al., 2006), and there is a complex interplay between FLI1 and the TGF-β signaling pathway in regulating collagen deposition and fibrosis during the wound healing process. Further functional work will be required to evaluate the mechanisms of expression of these genes either in biopsies or cells from patients infected by L. braziliensis to determine their potential as therapeutic targets.
In brief, our data contribute to the dissection of an important pathway associated with development of CL, identifying host molecular biomarkers for this disease. This contributes to our further understanding of the role of wound healing in the resolution of CL disease, providing potential for therapies modulating COL1A1 via drugs acting on FLI1.
Supplementary Material
Highlights.
First demonstration that COL1A1 controls cutaneous leishmaniasis development.
Dissection of the FLI1 gene pathway identifying host biomarkers in leishmaniasis.
Validation of the mouse-to-human approach to identify genes of disease in humans.
Acknowledgements
We acknowledge Ednaldo Lago for the support in the field work in Brazil.
Funding
This work was supported by the NIH Grant AI 30639 and CNPq/INCT
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
Authors' contributions
LC, LA and JO carried out the field collection and preparation of the samples. LC supervised the laboratory work and statistical analyses. LA performed the genotyping. LA and LC undertook the statistical analyses, interpretation of the data, and preparation of the draft manuscript. LHG participated in the field collection of data. EMC helped conceive the study, initial selection of cases from the health post, and provided the logistical support to make the study possible. JMB participated in the design of the study, conceived the specific hypothesis to be tested, made the final interpretation of the data, and finalized preparation of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Lucas Almeida, Email: lucasfedrigo@hotmail.com.
Joyce Oliveira, Email: joycemouraoliveira@yahoo.com.br.
Luiz Henrique Guimarães, Email: luizhenriquesg@yahoo.com.br.
Edgar M Carvalho, Email: edgar@ufba.br.
Jenefer M Blackwell, Email: jmb37@cam.ac.uk, jblackwell@ichr.uwa.edu.au.
Léa Castellucci, Email: leacastel@hotmail.com.
References
- Alexakis C, Maxwell P, Bou-Gharios G. Organ-specific collagen expression: implications for renal disease. Nephron Exp Nephrol. 2006;102:e71–e75. doi: 10.1159/000089684. [DOI] [PubMed] [Google Scholar]
- Amalinei C, Caruntu ID, Giusca SE, Balan RA. Matrix metalloproteinases involvement in pathologic conditions. Rom J Morphol Embryol. 2010;51:215–228. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Border WA, Yamamoto T, Noble NA. Transforming growth factor beta in diabetic nephropathy. Diabetes Metab Rev. 1996;12:309–339. doi: 10.1002/(SICI)1099-0895(199612)12:4<309::AID-DMR171>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Almeida L, Oliveira J, Guimaraes LH, Lessa M, Fakiola M, Jesus AR, Nancy Miller E, Carvalho EM, Blackwell JM. Wound healing genes and susceptibility to cutaneous leishmaniasis in Brazil. Infect Genet Evol. 2012;12:1102–1110. doi: 10.1016/j.meegid.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Miller EN, de Almeida LF, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, Lago E, de Jesus AR, Carvalho EM, Blackwell JM. FLI1 polymorphism affects susceptibility to cutaneous leishmaniasis in Brazil. Genes Immun. 2011;12:589–594. doi: 10.1038/gene.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Miller EN, Menezes E, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, de Jesus AR, Carvalho EM, Blackwell JM. CXCR1 and SLC11A1 polymorphisms affect susceptibility to cutaneous leishmaniasis in Brazil: a case-control and family-based study. BMC Med Genet. 2010;11:10. doi: 10.1186/1471-2350-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Menezes E, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, Ribeiro S, Reale J, Noronha EF, Wilson ME, Duggal P, Beaty TH, Jeronimo S, Jamieson SE, Bales A, Blackwell JM, de Jesus AR, Carvalho EM. IL6 -174 G/C promoter polymorphism influences susceptibility to mucosal but not localized cutaneous leishmaniasis in Brazil. J Infect Dis. 2006;194:519–527. doi: 10.1086/505504. [DOI] [PubMed] [Google Scholar]
- Curran S, Murray GI. Matrix metalloproteinases in tumour invasion and metastasis. J Pathol. 1999;189:300–308. doi: 10.1002/(SICI)1096-9896(199911)189:3<300::AID-PATH456>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Erdei E, Luo L, Sheng H, Maestas E, White KA, Mackey A, Dong Y, Berwick M, Morse DE. Cytokines and tumor metastasis gene variants in oral cancer and precancer in Puerto Rico. PLoS ONE. 2013;8:e79187. doi: 10.1371/journal.pone.0079187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget MA, Desrosiers RR, Beliveau R. Physiological roles of matrix metalloproteinases: implications for tumor growth and metastasis. Can J Physiol Pharmacol. 1999;77:465–480. [PubMed] [Google Scholar]
- Friedman SL. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N Engl J Med. 1993;328:1828–1835. doi: 10.1056/NEJM199306243282508. [DOI] [PubMed] [Google Scholar]
- Imai K, Sato T, Senoo H. Adhesion between cells and extracellular matrix with special reference to hepatic stellate cell adhesion to three-dimensional collagen fibers. Cell Struct Funct. 2000;25:329–336. doi: 10.1247/csf.25.329. [DOI] [PubMed] [Google Scholar]
- Knapp M. A note on power approximations for the transmission disequilibrium test. Am. J. Hum. Genet. 1999;64:1177–1185. doi: 10.1086/302334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Larsen M, Artym VV, Green JA, Yamada KM. The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr Opin Cell Biol. 2006;18:463–471. doi: 10.1016/j.ceb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Lira R, Rosales-Encina JL, Arguello C. Leishmania mexicana: binding of promastigotes to type I collagen. Exp Parasitol. 1997;85:149–157. doi: 10.1006/expr.1996.4127. [DOI] [PubMed] [Google Scholar]
- Malemud CJ, Goldberg VM. Future directions for research and treatment of osteoarthritis. Front Biosci. 1999;4:D762–D771. doi: 10.2741/malemud. [DOI] [PubMed] [Google Scholar]
- McGwire BS, Chang KP, Engman DM. Migration through the extracellular matrix by the parasitic protozoan Leishmania is enhanced by surface metalloprotease gp63. Infect Immun. 2003;71:1008–1010. doi: 10.1128/IAI.71.2.1008-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metlapally R, Li YJ, Tran-Viet KN, Abbott D, Czaja GR, Malecaze F, Calvas P, Mackey D, Rosenberg T, Paget S, Zayats T, Owen MJ, Guggenheim JA, Young TL. COL1A1 and COL2A1 genes and myopia susceptibility: evidence of association and suggestive linkage to the COL2A1 locus. Invest Ophthalmol Vis Sci. 2009;50:4080–4086. doi: 10.1167/iovs.08-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakerakanti SS, Kapanadze B, Yamasaki M, Markiewicz M, Trojanowska M. Fli1 and Ets1 have distinct roles in connective tissue growth factor/CCN2 gene regulation and induction of the profibrotic gene program. J Biol Chem. 2006;281:25259–25269. doi: 10.1074/jbc.M600466200. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am.J.Hum.Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropolis DB, Rodrigues JC, Viana NB, Pontes B, Pereira CF, Silva-Filho FC. Leishmania amazonensis promastigotes in 3D Collagen I culture: an in vitro physiological environment for the study of extracellular matrix and host cell interactions. PeerJ. 2014;2:e317. doi: 10.7717/peerj.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Curtis JM, Elso C, Kumar B, Baldwin TM, Lopaticki S, Kedzierski L, Smyth GK, Foote SJ, Handman E. Fine mapping of Leishmania major susceptibility Locus lmr2 and evidence of a role for Fli1 in disease and wound healing. Infect Immun. 2010;78:2734–2744. doi: 10.1128/IAI.00126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Elso CM, Simpson K, Curtis JM, Kumar B, Speed TP, Handman E, Foote SJ. The wound repair response controls outcome to cutaneous leishmaniasis. Proc Natl Acad Sci U S A. 2005;102:15551–15556. doi: 10.1073/pnas.0505630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Foote SJ, Handman E. The role of host genetics in leishmaniasis. Trends Parasitol. 2009;25:383–391. doi: 10.1016/j.pt.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Stamenkovic I. Extracellular matrix remodelling: the role of matrix metalloproteinases. J Pathol. 2003;200:448–464. doi: 10.1002/path.1400. [DOI] [PubMed] [Google Scholar]
- Trojanowska M, LeRoy EC, Eckes B, Krieg T. Pathogenesis of fibrosis: type 1 collagen and the skin. J Mol Med (Berl) 1998;76:266–274. doi: 10.1007/s001090050216. [DOI] [PubMed] [Google Scholar]
- Vincenti MP, Clark IM, Brinckerhoff CE. Using inhibitors of metalloproteinases to treat arthritis. Easier said than done? Arthritis Rheum. 1994;37:1115–1126. doi: 10.1002/art.1780370802. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–2279. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.