Abstract
Autophagy is an intracellular self-digestion mechanism, by which cellular components are sorted into double-membrane autophagosomes and delivered to lysosomes for degradation. Cells utilize autophagy to dispose of wastes and eliminate hazards, while recycling nutrients and tuning metabolism in the process. Through these functions, autophagy promotes cell fitness, genome integrity, tissue homeostasis, and cell survival and growth under stress. Autophagy up- and down-regulation have both been found in human cancers, suggesting a complex role in tumor development. Accumulating results from autophagy-deficient mice and mouse models of human cancers have demonstrated that autophagy generally suppresses tumor initiation, but promotes tumor progression, in a manner that is dependent on timing and context and modified by specific tumorigenic events. Given the role of autophagy in facilitating tumor growth, autophagy inhibition has gained wide attention as a potential anticancer therapy. Here, we summarize relevant genetic, preclinical and clinical studies and discuss the multi-faceted role of autophagy in cancer, as well as the prospects of autophagy inhibition for cancer therapy.
Background
Autophagy as an intracellular self-digestion mechanism
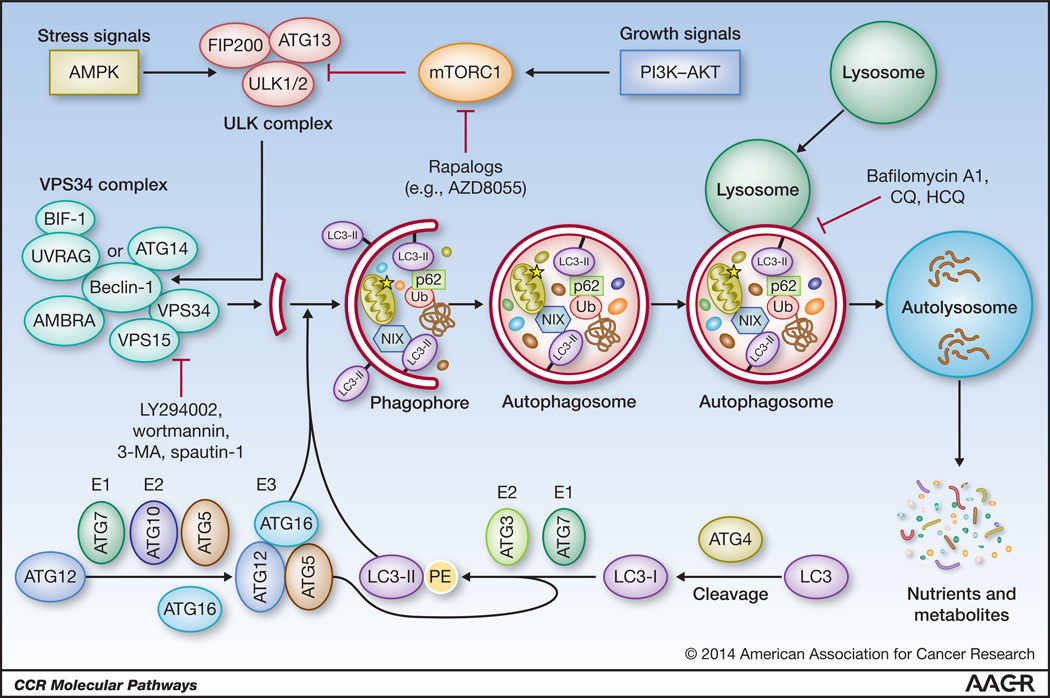
Macroautophagy (autophagy hereafter) is a catabolic process whereby cellular material is enclosed in the double-membrane autophagosomes and delivered to lysosomes for degradation (1–4). Autophagy begins with the formation of a crescent-shaped phagophore (or isolation membrane) (Fig. 1). This highly regulated process involves 2 key kinases, the UNC-51-like kinase (ULK) and the Class III phosphatidylinositol 3-kinase VPS34, their associated regulatory factors, such as FIP200, Beclin-1, UVRAG and BIF-1, and several other autophagy-related (ATG) proteins (Fig. 1). Upon induction, ULK (ATG1) phosphorylates Beclin-1 (ATG6) and activates the VPS34 complex (5). VPS34 generates phosphatidylinositol 3,4,5-triphosphate (PI3P) on the membrane destined to become a phagophore, and PI3P recruits proteins required for phagophore elongation. Phagophore elongation requires the incorporation of phosphatidylethanolamine (PE)-lipidated LC3 (ATG8), whose formation is catalyzed by two “ubiquitin–like conjugation systems” composed of multiple other ATG proteins such as ATG5 and ATG7, etc. (2). The phagophore elongates until its membranes fuse, generating an autophagosome, which eventually fuses with the lysosome forming an autolysosome, where the resident lysosomal hydrolases breakdown the cargo.
Figure 1.
The autophagic process. Upon deprivation of nutrients or growth factors, activation of AMPK and/or inhibition of mTOR lead to activation of ULK, which phosphorylates Beclin-1, leading to VPS34 activation and phagophore formation. ULK functions in a complex with FIP200 and ATG13, while VPS34 function requires a regulatory subunit, VPS15 (p150), and Beclin-1, which further mediates the association of other regulatory factors such as AMBRA, ATG14, UVRAG and BIF-1. Multiple ATG proteins such as ATG5 and ATG7 constitute two “ubiquitin–like conjugation systems” that catalyze the formation of phosphatidylethanolamine (PE)-conjugated LC3 (LC3-II) and direct its proper incorporation into the phagophore membrane, where it serves as docking site of adaptor proteins (and bound cargos). The closure of an elongated phagophore marks the formation of a mature autophagosome, which eventually fuses with a lysosome, leading to cargo degradation and recycling of nutrients and metabolites. Ub, ubiquitin.
Basal autophagy uses adaptor proteins, such as p62/SQSTM1 and NIX, to identify and deliver misfolded or aggregated proteins and damaged organelles to the autophagosome for degradation (6, 7), thereby preserving cellular fitness. Key to this selective cargo delivery are the specific interactions between the adaptor proteins and the phagophore membrane-bound LC3 (LC3-II), which serves as a cargo receptor Under stress conditions, such as oxygen and/or nutrient deprivation, autophagy is induced as a survival mechanism to recycle cytoplasmic constituents and generate fresh nutrients for cellular metabolism, e.g. macromolecule biosynthesis and energy production (8, 9). Stress-induced autophagy relies on non-selective engulfment of cytoplasmic material by the phagophore for degradation.
Autophagy suppresses oxidative stress and genome instability
Many studies have demonstrated the importance of functional autophagy in limiting oxidative stress (10–14). Electron leakage from mitochondrial electron transport to molecular oxygen is a main source of intracellular reactive oxygen species (ROS). As damaged mitochondria are mainly eliminated by autophagy (i.e. mitophagy) (15), autophagy-deficient cells accumulate dysfunctional mitochondria (12, 16), which are thought to be leaky and produce ROS. In addition, autophagy potentially may also reduce ROS levels by generating reducing powers through its metabolic function or by regulating proteins involved in antioxidant production.
Increased genome instability is a common feature of autophagy-deficient cells (10–12). This has been reviewed recently (17). Since ROS can cause mutations and DNA strand breaks, elevated ROS production in autophagy-deficient cells is considered as a key factor causing genome instability. Autophagy can also promote DNA repair and regulate cell division. During starvation, autophagy via its nutrient recycling function maintains the levels ATP and dNTP (8), both of which are important for DNA repair. Moreover, autophagy regulates specific proteins involved in lesion processing and nucleotide production, e.g. Sae2 and Rnr1 (18, 19). Finally, autophagy deficiency in yeast causes premature nuclear division in starvation, leading to aneuploidy upon nutrient replenishment (20).
Autophagy deregulation in human cancers
Autophagy was first linked to cancer when 40%, 50%, and 70% of prostate, breast, and ovarian cancers, respectively, were reported to have allelic loss of BECN1, which encodes the essential VPS34 complex component Beclin-1 (21). This finding suggests that Beclin-1, and autophagy in general, may be a tumor suppressor. However, recent investigation of BECN1 mutational status in The Cancer Genome Atlas (TCGA) has called into question the significance of allelic BECN1 deletions in cancers (22), since they occur in conjunction with deletion of the breast and ovarian tumor suppressor gene BRCA1, located only ~150 KB away on chromosome 17q21. Notably, hypermethylation of the BECN1 promoter has been observed in up to 70% of breast cancers (23), and low BECN1 mRNA levels have been reported in ovarian (24), colon (25), brain (26), and liver (27) malignancies. Thus, defective autophagy due to epigenetic silencing and/or transcriptional inhibition of BECN1 may play a role in the etiology and/or progression of these tumors.
Other than BECN1, allelic UVRAG loss is noted in colon (28) and gastric (29) cancers; whereas LAPTM4B, which maintains lysosomal pH and allows autophagosome-lysosome fusion, is frequently amplified and overexpressed in breast cancer (30). Furthermore, cancers often exhibit functional suppression of autophagy, especially upon activation of the PI3K-AKT-mTOR pathway, which inhibits autophagy (31). For example, approximately 25% of breast cancers have amplification of HER2 (ERBB2), which encodes a receptor tyrosine kinase that activates the above pathway.
Autophagy-deficient mice and allograft/xenograft studies
Homozygous Becn1 deletion leads to embryonic lethality, while Becn1+/− mice are viable, display partial autophagy defect and develop liver and lung tumors, lymphomas, and mammary hyperplasias (32, 33). Monoallelic Becn1 loss-associated mammary hyperplasias progress to tumors following parity in FVB background, but not in C57BL/6 background (34). Mice with mosaic Atg5 deletion or liver-specific Atg7 deletion develop benign hepatomas (35), and Bif-1 knockout mice show increased incidence of lymphomas (36). Collectively, these results indicate that basal autophagy suppresses tumor development. Consistent with this notion, Becn1+/− immortalized mouse mammary epithelial cells (iMMECs) and baby mouse kidney (iBMK) cells are more tumorigenic than their Becn1+/+ counterparts in nude mice (10, 13). Also, EGFR- and AKT-mediated Beclin-1 phosphorylation, which both attenuate autophagy, promote the growth of lung and breast cancer xenografts, respectively (37, 38).
Genetic modulation of autophagy in mouse tumor models
Complete or partial autophagy deficiency has been introduced into several mouse tumor models (Table 1). As aging Becn1+/− mice develop spontaneous lymphomas (32, 33), the role of autophagy has been assessed in mouse lymphoma models. In Atm−/− mice, which spontaneously develop lymphomas, monoallelic Becn1 loss delayed tumor development and increased survival (T50=262 vs 137 days, p=0.006) (39). In contrast, monoallelic Becn1 loss in an Eµ-MYC-driven lymphoma model led to faster tumor development and decreased survival (T50=80 vs 142 days, p=0.007) (39). Bif-1−/− and Bif-1+/− mice also showed accelerated Eµ-MYC-driven lymphomagenesis (T50=65 and 75 days vs 107 days, p=0.0006 and p<0.0001, respectively) (12). These studies demonstrate that the role of autophagy in lymphoma depends on the specific drivers of tumor development.
Table 1.
Tumor phenotypes of autophagy-deficient mice and the effect of autophagy deficiency on tumor development in established mouse tumor models.
| Autophagy defect | Oncogenic driver | Genetic background |
Lesion or tumor type | Effect | Ref. |
|---|---|---|---|---|---|
| Becn1+/− | N/A | C57BL/6 | Liver/lung tumors, lymphomas, mammary hyperplasias (independent of parity) |
|
32, 33 |
| Becn1+/− | N/A | FVB/N | Mammary hyperplasias in nulliparous mice Mammary tumors after parity |
|
34 |
| Atg5F/F;CAG-Cre | N/A | C57BL/6 | Hepatomas | 35 | |
| Mosaic Atg7 deletion | N/A | C57BL/6 | Hepatomas | 35 | |
| Bif1−/− | N/A | mixed | Lymphomas | 36 | |
| Becn1+/− | Atm−/− | C57BL/6 | Lymphoma | 39 | |
| Becn1+/− | Eu-Myc | C57BL/6 | Lymphoma | 39 | |
| Bif1−/− and Bif1+/− | Eu-Myc | C57BL/6 | Lymphoma | 12 | |
| FIP200F/F;MMTV-Cre | MMTV-PyMT | FVB/N | Mammary tumors Lung metastases |
|
40 |
| Becn1+/− | Palb2F/F;WAP-Cre | Mixed | Mammary tumors | 41 | |
| Becn1+/− | Palb2F/F;Trp5F/F;WAP-Cre | Mixed | Mammary tumors | Neutral | 41 |
| Becn1+/− | MMTV-Neu | Mixed | Mammary tumors | Neutral | 42 |
| Becn1+/− | MMTV-PyMT | FVB/N | Mammary tumors | Neutral | 42 |
| Becn1+/− | MMTV-Wnt1 | FVB/N | Mammary tumors (basal-like) | 34 | |
| Atg5F/F;Adeno-Cre* | LSL-Kras(G12D) | ND | Early lung tumors Late lung tumors*** |
|
43 |
| Atg5F/F;Adeno-Cre* | LSL-Kras(G12D);Trp53F/F** | ND | Early lung tumors Late lung tumors*** |
Neutral Neutral |
43 |
| Atg7F/F;Adeno-Cre* | LSL-Kras(G12D) | C57BL/6 | Early lung tumors Late lung tumors*** |
ND |
45 |
| Atg7F/F;Adeno-Cre* | LSL-Kras(G12D);Trp53F/F** | ND | Early lung tumors Late lung tumors*** |
ND |
45 |
| Atg7F/F;Adeno-Cre* | LSL-Braf(V600E) | mixed | Early lung tumors Late lung tumors*** |
|
44 |
| Atg7F/F;Adeno-Cre* | LSL-Braf(V600E);Trp53F/F** | mixed | Early lung tumors Late lung tumors*** |
|
44 |
| Atg5F/F;Pdx1-Cre | LSL-Kras(G12D) | ND | Low-grade PanIN High-grade PanIN and PDAC |
|
48 |
| Atg7F/F;Pdx1-Cre | LSL-Kras(G12D) | ND | Low-grade PanIN High-grade PanIN and PDAC |
|
48 |
| Atg5F/F;Pdx1-Cre | LSL-Kras(G12D);Trp53F/F** | ND | PDAC | 48 | |
| Atg7F/F;Pdx1-Cre | LSL-Kras(G12D);Trp53F/F** | ND | PDAC | 48 | |
| Atg5F/F;Pdx1-Cre | LSL-Kras(G12D);Trp53F/W | mixed | Low-grade PanIN High-grade PanIN and PDAC |
|
49 |
ND, not described;
deletion is driven by intranasal delivery of Adeno-Cre;
Trp53 is co-deleted with Atg5 or Atg7 in the same cells;
in autophagy-proficient setting, lung tumors (early and late) are adenomas and adenocarcinomas under Trp53+/+ and Trp53-null conditions, respectively, whereas in autophagy-deficient setting, late lung tumors are benign oncocytomas regardless of Trp53 status.
The role of autophagy has also been assessed in multiple breast cancer models. In the polyoma middle T-antigen (MMTV-PyMT) driven model, conditional biallelic Fip200 deletion delayed tumor development (T50=85 vs 62 days, p<0.01) (40). In a model of hereditary breast cancer, monoallelic Becn1 loss delayed and reduced tumor development induced by conditional loss of the PALB2 tumor suppressor (27% vs 66% penetrance, p=0.0035) (41). However, Becn1 heterozygosity did not affect mammary tumorigenesis driven by HER2 or PyMT oncogenes (42). Interestingly, monoallelic Becn1 loss accelerated WNT1-driven mammary tumorigenesis (T50=120 vs 219 days, p=0.004) (34). In this case, allelic BECN1 loss appears to promote mammary tumorigenesis by deregulating the mammary hierarchy and expanding the mammary progenitor cell (MaPC) population, thus cooperating with WNT1 activation, which drives MaPC transformation.
Recent studies using the KRASG12D- and BRAFV600E-driven models of non-small cell lung cancer (NSCLC) have revealed that autophagy defects both promote early tumorigenesis (43, 44) and impede tumor progression (44, 45). Conditional biallelic Atg7 deletion slowed the progression of KRASG12D–driven lung tumors, but was not associated with any survival benefit, as mice with Atg7-deficient pulmonary epithelium developed pneumonia and died at the same time as the control mice with higher tumor burden (45). Interestingly, Atg7 deletion in a BRAFV600E-driven NSCLC mouse model accelerated the onset of tumor development but delayed tumor progression and prolonged survival (44). The same dual effects were also observed when Atg5 was deleted in the KRASG12D-driven lung cancer model (43). An important finding from these studies is that autophagy impacts the histological fate, and likely the aggressiveness, of KRASG12D- and BRAFV600E-driven lung tumors, as either Atg5 or Atg7 deletion results in development of oncocytomas instead of adenocarcinomas (43–45), which are benign tumors characterized by massive accumulation of abnormal mitochondria (46).
RAS-mutant pancreatic cancer cells have high basal autophagy and seem to be “addicted” to autophagy for growth (47). Recently, the role of autophagy in pancreatic ductal adenocarcinoma (PDAC) has been thoroughly assessed in a KRASG12D-driven model in two separate studies (48, 49). Normally, these mice develop a small number of pre-malignant pancreatic intraepithelial neoplasia (PanIN) lesions, which stochastically evolve into PDAC over time. Interestingly, conditional biallelic deletion of either Atg5 or Atg7 led to increased incidence of PanIN, but blocked its progression to high-grade hyperplasia and PDAC (48, 49), reminiscent of the effect produced by Atg7 or Atg5 deletion in the above BRAF- and KRAS-driven lung cancer models, respectively.
Besides the timing-, context- and dosage-dependent role of autophagy in tumor development, the above studies also reveal p53 status as a key parameter impacting the role of autophagy in cancer. Specifically, Becn1 haploinsufficiency delayed Palb2-associated mammary tumorigenesis in Trp53-wild type mice, but not when Trp53 was co-deleted with Palb2 in the mammary epithelium (41). Similarly, co-deletion of Trp53 diminished or eliminated the inhibitory effect of either Atg5 or Atg7 deletion on tumor progression in both KRAS- and BRAF-driven lung cancer models (43–45). These results indicate that a key function of autophagy may be to limit p53 induction and/or overcome the barrier imposed by p53 activation to tumorigenesis, and they also suggest that autophagy may be less relevant in p53-mutant cancers. However, in the KRASG12D-driven PDAC model, Atg5 or Atg7 ablation accelerated tumor progression and reduced survival when Trp53 was also deleted simultaneously (48). Thus, how p53 status influences the role of autophagy in cancer may vary from tissue to tissue. A possible caveat of the above models is that they all involve biallelic Trp53 deletion at the time of another oncogenic event, whereas in human cancers, TP53 status generally changes by acquisition of point mutations and loss of heterozygosity (LOH) over time. Nonetheless, given its key role in cell cycle control, apoptosis and metabolism, the final TP53 status in established tumors is still likely to modify the impact of autophagy defect or inhibition on tumor growth and progression.
Finally, studies using tumor-derived cell lines from the above models have revealed novel functions of autophagy in metabolism. In particular, Atg7–null KRAS- and BRAF-driven lung cancer cells showed defects in mitochondrial lipid oxidation and low levels of tricarboxylic acid (TCA) cycle intermediates and glutamine, which was found to be critical for the survival of the cancer cells (44, 45, 50). Also, decreased glycolysis was observed in Fip200-deficient mammary tumor cells (40), potentially connecting autophagy to increased aerobic glycolysis in cancer, known as the “Warburg effect”.
Clinical-Translational Advances
Chemical autophagy inhibition in vitro and in mouse tumor models
Autophagy inhibitors can be broadly divided into two groups, with one group inhibiting autophagosome formation (early-stage inhibitors) and the other blocking autophagosome-lysosome fusion (late-stage inhibitors). The currently available early-stage inhibitors, including 3-Methyladenine (3-MA), wortmannin, LY294002 and the newly identified spautin-1, all target the VPS34 complex. The first three interfere with its membrane recruitment (51), whereas spautin-1 promotes its degradation (52). Late-stage inhibitors include bafilomycin A1, chloroquine (CQ) and hydroxychloroquine (HCQ). These drugs effectively inhibit lysosome acidification, block autophagosome-lysosome fusion and, thus, cargo degradation (51).
The efficacy of pharmacologic autophagy inhibition in killing cancer cells has been assessed in numerous studies. As single agents, autophagy inhibitors generally inhibit the growth and/or survival of cancer cells with high basal autophagy, such as RAS-driven cancer cells, which markedly upregulate and are addicted to autophagy (47, 50). CQ/HCQ treatment also delays tumorigenesis and/or improved survival in lymphomas caused by either Atm deficiency or MYC activation (53, 54). Importantly, dozens of anticancer agents as well as radiation have been found to induce autophagy, and cancer cells not relying on autophagy under normal growth conditions may induce autophagy as a survival mechanism in response to anticancer therapies (55–57). Thus, combining autophagy inhibition with autophagy-inducing therapeutics may achieve better tumor cell killing. For example, CQ sensitized HER2-positive and hormone-refractory estrogen receptor (ER)-positive breast cancer cells to HER2-targeting therapies (42) and tamoxifen (58), respectively. This strategy has seen promise in pre-clinical studies and is currently under clinical investigation. However, it should be noted that there exists evidence that suggests that therapy-induced autophagy may contribute to cell killing (56, 57). Thus, the role of autophagy in cancer therapy needs to be determined case by case in different cancer-drug combinations.
Clinical trials involving autophagy modulation by HCQ
There are currently 27 NCI-registered clinical trials actively evaluating the therapeutic efficacy of pharmacologic autophagy inhibition in different tumor types. In all trials, autophagy is modulated by HCQ given in combination with standard chemotherapy (and concurrent radiation therapy in one trial). In an earlier Phase I study, addition of HCQ to erlotinib produced partial response (PR) in 1 out of 19 NSCLC patients with EGFR-mutant tumors (59). The responding patient received 600 mg HCQ daily. Results of several other studies have recently become available. In a phase I/II trial of HCQ in conjunction with radiation therapy and concurrent/adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme (GBM), the maximum tolerated dose (MTD) for HCQ was 600 mg daily, which resulted in inconsistent autophagy inhibition, as evaluated by autophagic vacuole (AV) formation in peripheral blood mononuclear cells (PBMCs), and did not prolong overall survival (60). In a phase I study combining the HDAC inhibitor vorinistat with escalating doses of HCQ in patients with advanced solid tumors, the MTD for HCQ was again 600 mg daily; although treatment-related changes in AV numbers in PBMCs were not seen, increases in the expression of CDKN1A and CTSD were reported and were more pronounced in tumor biopsies than PBMCs; out of 24 evaluable patients, 1 patient with renal cell carcinoma (RCC) had confirmed durable PR and 2 patients with colorectal cancer have prolonged stable disease (SD) (61). In a phase I trial combining bortezomib and HCQ for relapsed/refractory myeloma, PR, minor response and SD were observed in 14%, 14% and 45% of patients, respectively (62). In this study, HCQ was given at 600 mg twice daily with standard doses of bortezomib and resulted in therapy-associated AV number increases in bone marrow plasma cells. In another phase I study, combination of HCQ with dose-intense temozolomide resulted in 14% PR and 27% SD in patients with metastatic melanoma and induced significant AV accumulation in PBMCs; the recommended phase II dose was HCQ 600 mg twice daily (63). Finally, combined MTOR and autophagy inhibition in a phase I/II trial of HCQ and temsirolimus resulted in SD in about 70% of patients with melanoma; the HCQ dose in the phase II part of the study was 600 mg twice daily and at that, but not lower, dose, autophagy inhibition was documented in PMBCs and tumor biopsies (64).
Taken together, the results of the above studies indicate that, when tolerated, the combination of HCQ at higher dose (600 mg twice daily) with standard chemotherapy regimens modulates autophagy in patients and has antitumor activity. Treatment-related toxicities limit the use of high-dose HCQ in combination with vorinostat and during adjuvant chemoradiotherapy for newly resected GBM, thus indicating a need for the development of lower toxicity compounds that inhibit autophagy more consistently than HCQ.
CONCLUSIONS AND PERSPECTIVES
The role of autophagy in cancer is complex and varies depending on the timing and context of tumor development. Yet, despite the seemingly paradoxical roles of autophagy under different settings, two principles have been emerging from the mouse models studied so far. First, basal autophagy generally suppresses tumor initiation. This may be achieved by suppressing ROS, which causes DNA damage and genome instability, thereby promoting age-associated, spontaneous tumor development, as seen in autophagy deficient mice. Also, modestly increased ROS can directly stimulate cell growth, which may contribute to the initial acceleration of tumor (or pre-cancerous lesion) formation observed in the above oncogene-driven models. Second, in contrast to its suppressive role in tumor initiation, autophagy facilitates tumor progression in most cases examined so far. This function may be exerted by autophagy-mediated mitigation of excessive ROS, suppression of DNA damage response, recycling of nutrients for biosynthesis and energy production, maintenance of mitochondrial and lipid metabolism, and possibly degradation of key regulators of cell growth.
How the functional status of autophagy impacts tumorigenesis likely depends on how ROS levels, DNA damage and autophagy-related metabolism affect the fate of a given tumor at a particular stage of its trajectory. The outcome may be decided by the specific oncogene activation and/or tumor suppressor loss that drives cancer initiation and progression, perhaps in conjunction with the tissue/cell type of origin. It is important to appreciate the genetic/epigenetic and physiological differences between early and late stage tumors. Early lesions tend to be more “authentic” and uniform, therefore may be affected more readily by changes in autophagy status. Late stage tumors have often acquired additional alterations in key regulatory genes, such as TP53 and NRF2, which may dramatically alter their requirement for autophagy and therefore its role in tumor progression.
Given the role of autophagy in generally facilitating tumor progression, targeting autophagy has considerable potential for cancer therapy. To better harness this potential, it is critical to determine the exact “context” for each tumor, particularly the status of key oncogenes and tumor suppressors, such as the ones noted before. As tumors with high levels of basal autophagy, ROS and DNA damage or under considerable metabolic stress are more likely reliant on autophagy for survival and growth, and should thus be more sensitive to drugs that inhibit autophagy, a set of effective biomarkers to diagnose these autophagy-addicted malignancies may be useful. Also, given the context-dependent tumor-suppressive function of autophagy, the long-term consequences of chronic or periodic pharmacologic autophagy inhibition need to be determined. In the same vein, it is imperative to develop novel agents that specifically target mechanisms upregulating autophagy in cancer cells, while leaving basal autophagy in normal cells intact.
Acknowledgments
Grant Support
This work was supported by the National Cancer Institute (R01CA138804; to B.X.), the American Cancer Society (RSG #TBG-119822; to B.X.), the Damon Runyon Cancer Research Foundation (Clinical Investigator Award; to V.K.), and the New Jersey Commission on Cancer Research (predoctoral fellowship; to M.C.)
Footnotes
Disclosure of Potential Conflicts of Interest
V. Karantza was an employee of Rutgers University at the time of article submission; she is now an employee of Merck. No potential conflicts of interest were disclosed by the other authors.
REFERENCES
- 1.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Molecular Biol. 2014;21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 7.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Devel. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Deve. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi Y, Hori T, Cooper TK, Liao J, Desai N, Serfass JM, et al. Bif-1 haploinsufficiency promotes chromosomal instability and accelerates Myc-driven lymphomagenesis via suppression of mitophagy. Blood. 2013;121:1622–1632. doi: 10.1182/blood-2012-10-459826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng D, Liu L, Zhu Y, Chen Q. Molecular signaling toward mitophagy and its physiological significance. Exp Cell Res. 2013;319:1697–1705. doi: 10.1016/j.yexcr.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 16.Guo JY, White E. Autophagy is required for mitochondrial function, lipid metabolism, growth and fate of KRAS(G12D)-driven lung tumors. Autophagy. 2013;9:1636–1638. doi: 10.4161/auto.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vessoni AT, Filippi-Chiela EC, Menck CF, Lenz G. Autophagy and genomic integrity. Cell Death Differ. 2013;20:1444–1454. doi: 10.1038/cdd.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyavaiah M, Rooney JP, Chittur SV, Lin Q, Begley TJ. Autophagy-dependent regulation of the DNA damage response protein ribonucleotide reductase 1. Mol Cancer Res. 2011;9:462–475. doi: 10.1158/1541-7786.MCR-10-0473. [DOI] [PubMed] [Google Scholar]
- 19.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsui A, Kamada Y, Matsuura A. The role of autophagy in genome stability through suppression of abnormal mitosis under starvation. PLoS Genet. 2013;9:e1003245. doi: 10.1371/journal.pgen.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 22.Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res. 2014;12:485–490. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Chen B, Wu Y, Jin F, Xia Y, Liu X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. doi: 10.1186/1471-2407-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Chen S, Gou WF, Xiao LJ, Takano Y, Zheng HC. Aberrant Beclin 1 expression is closely linked to carcinogenesis, differentiation, progression, and prognosis of ovarian epithelial carcinoma. Tumour Biol. 2014;35:1955–1964. doi: 10.1007/s13277-013-1261-6. [DOI] [PubMed] [Google Scholar]
- 25.Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A. Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 2007;27:1453–1457. [PubMed] [Google Scholar]
- 26.Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30:429–436. [PubMed] [Google Scholar]
- 27.Daniel F, Legrand A, Pessayre D, Borrega-Pires F, Mbida L, Lardeux B, et al. Beclin 1 mRNA strongly correlates with Bcl-XLmRNA expression in human hepatocellular carcinoma. Cancer Invest. 2007;25:226–231. doi: 10.1080/07357900701206323. [DOI] [PubMed] [Google Scholar]
- 28.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 29.Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Human Pathol. 2008;39:1059–1063. doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Iglehart JD, Richardson AL, Wang ZC. The amplified cancer gene LAPTM4B promotes tumor growth and tolerance to stress through the induction of autophagy. Autophagy. 2012;8:273–274. doi: 10.4161/auto.8.2.18941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cicchini M, Chakrabarti R, Kongara S, Price S, Nahar R, Lozy F, et al. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. doi: 10.4161/auto.34398. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nature Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valentin-Vega YA, Maclean KH, Tait-Mulder J, Milasta S, Steeves M, Dorsey FC, et al. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov. 2013;3:894–907. doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozy F, Cai-McRae X, Teplova I, Price S, Reddy A, Bhanot G, et al. ERBB2 overexpression suppresses stress-induced autophagy and renders ERBB2-induced mammary tumorigenesis independent of monoallelic loss. Autophagy. 2014;10:662–676. doi: 10.4161/auto.27867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 44.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta. 2011;1807:633–642. doi: 10.1016/j.bbabio.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 49.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen S, Kepp O, Michaud M, Martins I, Minoux H, Metivier D, et al. Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study. Oncogene. 2011;30:4544–4556. doi: 10.1038/onc.2011.168. [DOI] [PubMed] [Google Scholar]
- 56.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Molecular Pharmacology. 2014;85:830–838. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112:389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 59.Goldberg SB, Supko JG, Neal JW, Muzikansky A, Digumarthy S, Fidias P, et al. A phase I study of erlotinib and hydroxychloroquine in advanced non-small-cell lung cancer. J Thorac Oncol. 2012;7:1602–1608. doi: 10.1097/JTO.0b013e318262de4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, et al. Combined autophagy and HDAC inhibition: A phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10:1403–1414. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vogl DT, Stadtmauer EA, Tan KS, Heitjan DF, Davis LE, Pontiggia L, et al. Combined autophagy and proteasome inhibition: A phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, et al. Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]