Abstract
Recent findings identified the minor A allele present in the single nucleotide polymorphism rs3865444 in the CD33 gene as being associated with reduced risk of developing Alzheimer’s disease (AD). CD33 (Siglec-3) is an immune function protein with anti-inflammatory signaling, cell adhesion and endocytosis functions with sialic acid-modified proteins or lipids as ligands. Its involvement in AD pathological mechanisms is still unclear, so the goal of this study was to investigate if the rs3865444 polymorphism affects development of AD pathology and the expression of CD33 mRNA and protein. For this study, we utilized DNA from 96 non-demented (ND) and 97 AD neuropathologically diagnosed cases to identify the different rs3865444 alleles and correlate with different measures of AD pathology. Using semi-quantitative histological measures of plaque and tangle pathology, we saw no significant differences between the different genotypes within these disease groups. However, increased expression of CD33 mRNA was associated with increasing AD pathology in temporal cortex brain samples. We also showed that cases with A/A alleles had reduced levels of CD33 protein in temporal cortex, but increased levels of the microglia protein IBA-1. Using immunohistochemistry on temporal cortex sections, CD33 was selectively localized to microglia, with greater expression in activated microglia. The factors causing increased CD33 expression by microglia in brain are still unclear, although both genetic and disease factors are involved. Treatment of human microglia isolated from autopsy brains with amyloid beta (Aβ) peptide and a range of other inflammatory activating agents resulted in reduced CD33 mRNA and protein levels.
Keywords: Microglia, Siglec-3, Immune signaling, Immunoreceptor tyrosine-based inhibitory motif, Amyloid beta, Western blot, Immunohistochemistry, Cytokines
1. Introduction
Alzheimer’s disease (AD) is a significant and growing public health issue amongst elderly populations (Carrillo, 2013). Unless effective therapies can be identified, it will become a problem of epidemic proportion for the progressively aging population. Prevention of amyloid buildup or promotion of its removal using various strategies have been tested in a range of clinical trials, but they have generally not shown significant effects (Golde, 2014;Li et al, 2013;O'Banion, 2013;Panza et al, 2014;Winblad et al, 2014). Disease-modifying strategies need to consider other factors beyond amyloid, particularly those that change early in the disease (Herrup et al, 2013). Inflammation as a feature of AD pathology has been widely characterized since the initial discoveries of activated microglia associated with AD hallmark features (recent reviews (Czirr and Wyss-Coray, 2012;Lucin and Wyss-Coray, 2009;Mosher and Wyss-Coray, 2014;Querfurth and LaFerla, 2010). There is much evidence that inflammation can accelerate neuronal cell death ongoing in AD, however it is also associated with amyloid removal by microglia and so modulation of inflammation has to be a balanced approach (Chakrabarty et al, 2010a;Chakrabarty et al, 2010b). To date, clinical trials of anti-inflammatory agents for treating AD have not proven to have significant disease-slowing effects, but epidemiological data suggest long term manipulation of inflammation may be neuroprotective (Szekely and Zandi, 2010;Vlad et al, 2008).
Understanding disease mechanisms of AD have been advanced by the identification of genetic risk factors. Certain mutations in the amyloid precursor protein (APP) or presenilin result in the autosomal dominant transmission of AD (Bertram et al, 2010;Schellenberg and Montine, 2012). Possession of the apolipoprotein E ε4 allele is a highly replicated factor associated with increased risk of developing AD (Liu et al, 2013;Piaceri et al, 2013). Recently, large scale genome wide association studies (GWAS) for disease-associated single nucleotide polymorphisms (SNPs) have identified genes associated with altered risks of AD that are also involved in inflammation. These genes include CR1 (complement receptor 1), CLU (clusterin), EPHA1 (Ephrin A1 receptor), MS4A4E/MS4A6A (membrane spanning 4A) and CD33 (Hollingworth et al, 2011;Naj et al, 2011). More recently, separate studies have shown that a rare SNP in the triggering receptor expressed by myeloid cells-2 (TREM-2) gene can increase the risk of developing AD (Guerreiro et al, 2013;Jonsson et al, 2013). How inflammatory genes alter the risk of AD is of intense interest as they could indicate new therapeutic targets.
CD33 (Siglec-3) is a myeloid differentiation marker with immune functions associated with anti-inflammatory signaling, cell adhesion and endocytosis (Crocker et al, 2012). It is a member of a family of genes with related structures and functions, whose only known ligands are sialic acid-modified proteins or lipids (Cao and Crocker, 2011). The minor A allele form of the rs3865444 SNP polymorphism has been associated with reduced risk of AD with an overall odds ratio (OR) of 0.89 (Hollingworth et al, 2011;Naj et al, 2011;Yuan et al, 2012). This polymorphism is just 372 base pair acids upstream of the first exon of CD33 indicating the possibility that it could affect promoter activity and expression of the CD33 gene. Recent studies have shown significantly reduced expression of CD33 in brain tissue and peripheral monocytes from subjects with rs3865444 A/A alleles, but increased expression of CD33 in AD compared to non-demented (ND) brains (Bradshaw et al, 2013;Griciuc et al, 2013;Malik et al, 2013). Increased CD33 expression by microglia appears to result in significantly reduced amyloid beta (Aβ) peptide phagocytosis as microglia lacking CD33 were able to phagocytize significantly greater amounts of Aβ peptide than microglia expressing CD33 at normal levels (Griciuc et al, 2013).
In this study, we addressed the question of CD33 expression in human brains, in human microglia, and whether the rs3865444 SNP was associated with degree of AD pathology. We screened DNA from neuropathologically-diagnosed non-demented (ND) and AD cases for different rs3865444 alleles. In this population of genotyped cases, the CD33 rs3865444 genotype did not alter levels of plaque and tangle pathology, but, in agreement with the recent work of others (Griciuc et al, 2013;Malik et al, 2013), the major findings of this study were lower levels of CD33 protein in A/A cases, but with increased expression of CD33 v2 mRNA and higher levels of the microglia marker IBA-1 in the same group.
2. Methods
2.1. Human Brain Tissue Resources
All brain samples were from participants in the Arizona Study of Aging and Neurodegenerative Disorders and were autopsied by the Brain and Body Donation Program (BBDP) of the Banner Sun Health Research Institute, Sun City, Arizona. This longitudinal clinicopathological study has been running for 27 years with continuous Institutional Review Board approval. DNA samples prepared from fixed human cerebellum from neuropathologically confirmed cases in this program were available. These DNA samples had been used for apolipoprotein E (APOE) genotyping and then stored. All cases were diagnosed according to National Institutes on Aging/Reagan criteria for Alzheimer’s disease (AD) (Newell et al, 1999) and accepted clinicopathological criteria for Parkinson’s disease (PD) (Gelb et al, 1999;McKeith et al, 2005). The demographic data on cases used for genotyping are shown in Table 1A. To identify CD33 rs3865444 polymorphisms, we analyzed 96 non demented (ND) elderly subjects and 97 Alzheimer’s disease (AD) cases. We excluded AD cases that were APOE ε4/ε4 from the genotyping study as its dominant effect on AD risk and AD pathology might mask the weaker disease effect of the CD33 rs3865444 A allele. We used temporal cortex frozen brain tissue from many of the CD33 rs3865444 SNP genotyped cases for RNA and protein expression analyses. With the exception of cases diagnosed as mild cognitive impairment (MCI) (Table 1B), all ND and AD cases used for biochemical analysis were a subset of the genotyped cases listed in Table 1A. As part of the diagnostic process, each brain was assessed for plaque and tangle load using histological procedures. This ranking method gives a score (0–3) for each of 5 brain regions (entorhinal cortex, hippocampus, temporal cortex, parietal cortex and frontal cortex) for a potential summary score (0–15) and is based on the histological assessment of frequency of plaques and tangles in Thioflavin S stained tissue sections (Beach et al, 2012). These overall ranking scores were used as non-parametric data for correlation analyses to assess effects of CD33 rs3865444 SNP and CD33 expression levels on severity of pathology.
Table 1.
| A. Demographics of cases used for CD33 rs386544 genotyping for neuropathology analyses | |||||
|---|---|---|---|---|---|
| Disease Groups (n) | M/F | Age (yrs) | PMI (hrs) | Plaque Load | Tangle Load |
| Non-demented (96) | 50/46 | 84.9±7.9 | 4.0±7.4 | 5.1±5.5 | 4.9±2.6 |
| Alzheimer’s Disease (97) | 49/48 | 82.2±8.5 | 3.6±3.1 | 13.2±2.2 | 12.4±3.5 |
| B. Demographics of Mild Cognitive Impairment cases used in mRNA study | |||||
|---|---|---|---|---|---|
| Disease Groups (n) | M/F | Age (yrs) | PMI (hrs) | Plaque Load | Tangle Load |
| MCI (14) | 10/4 | 89.1±6.7 | 2.7±0.7 | 9.8±2.9 | 6.1±2.6 |
Abbreviations: M: number of male; F: number of female; yrs: years; PMI: postmortem interval; hrs: hours.
For isolation of human microglia, endothelial cells and astrocytes, samples of frontal cortex were used. These were provided within 3 hours of death by the BBDP from consented donors.
2.2. CD33 rs3865444 polymorphism genotyping
A polymerase chain reaction (PCR) — restriction fragment length polymorphism technique was developed to identify the three different allelic genotypes of CD33 rs3865444. Approximately 0.5 µg of DNA from neuropathologically diagnosed cases was amplified using the following primers - (Sense) GTGAATGAATGAATAAATGAATGG and (Antisense) GAGAGATGGGAGGAGATGGA to produce a fragment of 317 base pairs (bp). PCR was carried out using the following conditions with Promega Hot Start DNA polymerase (1× Promega Green reaction buffer, primers (0.5 µM), deoxynucleotides (0.2 µM), MgCl2 (1.5 mM) and DNA polymerase (0.125 units/reaction)). PCR amplification involved 35 cycles of 94°C for 30 seconds, 58°C for 30 seconds and 72°C for 1 minute after a 2 minute step at 94°C to activate the enzyme. To discriminate between C and A alleles, amplified DNA products were digested with the restriction enzyme NlaIII (New England Biolabs, MA) (10 u/reaction for 3 hours). Digested fragments were separated through a 9% polyacrylamide gel and imaged after staining with GelRed stain (Biotium). A representative gel pattern of the different genotypes of CD33 rs38465444 is shown in Supplemental Fig 1.
2.3. RNA isolation and Quantitative Real Time Polymerase Chain Reaction
RNA was prepared from human brain tissue samples, and cultured human microglia, human brain endothelial cells and astrocytes using RNAeasy Plus Mini kits (Qiagen, Valencia, CA) according to the manufacturer’s directions. Concentrations of RNA samples were determined using a Nanodrop 1000 spectrophotometer, and integrity using an Agilent Bioanalyzer and Nano 6000 kits (Agilent). Samples used for qPCR had RIN values greater than 8.0. RNA from each brain sample (0.5 µg) and cultured cell sample (0.2 µg) was reverse transcribed using the Quantitect reverse transcription kit (Qiagen). Prior to use in quantitative PCR (qPCR), each cDNA was diluted 1:1 with water. Appropriate numbers of no reverse transcriptase controls were prepared in parallel for each batch of samples. For qPCR, cDNA samples were amplified using Perfecta SYBR Green Fast Mix 2× reaction mixture (Quanta Biosciences, Gaithersburg, MD) using CD33 mRNA specific primers - (Sense) CCTGCTCGCTCTTTGTCTCT and (Antisense) GCTCCTCATCCATCTCCAC (Exon 6 and Exon 8 on CD33 transcript variant 1 (v1) – Reference sequence NM_001772.3). These primers produce a single amplicon of 188 bp, and will also recognize common sequences present in CD33 transcript variant 2 (v2) (Reference sequence NM_001082618.1). To specifically detect CD33 transcript v2, which has a deletion of exon2, the following primers were used – (Sense) CTGTGGGCAGACTTGAC and (Antisense) TGATTATGAGCACCGAGGAG - that produce a single amplicon of 189 bp. QPCR was carried out using a Stratagene Mx3000p machine and abundance of gene expression quantified relative to a standard curve. All CD33 PCR values were normalized against values for β-actin mRNA expression as described previously (Walker et al, 2009). QPCR analyses followed most recommended criteria for minimal information for publication of quantitative real time PCR experiments (MIQE) (Bustin et al, 2009)(Bustin et al, 2009)(Bustin et al, 2009)(Bustin et al, 2009)(Bustin et al, 2009)(Bustin et al, 2009).
2.4. Western Blot Analysis
Protein extracts from temporal cortex from rs3865444 genotyped cases were analyzed by western blot methodology for levels of CD33 protein using our published protocols (Walker et al, 2009;Walker et al, 2013). Samples were dissolved at a concentration of 1 µg/µl protein in western blot sample buffer (NUPAGE LDS—Life Technologies, Carlsbad, CA) containing 0.1 M DTT and heated at 70 °C for 10 minutes. Samples we re separated on 4–12% NuPAGE Bis– Tris Mini gels using MOPS-SDS or MES-SDS running buffer (Life Technologies). Proteins were transferred to nitrocellulose membranes at 30 V for 90 minutes. Following drying, membranes were blocked in 5% skim milk solution dissolved in Tris-buffered saline (TBST—50 mM Tris–HCl (pH 8.0), 250 mM NaCl, 0.05% (w/v) Tween 20), and then reacted for 18 hours in appropriate dilutions of antibodies in TBST containing 2% milk. Washes between incubation steps used TBST. Bound antibodies were detected by reaction for 2 hours with the appropriate horseradish peroxidase (HRP) labeled anti-immunoglobulin (Thermo-Fisher - 1:10,000 dilution) followed by reaction of membranes with HRP chemiluminescent substrate (Advansta Western blot Bright chemiluminescent substrate, Advansta, Menlo Park, CA) with direct imaging using a FluorochemQ imaging system (Cell Biosciences). Intensities of chemiluminescence bands were quantified using Fluorochem Q SA software (Cell Biosciences). To ensure reproducible measurements across multiple blots, each set of gels included repeated samples that could be used for correction between blots. Each set of membranes was imaged for different times to obtain exposure of optimal images without signal saturation. The antibody used for detection of CD33 was the rabbit monoclonal EPR4423 (Abcam, Cambridge, MA). This antibody was prepared using a peptide sequence located between amino acids 30–80 in the sialic acid binding immunoglobulin domain of CD33 (NP_001763.3). After stripping (Restore Plus, Thermo Fisher), western blots were reprobed using antibodies to the microglia protein IBA-1 (1:3000, Wako USA, Richmond, VA), the presynaptic protein synaptophysin (1:3000, SYP38; Abcam) and β-actin for normalization purposes (mouse monoclonal: 1:5000, Sigma (St. Louis, MO))..
2.5 Immunohistochemistry of Brain Tissue Sections
Formaldehyde fixed tissue sections from temporal cortex were used for cellular localization of CD33. Tissue sections (25 µm) were heated in 1 ml of 20 mM citrate buffer (pH 8.0) at 80°C for 30 minutes as antigen retrieval produced optimal results (Jiao et al, 1999). Cellular localization of bound antibody was visualized using avidin-biotin horseradish peroxidase enzyme complex (ABC-Vector Laboratories, Burlingame, CA) histochemistry and nickel ammonium sulfate-enhanced diaminobenzidine as substrate to produce a purple reaction product (Walker et al, 2009). In certain cases, dual-color histochemistry was carried out using the mouse monoclonal antibody LN3 (MBL International, Woburn, MA) that identifies HLA-DR, a marker of activated microglia. This second antibody was detected using the same procedure, but with diaminobenzidine without nickel ammonium sulfate as substrate to produce a brown reaction product.
2.6. Human Autopsy Brain Cultures and Experimental Treatments
Human autopsy brain microglia were isolated from frontal cortex according to our standard protocols (Walker et al, 2006;Walker et al, 2009). After isolation, microglia were cultured for 10–14 days prior to use in experiments. From this digested autopsy brain material, we were also able to isolate human brain endothelial cells. Prior to final plating of isolated cells, the cell supernatants were incubated with 100 ml of Ulex-Europaeus (UEA)-conjugated magnetic beads (Life Technologies). Brain microvascular endothelial cells (EC), which bind to the UEA lectin, were magnetically removed, washed and placed in endothelial cell growth media (EBM-2 media supplemented with growth factor mixture-Lonza, Walkersville, MD). Astrocytes used in this study were from a single case.
Microglia isolated from 3 separate ND cases were used in this study. Each of these cases possessed the C/C rs3865444 CD33 genotype. Microglia (n=3 per treatment) were stimulated with fibril/oligomer containing aggregated Aβ42 peptide (0.5, 2 and 5 µM) (CPC Scientific, Sunnyvale, CA), polyinosinic:polycytidylic acid (pIC)(Sigma) (0.25 to 25 µg/ml), interferon-gamma (IFN-γ), interleukin (IL)-4, IL-6 (10 ng/ml) (all cytokines from R&D Systems, Minneapolis, MN) or lipopolysaccharide (LPS)(Sigma)(100 ng/ml) for 24 hours. RNA prepared from stimulated microglia were used for CD33 mRNA qPCR analyses. For measurements of CD33 protein levels in microglia by western blot analyses, cells were stimulated for 24 hours with Aβ (2 µM), pIC (10 µg/ml) or a combination of Aβ and pIC at the same concentrations.
2.7. Statistical Analysis
All statistical analyses were performed using Graphpad Prism version 6 (Graphpad software, San Diego, CA). The Kruskall-Wallis test with the Dunn’s test for post-hoc comparison between groups was used for comparing more than two groups with non-parametric measures (plaque and tangle scores), two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test for comparing effects of disease and genotypes on biochemical measures or one-way ANOVA followed by the Fisher LSD test for post-hoc comparison between groups. Correlation analyses used the Spearman method when non-parametric measures were involved, or the Pearson method for correlation analysis between continuous measures. The significance level was defined as P<0.05.
3. Results
3.1. CD33 rs3865444 genotyping
We developed a simple method to genotype the different alleles of the CD33 rs3865444 single nucleotide polymorphism (SNP) that involved polymerase chain reaction (PCR) amplification of a 317 bp fragment spanning either side of this SNP followed by digestion of amplified DNA with the restriction endonuclease NlaIII. The presence of the A allele abolished the NlaIII restriction site present within the C allele sequence of rs3865444. In amplified DNA with a C/C genotype, there are 3 NlaIII restriction sites within the 317 bp sequence producing fragments of 116 bp, 103 bp, 87 bp and 11 bp upon cleavage. In DNA with a A/A genotype, there are 2 NlaIII restriction sites resulting in fragments of 219 bp, 87 bp and 11 bp upon cleavage. A representative gel showing the patterns of each genotype (C/C, C/A and A/A) is shown (Supplemental Fig. 1). The 11 bp fragment is not visible on the gel. This method was used to genotype 96 non-demented (ND) cases and 97 Alzheimer’s disease cases (AD) from which neuropathology and tissue was available. The demographics of these cases are shown in Table 1A. The distribution of rs3865444 genotypes in these cases is shown in Table 2. We identified 10 ND and 11 AD cases homozygous for A/A alleles. We were now able to assess if there were significant differences in histological measures of AD pathology with different CD33 genotypes in these cases, or biochemical differences in CD33 expression levels with genotype or disease.
Table 2.
Distribution of CD33 rs386544 genotypes of cases used in study
| Non-Demented | Alzheimer’s Disease | (disease) | ||||
|---|---|---|---|---|---|---|
| C/C | C/A | A/A | C/C | C/A | A/A | (genotype) |
| 39 | 47 | 10 | 35 | 51 | 11 | (no. of cases) |
3.1.1. Distribution of CD33 rs3865444 genotypes with pathological features of Alzheimer’s disease
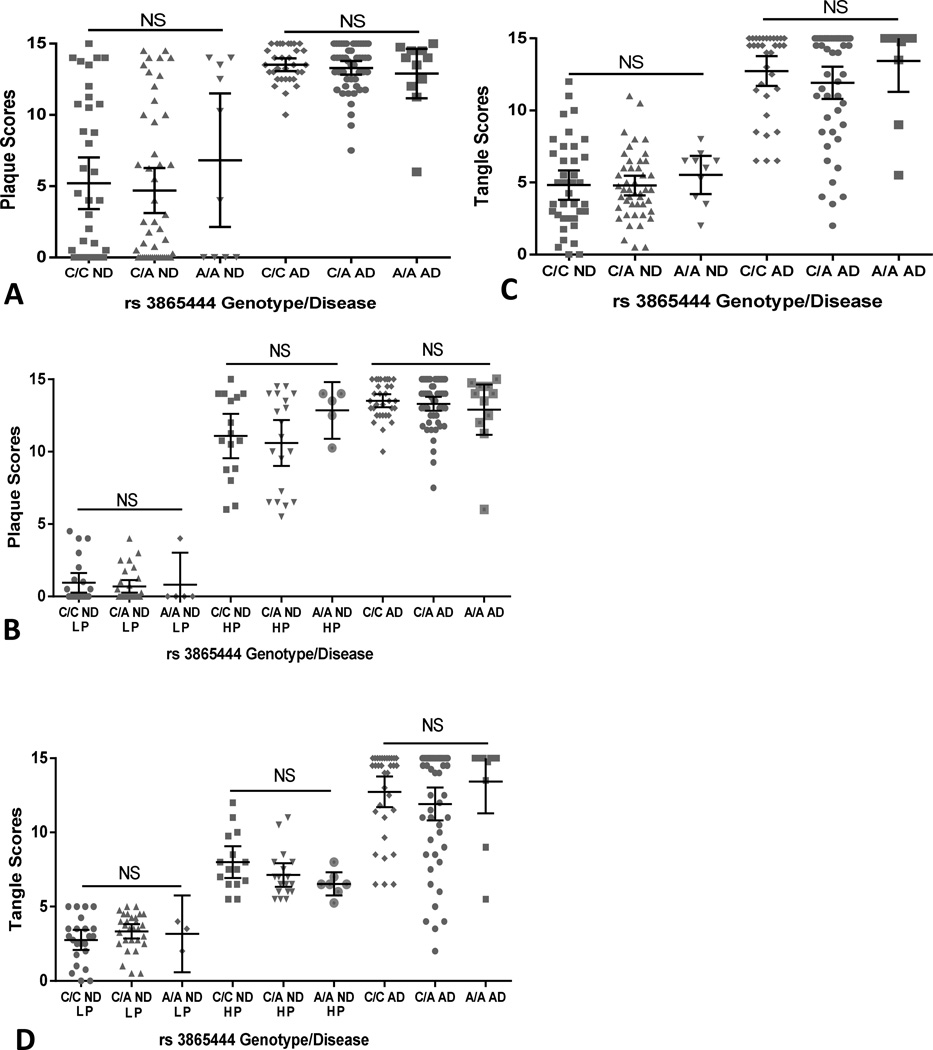
We next addressed the question whether possession of A/A alleles had an effect on the extent of pathological features of plaques and tangles in these cases. In our population (Table 1A, mean age of 84.9 years in the ND cases and 82.2 years old in the AD cases), there were significant levels of age-associated plaque (Fig 1A and 1B) and tangle (Fig 1C and 1D) pathology in many of the ND cases even though these cases had no record of significant cognitive decline and did not meet the accepted criteria for a diagnosis of AD. Analysis of genotypes compared to histological plaque and tangle scores demonstrated no significant differences between the genotyped ND cases or between the genotyped AD cases (Fig 1A and Fig.1C). To investigate this further, the plaque and tangle data of the ND cases were subdivided into low pathology (LP) non-demented (plaque and tangle scores of 5.0 or less) and high pathology (HP) non-demented (plaque and tangle scores of 5.0 or greater) (Fig 1B and Fig 1D). Similar analysis using Kruskal-Wallis non-parametric test showed no genotype effects between the LP groups or between the HP groups for plaques or tangles.
Fig. 1.
Effect of different CD33 rs3865444 genotypes on disease incidence and disease pathology. (A). Scatter plot showing the relative distributions of histological plaque scores with different disease states and genotypes. Plaque scores represent the cumulative ranking of plaque load in 5 affected brain regions of each case. (Non-significant rank differences between genotypes within ND groups or within AD groups, Kruskal-Wallis test). (B). Scatter plot showing the relative distributions of histological plaque scores within the ND low pathology (LP) groups (plaque score < 5), ND high plaque (HP) groups (plaque score >5) and AD groups with the different genotypes. (Non-significant rank differences between genotypes within ND-LP groups, ND-HP groups or within AD groups - Kruskal-Wallis test). (C). Scatter plot showing the relative distributions of histological tangle scores with different disease states and genotypes. Tangle scores represent the cumulative ranking of tangle load in 5 affected brain regions of each case. (Non-significant rank differences within ND groups or within AD groups, Kruskal-Wallis test). (D). Scatter plot showing the relative distributions of histological tangle scores within the ND low pathology (LP) groups (tangle score < 5), ND high pathology groups (tangle score >5) and AD groups different disease states and genotypes. (Non-significant rank differences within ND-LP groups, ND-HP groups or within AD groups- Kruskal-Wallis test).
Bars in panels represent mean ± 95% confidence intervals. NS – Not significant (P>0.05)
3.2 Effect of CD33 rs3865444 on CD33 mRNA expression in human brain
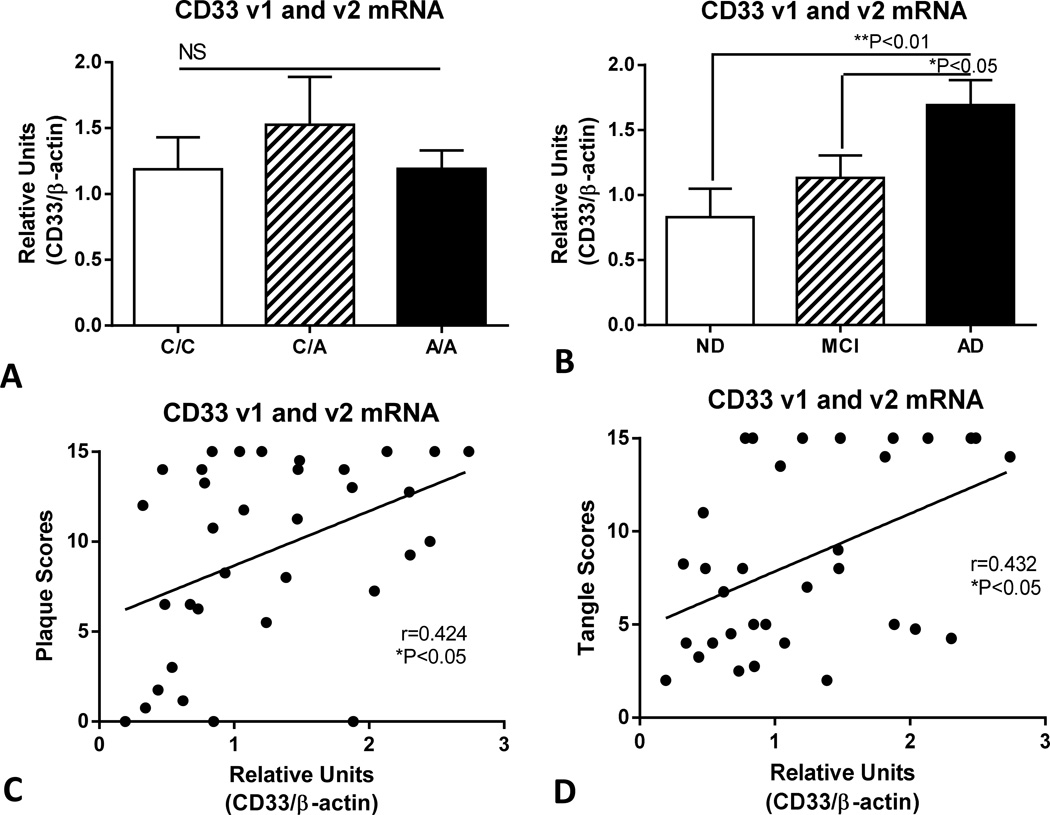
Analyzing a subset of genotyped samples from which acceptable quality RNA could be isolated, qPCR methodology was used to measure the relative levels of total CD33 mRNA (CD33 transcript variant 1 (v1) and 2 (v2) combined) (Fig. 2) and CD33 transcript v2 alone (Fig. 3). CD33 v2 mRNA codes for an alternatively spliced form of CD33 that lacks exon 2, the sequence for the sialic acid binding domain (Hernandez-Caselles et al, 2006). We demonstrated no significant differences in total CD33 mRNA (v1 and v2) levels between the different genotypes (Fig. 2A), but increased expression in AD samples compared to those with mild cognitive impairment (MCI) or ND (Fig. 2B). Combining data from all of these cases allowed correlation analyses between CD33 mRNA levels and plaque scores (Fig. 2C) and tangle scores (Fig. 2D). The results demonstrated significant positive correlation between CD33 mRNA levels and plaque scores (Spearman r = 0.424, P<0.05), and between CD33 mRNA levels and tangle scores (Spearman r=0.432, P<0.05).
Fig. 2.
Expression of CD33 mRNA (transcript variant 1 and 2) increases with AD pathology but not with CD33rs385444 polymorphism in human brains. (A). Absence of changes in relative levels of CD33 (v1 and v2) mRNA in temporal cortex samples of samples with designated genotypes. No significant differences in levels of CD33 mRNA between samples from different rs3865444 genotypes. (P=0.6903, One way ANOVA). Each genotype contained ND, MCI and AD cases. NS – Not significant (P>0.05). Columns represent mean values ± standard error of mean (SEM). (B). Increased in relative levels of CD33 (v1 and v2) mRNA in temporal cortex samples of designated disease groups. Significant increased levels of CD33 mRNA in AD cases compared to ND (**P<0.01) and MCI (*P<0.05) cases (One way ANOVA with Fisher LSD test for between group significance). (n=34). ND – Non-demented; MCI – mild cognitive impaired; AD-Alzheimer’s disease. Columns represent mean values ± SEM. (C). Positive correlations between CD33 mRNA levels and plaque scores (Spearman rank correlation -r=0.424, *P=0.0125). (D). Positive correlations between CD33 mRNA levels and tangle scores (Spearman rank correlation – r=0.432, *P=0.0120). Bars in figures represent mean ± standard error of mean.
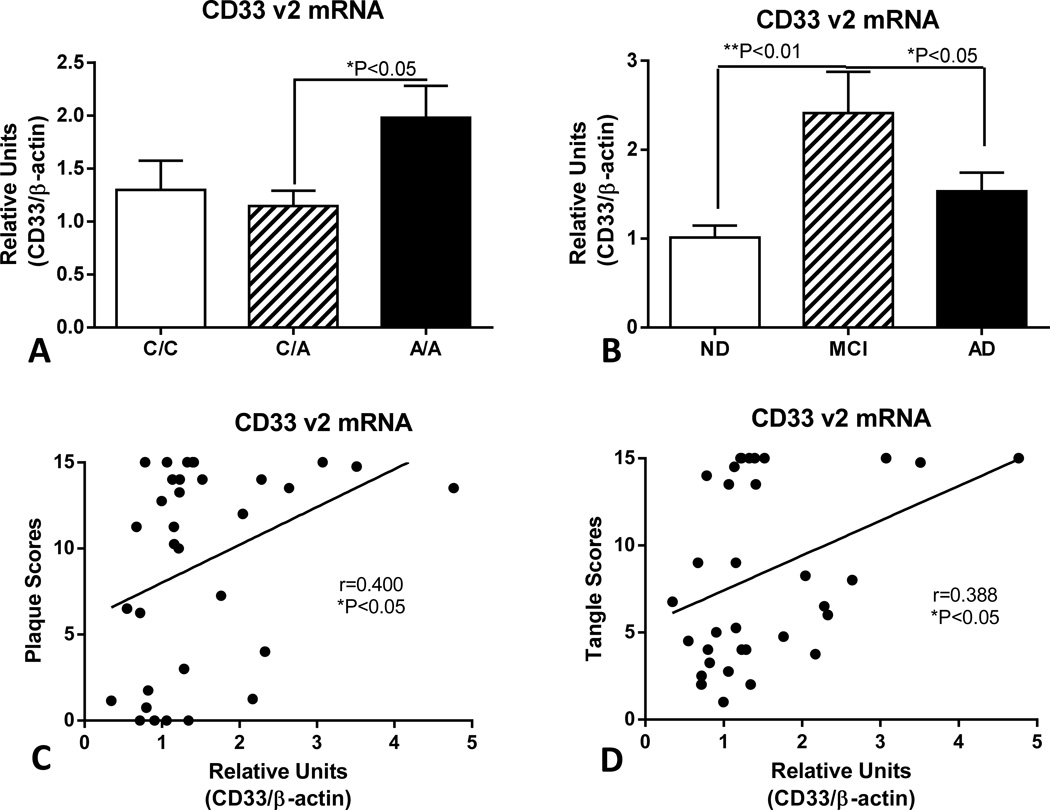
Fig. 3.
Expression of CD33 mRNA (transcript variant 2) in human brains differs with genotype and disease state. (A). Relative levels of CD33 (v2) mRNA in temporal cortex samples of designated genotypes. Significant increased levels of CD33 mRNA v2 in samples with A/A rs3865444 genotypes (P< 0.05 - One way ANOVA with Fisher PLD post hoc test) (n=34). Each genotype contained ND, MCI and AD cases. Columns represent mean values ± SEM. (B). Significant increase in levels of CD33 mRNA v2 in MCI cases compared to ND and AD cases. (One way ANOVA with Fisher LSD test of between group significance) (n=34). Columns represent mean values ± SEM. (C). Positive correlations between CD33 v2 mRNA levels and plaque scores (Spearman rank correlation - r=0.400, *P=0.021) and (D) tangle scores (Spearman rank correlation – r=0.387, *P=0.026).
Different trends were obtained when measuring levels of CD33 v2 mRNA expression (Fig. 3). CD33 v2 is a minor abundance transcript compared to CD33 (v1 and v2) mRNA. There was significantly increased expression of CD33 v2 mRNA in the A/A allele samples compared to C/A samples (Fig. 3A). The pattern of expression for CD33 v2 mRNA with disease classification was also different from CD33 (v1 and v2) mRNA with significantly increased expression in the MCI cases compared to ND and AD cases (Fig. 3B). Similar degrees of positive correlation were detected between CD33 v2 mRNA levels and plaque scores (Figure 3C–Spearman r=0.400, P<0.05) and tangle scores (Fig. 3D–Spearman r=0.388, P<0.05) to those obtained with total CD33 RNA measurements (Fig. 2C and Fig. 2D).
3.3. CD33 cellular expression in human brains
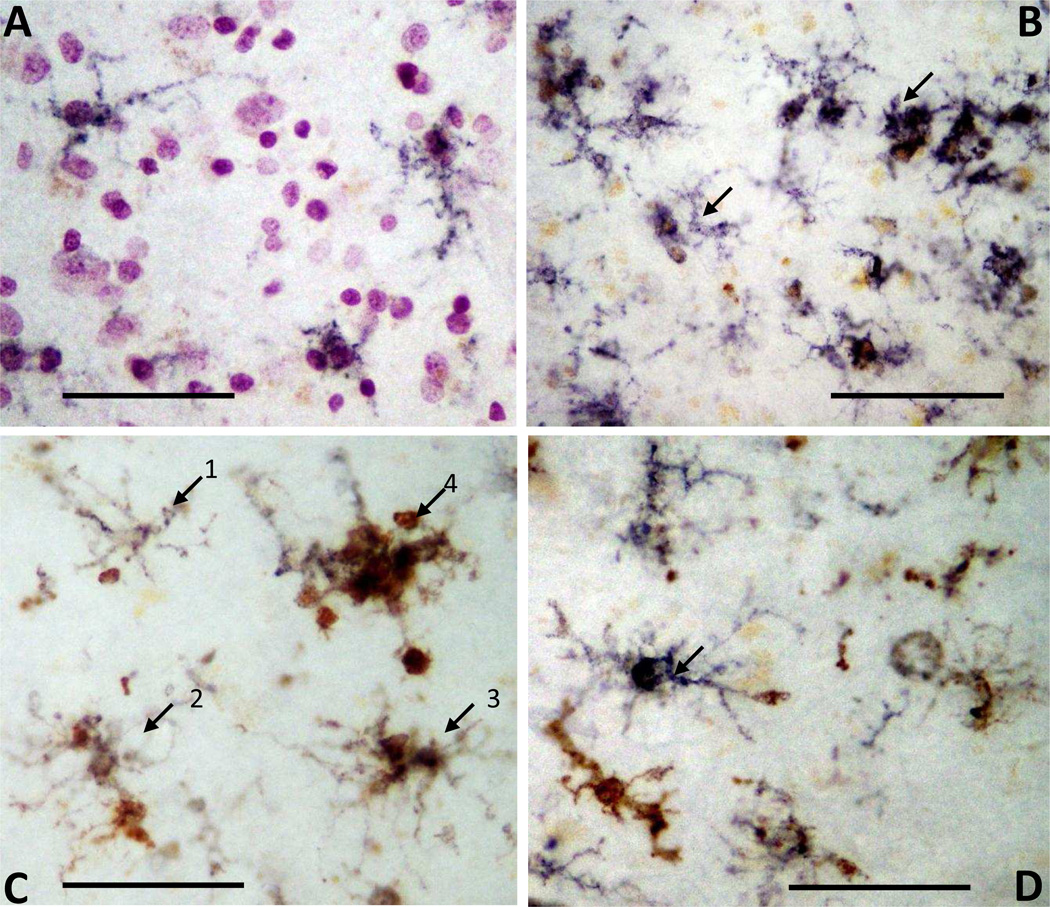
Immunohistochemistry was carried out using tissue sections of temporal cortex from a series of ND and AD cases to show the cellular localization of CD33. Representative photomicrographs demonstrate CD33 in cells with the morphology of microglia (Fig. 4A, ND case and Fig. 4B, AD Case). In the AD case, there is progressively increasing amounts of CD33 immunoreactivity in microglia with more activated morphologies (Fig. 4B). Double staining of tissue sections to identify colocalization of CD33 (purple) and HLA-DR (brown) is shown for two different AD cases (Fig. 4C and Fig 4D). HLA-DR is an accepted marker for identifying activated microglia in human brains. CD33 staining was present in each of the identified cells showing progressively increasing amounts of HLA-DR immunoreactivity (arrows 1–4) (Fig. 4C). A similar pattern can be seen in another AD case (Fig. 4D).
Fig. 4.
Immunohistochemistry to demonstrate cellular localization of CD33 in microglia in human brain sections. (A and B) Single color immunohistochemistry using antibody to CD33 in ND (A) and AD (B) cases identifies cells with morphology of microglia (purple color). Weak CD33 immunoreactivity seen in microglial like cells in ND case (A) and increased CD33 was apparent in clusters of microglia with activated morphology in AD case (B - arrows). Brown color in A and B is lipofuscin. (C and D) Dual-color immunohistochemistry (purple CD33: brown HLA-DR) identifies a range of cells with progressively activated morphologies showing colocalization of CD33 with increased amounts of HLA-DR in AD sections. The progressive increase in HLA-DR immunoreactivity (C: 1–4 labeled arrows) is seen with changes in morphology. (D) Arrow indicates microglia with strong CD33 reactivity and limited HLA-DR reactivity in an AD section, while other cells show differing degrees of combined reactivities. All cases examined showed similar results. Scale bar represent 100 µm.
3.4 Effect of CD33 rs3865444 on CD33 protein expression in human brains
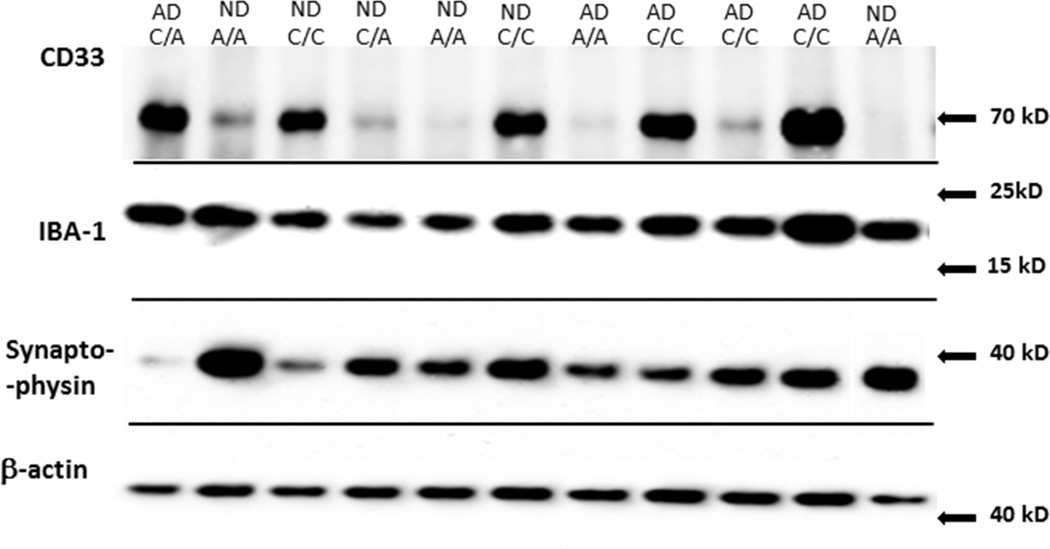
We carried out western analysis with a larger series of genotyped ND and AD cases than was used for the RNA studies. For this study, we sought to confirm the significant finding that subjects with A/A alleles have lower levels of CD33 protein in brain (Griciuc et al, 2013). We also measured levels of the microglia protein IBA-1 and the synaptic protein synaptophysin in this series of samples. A representative western blot panel of results of some of these cases with the antibodies used in this study is shown (Fig. 5). For CD33, a major reactive band of approximately 67 kD (CD33) could be detected in each sample, but at varying intensities depending on genotype. The molecular weights of the bands detected with the antibodies to IBA-1, synaptophysin and β-actin were of the expected size.
Fig. 5.
Representative Western blots of protein bands for CD33, IBA-1, synaptophysin and b-actin in CD33 rs3865444 genotyped brain samples. Composite figure showing representative gels of proteins to be measured in series of rs3865444 genotyped temporal cortex samples. Figure legend indicates genotype (C/C, C/A, A/A) or disease state (ND or AD).
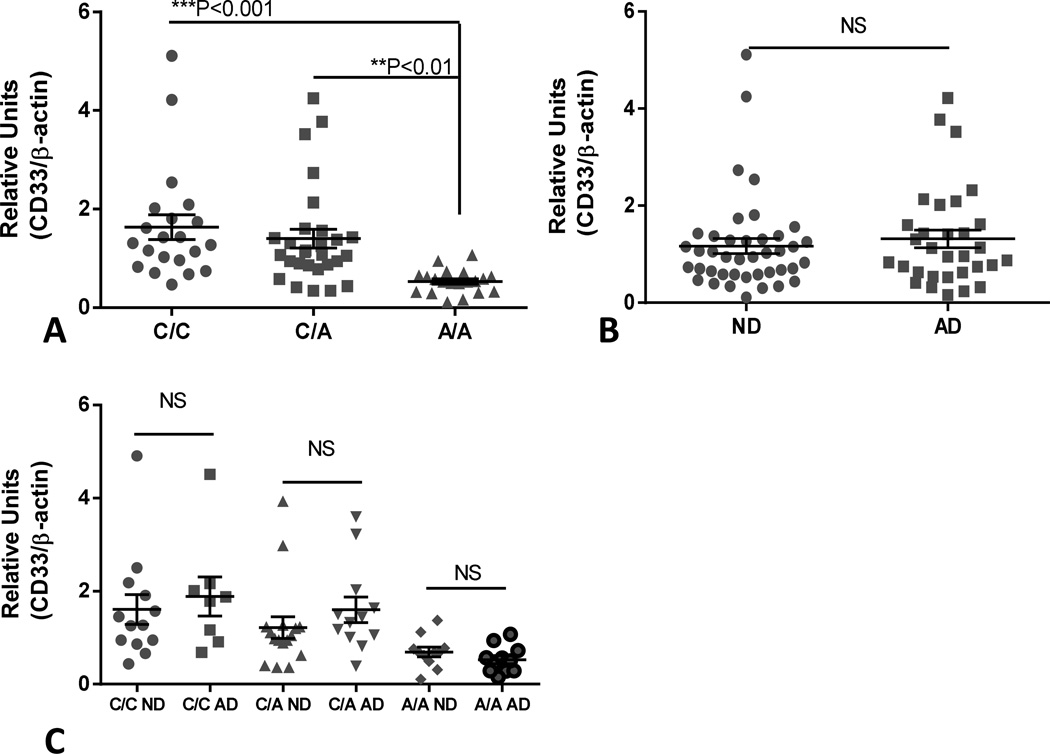
Our results show significantly lower expression of full length CD33 in A/A cases compared to the other genotypes (Fig 6A). These measurements for each genotype are based on the combined results from ND and AD cases. In this series of cases, we could not demonstrate a significant difference in CD33 levels between ND and AD cases (Fig 6B). The distribution of CD33 levels for the different genotypes and disease states is shown in Figure 6C. Two-way ANOVA confirmed that CD33 levels were significantly affected by genotype (F=9.489, SS = 15.49, ***P=0.0002), but not by disease state.
Fig. 6.
Effect of CD33 rs3865444 genotype and disease state on levels of CD33 protein in human brain samples. (A). Scatter plot showing relative levels of CD33 protein with CD33 rs3865444 genotype in temporal cortex samples. Significant differences were detected between C/C genotyped samples (n=21) and A/A samples (n=20)(P<0.01, One way ANOVA and Fisher LSD test of group significance) and between C/A samples (n=28) and A/A sample (P<0.001, One way ANOVA and Fisher LSD test of group significance). (B). Scatter plot showing relative levels of CD33 protein in temporal cortex samples categorized by disease state. No significant difference (P=0.5425, unpaired t test) between ND and AD cases. NS – Not significant. (C). Scatter plot showing relative levels of CD33 protein in samples categorized into CD33 rs3865444 genotype and ND or AD disease state. There were no significant (NS) differences between each genotype and disease state but overall genotypes had significant effect on CD33 levels (SS 15.69, ***P=0.0002) (Two way ANOVA with Sidak’s multiple comparison test). Lines indicate mean values ± SEM.
3.5 Effect of CD33 rs3865444 on levels of microglial protein IBA-1 and synaptophysin expression in human brain
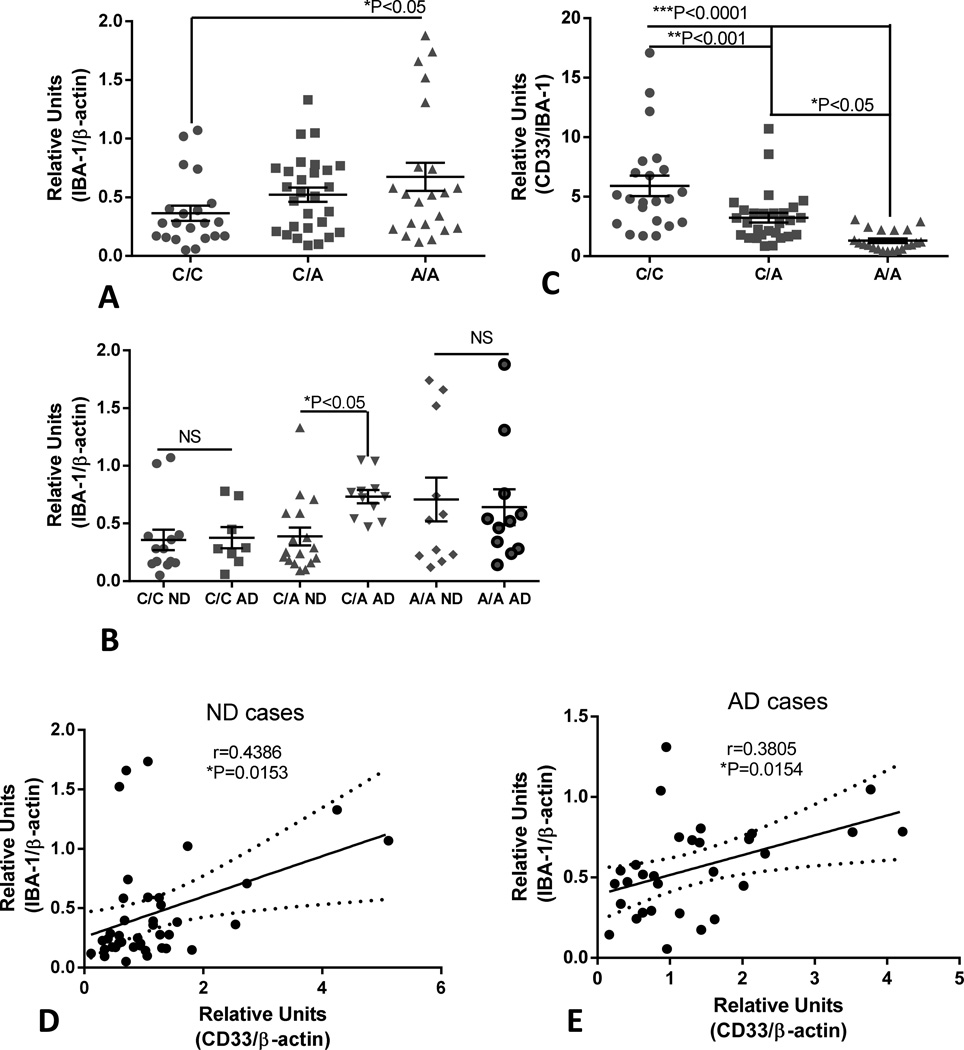
As CD33 expression appears to be restricted to microglia in human brain, we used these temporal cortex samples to address the question whether CD33 protein levels varied with abundance of microglia. IBA-1 protein levels is used as a measure of microglia numbers, though increased levels can also give an indication of microglia activation. We showed significantly increased IBA-1 levels in A/A cases compared to the C/C cases (Fig 7A). If these results were separated into disease groups as well as CD33 genotypes, significantly increased levels of lBA-1 were only detected in the C/A AD group compared to the C/A ND group, not for the other genotypes (Fig 7B). If expression levels of CD33 were normalized with matched values for the microglial protein lBA-1 rather than β-actin, differences in CD33 levels between the different genotypes became larger (Fig 7C), however, by two-way ANOVA, CD33 levels normalized for IBA-1 did not show statistically significant difference between disease groups and genotype groups (data not shown). Further analyses showed significant positive correlations between CD33 levels (normalized for β-actin) with lBA-1 levels for ND cases (Fig 7D) (Pearson r=0.4386, P=0.0153) and for AD cases (Pearson r=0.3805, P=0.0154) (Fig 7E).
Fig 7.
Increased levels of microglial marker protein IBA-1 in temporal cortex samples with A/A CD33 rs3865444 genotype and disease and correlations between IBA-1 and CD33 levels.
(A). Scatter plot showing relative levels of IBA-1 protein with CD33 rs3865444 genotype in temporal cortex samples. Significant differences were detected between C/C genotyped samples (n=23) and A/A samples (n=21)(P<0.05, One way ANOVA and Fisher LSD test of group significance). (B). Scatter plot showing relative levels of IBA-1 protein in samples categorized into CD33 rs3865444 genotype and ND or AD disease state. There was only a significant difference between C/A genotype and disease state for IBA-1 levels (*P<0.05)(Two way ANOVA with Sidak’s multiple comparison test). (C). Scatter plot showing relative levels of CD33 between CD33rs3865444 genotype when CD33 values were normalized against lBA-1 values. Significant differences between all groups became apparent (One way ANOVA and Fisher LSD test of group significance). Lines indicate mean values ± SEM. (D). Correlation analysis for levels of CD33 and IBA-1 in ND cases. Significant correlation (Pearson r test: r=0.4386, P=0.0153) was detected. Figure shows linear regression line (solid line) with 95% confidence intervals (dotted lines). (E). Correlation analysis for levels of CD33 and IBA-1 in AD cases. Significant correlation (Pearson r test: r=0.3805, P=0.0154) was detected. Figure shows linear regression line (solid line) with 95% confidence intervals (dotted lines).
We also measured relative levels of the synaptic protein synaptophysin, another marker for neuropathology, to determine if there were significant differences between CD33 rs3865444 genotyped cases and/or disease state. There were no significant differences in synaptophysin protein levels between genotyped groups (supplemental Fig 2A) or between genotype and disease groups (supplemental Fig 2C), however, these samples showed the expected decreased levels of synaptophysin in the AD samples compared to the ND samples (supplemental Fig 2B).
3.6. CD33 mRNA and protein regulation in cultured human brain microglia
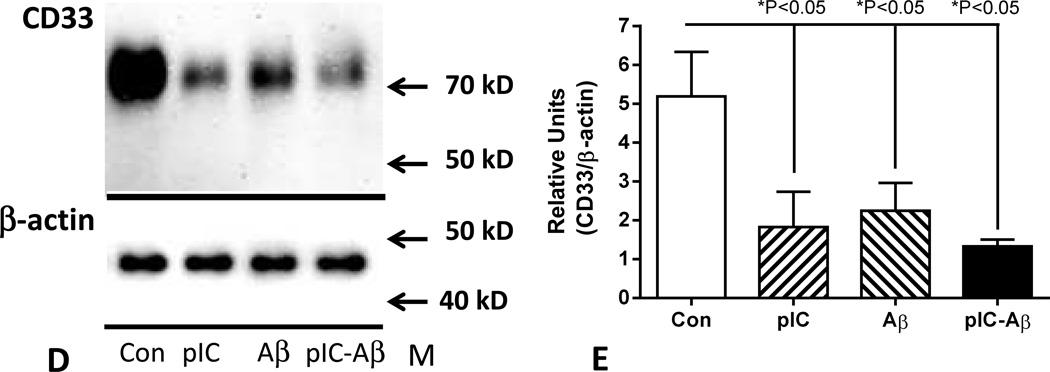
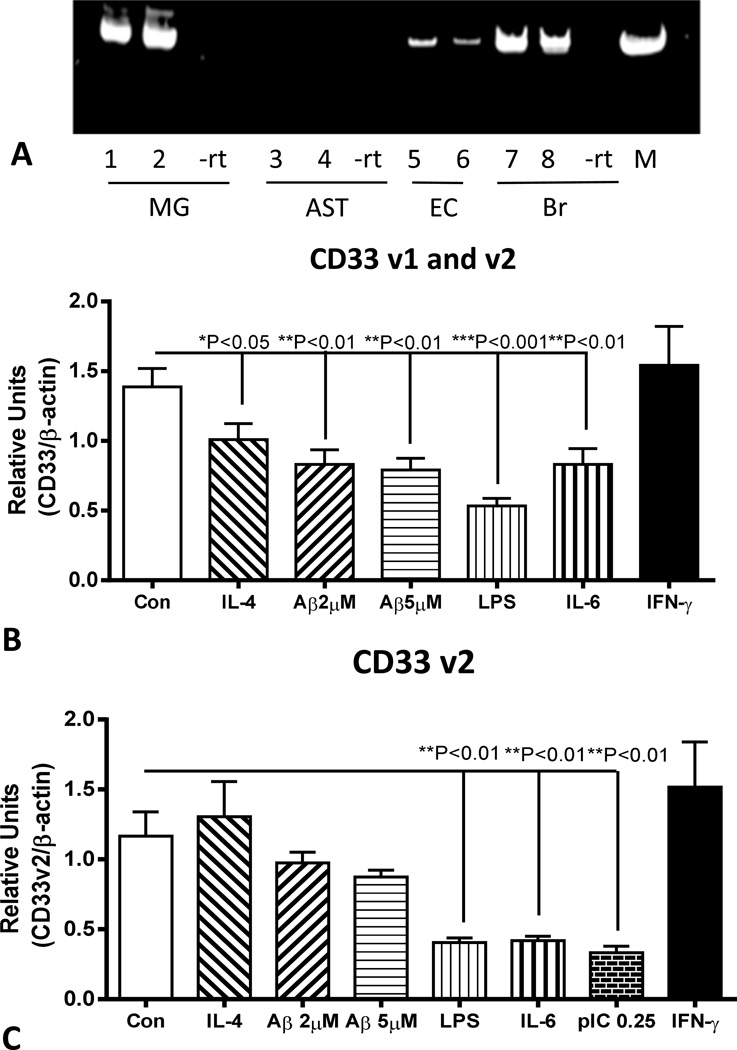
We also sought to model CD33 expression in brain by using microglia isolated from human brains in order to understand how inflammatory factors might be affecting expression of this gene. Initial experiments used reverse transcription-polymerase chain reaction (RT-PCR) amplification of cDNA from microglia, astrocytes, microvascular endothelial cells and brain to define cell types expressing CD33 mRNA (Fig 8A). In addition to microglia, CD33 mRNA expression was detected in brain endothelial cells, but not in astrocytes. Quantitative gene expression analyses of CD33 mRNA was carried out using microglia isolated from different cases (n=3) (all with C/C genotypes). These measures demonstrated reduced CD33 (v1 and v2 mRNA) expression following 24 hour-treatment with Aβ peptide and certain inflammatory agents (Fig 8B). We also showed expression of CD33 v2 mRNA in human microglia, with similar downregulation of expression with inflammatory activation with LPS, IL-6 and the toll-like receptor (TLR)-3 ligand polyinosinic;polycytidylic (pIC)(Fig 8C). The effect of Aβ on expression of CD33 v2 mRNA was less than observed for CD33 v1 and v2 mRNA (Fig 8B) and did not reach statistical significance. Surprisingly, the potent cytokine IFN-γ had no effect on expression of CD33 mRNA.
Figure 8.
Inflammatory activation of in vitro cultured human microglia downregulation expression of CD33 mRNA and protein in human microglia. (A). Gel showing DNA bands demonstrating expression of CD33 mRNA detected by PCR amplification of cDNA from human microglia (MG) cases (1 and 2) and brain microvascular endothelial cells (EC) cases (5 and 6) but not astrocytes (AST)(3 and 4). CD33 mRNA expression was also detected in human brain samples (7 and 8). –rt – result in matching sample processed without reverse transcriptase.
(B). Quantitative polymerase chain reaction analyses of CD33 (v1 and v2) mRNA expression in microglia (n=3 cases) treated with the indicated agents. Significant downregulation of expression was observed with all treatments except interferon-γ (One way ANOVA, Fisher LSD post hoc test). (C). Quantitative polymerase chain reaction analyses of CD33 (v2) mRNA expression in microglia treated with the indicated agents. Significant downregulation of expression was observed with indicated proinflammatory treatments (One way ANOVA, Fisher LSD post hoc test). (D). Changes in microglia expression of CD33 protein with inflammatory treatments. Representative gel shows only a single band corresponding to full length (approximately 67 kD) CD33. (E). Relative changes in CD33 protein levels in treated microglia. Effects of treatment with inflammatory activators poly inosinic:polycytidylic acid (pIC)(10 µg/ml); amyloid beta peptide (Aβ)(2 µM) and both agents combined (pIC-Aβ) for 24 hours. (One way ANOVA, Fisher LSD post hoc test). Bars indicate mean values ± SEM.
Similar downregulation of CD33 following inflammatory stimulation of microglia was seen at the protein level. Treatment of microglia with Aβ and pIC, alone and in combination resulted in significant decrease in CD33 protein levels (Fig.8D and 8E).
4. Discussion
The major findings of this study was the reduced levels of CD33 protein in brain tissue of cases with A/A alleles compared to C/C and C/A alleles, and also the demonstration of downregulation of CD33 in human microglia treated with Aβ or certain inflammatory agents. It had been hypothesized that as the CD33 rs3865444 SNP was located just 373 bp upstream of exon 1 of CD33, it might affect gene transcription or RNA stability (Jiang et al, 2014;Malpass, 2013;Morgan, 2011).
CD33 had been studied for many years in relation to hematological diseases, but until recently there had been no studies of CD33 expression in brain. However, the association of CD33 SNPs, particularly rs3865444, with altered risk of AD has spurred an interest in CD33 protein function and CD33 expression by microglia and monocytes (Bradshaw et al, 2013;Griciuc et al, 2013;Hollingworth et al, 2011;Karch et al, 2012;Morgan, 2011;Naj et al, 2011;Raj et al, 2014;Yuan et al, 2012). Activation of CD33 by ligands results in phosphorylation of the immunoreceptor tyrosine-based inhibition motifs (ITIM) that can inhibit signaling of a number of receptors with immunoreceptor tyrosine-based activation motifs (ITAM) (Linnartz et al, 2010;Linnartz and Neumann, 2013). CD33 has been defined as a myeloid differentiation antigen, but despite its importance in hematological cancers, there is still limited information on the regulation of CD33 gene expression (Cowan et al, 2013). We had approached this topic from the point of view of considering how CD33 expression by human microglia could affect development of AD pathology.
4.1 CD33 genotypes and brain
From our brain bank DNA resource, we had selected a series of elderly cases with the diagnosis of non-demented (ND) or Alzheimer’s disease (AD) for rs3865444 genotyping using possession of homozygous apolipoprotein Eε4 (apoE4) as the sole exclusion criteria. Within these ND groups were a range of age-associated AD plaque and tangle pathology so this group could be subdivided into low pathology (LP) and high pathology (HP) classes. This series of showed no differences in the frequency of the different genotypes with severity of neuropathology, maybe because our selected populations for study contained many elderly ND cases with significant levels of plaque and tangle pathology. This might be considered a limitation, however as these cases had progressively increasing degrees of plaque and tangle pathology, they were used for correlation analyses with other measures. Temporal cortex brain tissue samples from many of these cases were used for biochemical analysis of CD33 mRNA and protein levels. Since initiating our study, there have been reports showing differential expression of CD33 mRNA or protein with rs3865444 genotypes in human AD and ND brains, or in blood-derived monocytes (Bradshaw et al, 2013;Griciuc et al, 2013;Malik et al, 2013;Raj et al, 2014).
4.2 CD33 mRNA expression in brain
For mRNA studies, we used a subgroup of the genotyped cases from which suitable quality RNA could be obtained. For comparison, we included a group of MCI cases to study disease progression. MCI is generally considered a precursor to AD with cases having mild cognitive decline and intermediate degrees of AD pathology (Mufson et al, 2012). These cases had a mean age of 89.1 compared to 84.1 and 82.2 years, the mean ages of the ND and AD groups (Table 1). We used primers that amplified a sequence common to the abundant transcript variant 1 (v1) and the less abundant transcript variant 2 (v2) to measure total levels of CD33 mRNA in brain samples. There were disease differences in CD33 (v1 and v2) mRNA levels, but not differences between genotyped groups. The approximate two-fold increased expression in our AD samples was less than the 6-fold increase in expression of CD33 mRNA in AD cases observed in another study (Griciuc et al, 2013). There were significant correlations between total CD33 (v1 and v2) mRNA levels and plaque and tangle scores suggesting that features of pathology were driving the increased expression of CD33 mRNA. Although we were measuring CD33 mRNA levels only in temporal cortex, we correlated these values with total brain plaque and tangle scores, which give a good indication of total load of AD pathology for each case. Using primers that spanned the exon boundaries between CD33 exon 1 and exon 3 to specifically detect CD33 transcript variant (v2), we showed significantly higher levels of CD33 v2 in samples with the A/A genotype. This was in general agreement with recent findings (Malik et al, 2013) that demonstrated an increase in the ratio of CD33 v2 to CD33 v1 mRNA in A/A allele cases. Increased CD33 v2 mRNA expression in the MCI cases suggested that changes in transcription patterns potentially resulting in higher levels of CD33 lacking the crucial sialic acid binding domain could be an early event in the development of AD pathology.
4.3 CD33 protein in brain
A larger series of brain samples were available for measuring CD33 protein levels in relation to genotype and disease than for the mRNA studies. This series was selected to have sufficient numbers of matched cases in each group, particularly of the A/A genotype. It is appreciated that the numbers of cases of each genotype analyzed did not reflect the normal population distribution of this SNP. After testing a number of antibodies to CD33, these studies were carried out with a rabbit monoclonal antibody that had been prepared against a peptide sequence found in CD33 exon 2 (Abcam, personal communication) so will not detect CD33 protein lacking the sialic acid binding domain. This antibody detected the same polypeptide of 67 kD in brains and human microglia.
In agreement with a recent study on effects of rs3865444 genotypes in AD brains (Griciuc et al, 2013), we detected significantly reduced protein levels of CD33 in cases with A/A alleles. By comparison, there were higher levels of the microglial protein IBA-1 in these A/A genotyped samples. When the CD33 western blot quantitative data were normalized for microglial content (IBA-1 levels), there were significantly greater differences in CD33 levels between genotypes. This might suggest that there are greater numbers of microglia in A/A cases, though this would not agree with a previous finding (Bradshaw et al, 2013); alternatively the microglia in the A/A cases might be more activated as a result of reduced CD33 anti-inflammatory activity. IBA-1 (ionized calcium binding adapter protein-1) is a widely used marker to assess microglial abundance, though its expression can also be upregulated with microglial activation (Koppel et al, 2014;Puli et al, 2012;Zotova et al, 2013). Reduced amounts of CD33 protein was a feature of peripheral blood monocytes from A/A subjects (Bradshaw et al, 2013).
4.4. Downregulation of CD33 expression in activated microglia
Our findings of downregulated CD33 expression by microglia activated in vitro might appear to be contrary to the findings of increasing levels of CD33 in AD brains. However, these should be considered in the context of results obtained using different CD33 genetic expression constructs (Griciuc et al, 2013), along with findings showing the need for a certain degree of inflammatory activation for microglial phagocytosis of Aβ to occur (Chakrabarty et al, 2010a;Chakrabarty et al, 2010b;Chakrabarty et al, 2011). CD33 is a receptor with anti-inflammatory signaling functions induced following sialic acid ligand binding that result in phosphorylation of its ITIM. Reduced CD33 expression that results in enhanced inflammation might also be detrimental in the inflammatory environment of the AD brain. In this context, lower expression of CD33 in monocytes from type-2 diabetic patients compared to controls was associated with increased expression of proinflammatory tumor necrosis factor (TNF)-α, interleukin (IL)-1β and IL-8 cytokines (Gonzalez et al, 2012). In the experimental paradigms we used, stimulation of microglia with Aβ resulted in a strong proinflammatory response, but at the same time downregulation of genes with anti-inflammatory properties (Walker et al, 2006). In situations of chronic inflammation, less CD33 activation might result in enhanced inflammatory damage but with increased Aβ phagocytosis. However, upregulated expression of CD33, as was seen on microglia in human AD brain sections or with C/C genotype, will result in reduced phagocytosis of Aβ. Although microglia cultured from CD33 gene deficient mice had increased capacity to phagocytose Aβ, and microglia that overexpressed CD33 had reduced levels of Aβ phagocytosis, the extent of classical inflammatory responses of these microglia need to be studied. Expressing CD33 lacking the sialic acid domain in rodent microglia showed that CD33 activation was required to modulate Aβ phagocytosis (Griciuc et al, 2013). Cells expressing this mutant form of CD33 did not show inhibition of Aβ 42 uptake. A recent study has demonstrated that CD33 interaction with sialic acid domains of CD14 can have a significant effect on downregulating TLR-4 signaling (Ishida et al, 2014). Microglial CD14 and TLR-4 can directly interact with Aβ and mediate cellular activation and phagocytosis (Reed-Geaghan et al, 2009). Microglia expressing higher levels of CD33 (e.g. with C/C genotype), when activated, could result in stronger induction of anti-inflammatory signals that inhibit this or other known phagocytic receptors that directly bind to Aβ.
4.5 CD33 protein metabolism
There was some discrepancy between the effects of rs3865444 genotype and disease state on CD33 mRNA expression compared to CD33 protein levels, which suggests that there could be additional factors that affect CD33 protein metabolism. There is supporting evidence for this as it has been shown that CD33 internalization and breakdown is an active process mediated by the suppressor of cytokine signaling-3 (SOCS-3) protein, whose expression is upregulated following inflammatory stimulation. SOCS-3 can bind to the activated ITIM domains of CD33 resulting in ubiquitination and accelerated proteosomal inactivation of this complex (Gonzalez et al, 2012;Orr et al, 2007;Walter et al, 2008;Walter, 2014). Monocytes from diabetic patients had reduced levels of CD33 but increased levels of SOCS-3. This effect could be replicated with monocytes cultured under high glucose conditions (Gonzalez et al, 2012). Thus, inflammatory stimulation of microglia might downregulate CD33 mRNA expression by one mechanism, but it also affects protein levels to a larger extent by increasing cellular breakdown mediated by SOCS-3. The chronic inflammatory conditions ongoing in AD brains that result in microglial activation, but also lead to increased levels of CD33, need to be further defined. It was noticeable that the potent proinflammatory cytokine interferon-γ did not have any significant effect on CD33 mRNA expression.
4.6 Conclusion
In conclusion, these studies have added to understanding of CD33 in the human AD brain and its expression by human microglia. The recent findings showing CD33 interactions with CD14 and TLR-4 suggest further studies should consider possible interactions of CD33 with TREM-2. Recent genetic studies of TREM-2 have shown a significant effect on AD disease risk (Guerreiro et al, 2013;Ruiz et al, 2014), but the AD disease risk is increased when TREM-2 loss of function occurs. Further studies are also needed to identify the putative sialic acid-modified ligands in human brain that activate CD33. Identifying these ligands might provide an approach to specifically targeting CD33 responses, though it needs to be appreciated that CD33 belongs to a related large family of sialic acid binding receptors (Siglecs), many of which have been shown also to be expressed by microglia (Claude et al, 2013;Linnartz and Neumann, 2013;Wang and Neumann, 2010).
Supplementary Material
Supplemental Figure 1. Identification of CD33 rs3865444 genotypes. The different pattern of DNA bands resulting from PCR amplification of DNA followed by restriction enzyme digestion with NlaIII used to identify the different CD33 rs3865444 genotypes (C/C, C/A, A/A). m- sizes of DNA marker bands (bp-base pairs).
Supplemental Figure 2. Effect of CD33 rs3865444 genotype and disease state on levels of synaptophysin in human brain samples. (A). Scatter plot showing relative levels of truncated synaptophysin with CD33 rs3865444 genotypes in temporal cortex samples (C/C: n=21; C/A: n=26; A/A: n=20). One way ANOVA showed no significant differences between genotypes. (B). Scatter plot showing significantly decreased levels of synaptophysin in AD cases (n= 25) compared to ND cases (n=42) (P<0.05, unpaired t test). (C). Scatter plot showing relative levels of synaptophysin in samples divided into CD33 rs3865444 genotype and disease state. Two-way ANOVA (with Sidak’s test for multiple comparisons) demonstrated no significant differences between genotypes for synaptophysin levels, or between genotype and disease groups, but a significant effect of disease state (P< 0.05). Lines indicate mean values ± SEM.
Figure 9.
Highlights.
We identified human cases with A/A alleles for CD33 SNP rs3865444
We showed changes of expression of CD33 mRNA in AD temporal cortex
We confirmed reduced levels of CD33 protein in A/A allele genotyped cases
We demonstrate changes in IBA-1 levels in A/A allele genotyped cases
We demonstrate reduced CD33 expression in activated human microglia
Acknowledgements
This study was supported a grant from the State of Arizona Alzheimer’s Research Consortium and matching funds provided by the Sun Health Foundation, and also in part by grants R21AG034409-A1 and R21AG044068-1 from the National Institutes of Health to DGW.
The Brain and Body Donation Program at BSHRI is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors have financial conflicts of interest.
References
- Beach TG, Sue LI, Walker DG, Sabbagh MN, Serrano G, Dugger BN, Mariner M, Yantos K, Henry-Watson J, Chiarolanza G, Hidalgo JA, Souders L. Striatal amyloid plaque density predicts Braak neurofibrillary stage and clinicopathological Alzheimer's disease: implications for amyloid imaging. J Alzheimers Dis. 2012;28:869–876. doi: 10.3233/JAD-2011-111340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von KA, Morris MC, Evans DA, Johnson K, Sperling RA, Schneider JA, Bennett DA, De Jager PL. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cao H, Crocker PR. Evolution of CD33-related siglecs: regulating host immune functions and escaping pathogen exploitation? Immunology. 2011;132:18–26. doi: 10.1111/j.1365-2567.2010.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo MC. Leveraging global resources to end the Alzheimer's pandemic. Alzheimers Dement. 2013;9:363–365. doi: 10.1016/j.jalz.2013.05.1768. [DOI] [PubMed] [Google Scholar]
- Chakrabarty P, Ceballos-Diaz C, Beccard A, Janus C, Dickson D, Golde TE, Das P. IFN-gamma promotes complement expression and attenuates amyloid plaque deposition in amyloid beta precursor protein transgenic mice. J Immunol. 2010a;184:5333–5343. doi: 10.4049/jimmunol.0903382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Herring A, Ceballos-Diaz C, Das P, Golde TE. Hippocampal expression of murine TNFalpha results in attenuation of amyloid deposition in vivo. Mol Neurodegener. 2011;6:16. doi: 10.1186/1750-1326-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Jansen-West K, Beccard A, Ceballos-Diaz C, Levites Y, Verbeeck C, Zubair AC, Dickson D, Golde TE, Das P. Massive gliosis induced by interleukin-6 suppresses Abeta deposition in vivo: evidence against inflammation as a driving force for amyloid deposition. FASEB J. 2010b;24:548–559. doi: 10.1096/fj.09-141754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude J, Linnartz-Gerlach B, Kudin AP, Kunz WS, Neumann H. Microglial CD33-related Siglec-E inhibits neurotoxicity by preventing the phagocytosis-associated oxidative burst. J Neurosci. 2013;33:18270–18276. doi: 10.1523/JNEUROSCI.2211-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan AJ, Laszlo GS, Estey EH, Walter RB. Antibody-based therapy of acute myeloid leukemia with gemtuzumab ozogamicin. Front Biosci (Landmark Ed) 2013;18:1311–1334. doi: 10.2741/4181. 1311–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, McMillan SJ, Richards HE. CD33-related siglecs as potential modulators of inflammatory responses. Ann N Y Acad Sci. 2012;1253:102–111. doi: 10.1111/j.1749-6632.2011.06449.x. Epub@2012 Feb 21.:102-111. [DOI] [PubMed] [Google Scholar]
- Czirr E, Wyss-Coray T. The immunology of neurodegeneration. J Clin Invest. 2012;122:1156–1163. doi: 10.1172/JCI58656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb DJ, Oliver E, Gilman S. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56:33–39. doi: 10.1001/archneur.56.1.33. [DOI] [PubMed] [Google Scholar]
- Golde TE. Open questions for Alzheimer’s disease immunotherapy. Alzheimers Res Ther. 2014;6:3. doi: 10.1186/alzrt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Y, Herrera MT, Soldevila G, Garcia-Garcia L, Fabian G, Perez-Armendariz EM, Bobadilla K, Guzman-Beltran S, Sada E, Torres M. High glucose concentrations induce TNF-alpha production through the down-regulation of CD33 in primary human monocytes. BMC Immunol. 2012;13:19. doi: 10.1186/1471-2172-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Caselles T, Martinez-Esparza M, Perez-Oliva AB, Quintanilla-Cecconi AM, Garcia-Alonso A, Alvarez-Lopez DM, Garcia-Penarrubia P. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J Leukoc Biol. 2006;79:46–58. doi: 10.1189/jlb.0205096. [DOI] [PubMed] [Google Scholar]
- Herrup K, Carrillo MC, Schenk D, Cacace A, Desanti S, Fremeau R, Bhat R, Glicksman M, May P, Swerdlow R, Van Eldik LJ, Bain LJ, Budd S. Beyond amyloid: getting real about nonamyloid targets in Alzheimer's disease. Alzheimers Dement. 2013;9:452–458. doi: 10.1016/j.jalz.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van BC, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O'Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida A, Akita K, Mori Y, Tanida S, Toda M, Inoue M, Nakada H. Negative Regulation of Toll-like Receptor-4 Signaling through the Binding of Glycosylphosphatidylinositol-anchored Glycoprotein, CD14, with the Sialic Acid-binding Lectin, CD33. J Biol Chem. 2014;289:25341–25350. doi: 10.1074/jbc.M113.523480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Yu JT, Hu N, Tan MS, Zhu XC, Tan L. CD33 in Alzheimer's Disease. Mol Neurobiol. 2014;49:529–535. doi: 10.1007/s12035-013-8536-1. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Sun Z, Lee T, Fusco FR, Kimble TD, Meade CA, Cuthbertson S, Reiner A. A simple and sensitive antigen retrieval method for free-floating and slide-mounted tissue sections. J Neurosci Methods. 1999;93:149–162. doi: 10.1016/s0165-0270(99)00142-9. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer's disease risk genes in control and Alzheimer's disease brains. PLoS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel J, Vingtdeux V, Marambaud P, d'Abramo C, Jimenez H, Stauber M, Friedman R, Davies P. CB2 receptor deficiency increases amyloid pathology and alters tau processing in a transgenic mouse model of Alzheimer’s disease. Mol Med. 2014;20:29–36. doi: 10.2119/molmed.2013.00140.revised. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu Y, Wang Z, Jiang Y. Clinical trials of amyloid-based immunotherapy for Alzheimer’s disease: end of beginning or beginning of end? Expert Opin Biol Ther. 2013;13:1515–1522. doi: 10.1517/14712598.2013.838555. [DOI] [PubMed] [Google Scholar]
- Linnartz B, Neumann H. Microglial activatory (immunoreceptor tyrosine-based activation motif)- and inhibitory (immunoreceptor tyrosine-based inhibition motif)-signaling receptors for recognition of the neuronal glycocalyx. Glia. 2013;61:37–46. doi: 10.1002/glia.22359. [DOI] [PubMed] [Google Scholar]
- Linnartz B, Wang Y, Neumann H. Microglial immunoreceptor tyrosine-based activation and inhibition motif signaling in neuroinflammation. Int J Alzheimers Dis. 2010:587463. doi: 10.4061/2010/587463. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucin KM, Wyss-Coray T. Immune activation in brain aging and neurodegeneration: too much or too little? Neuron. 2009;64:110–122. doi: 10.1016/j.neuron.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, Estus S. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33:13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpass K. Alzheimer disease: functional dissection of CD33 locus implicates innate immune response in Alzheimer disease pathology. Nat Rev Neurol. 2013;9:360. doi: 10.1038/nrneurol.2013.119. [DOI] [PubMed] [Google Scholar]
- McKeith IG, Dickson DW, Lowe J, Emre M, O'Brien JT, Feldman H, Cummings J, Duda JE, Lippa C, Perry EK, Aarsland D, Arai H, Ballard CG, Boeve B, Burn DJ, Costa D, Del ST, Dubois B, Galasko D, Gauthier S, Goetz CG, Gomez-Tortosa E, Halliday G, Hansen LA, Hardy J, Iwatsubo T, Kalaria RN, Kaufer D, Kenny RA, Korczyn A, Kosaka K, Lee VM, Lees A, Litvan I, Londos E, Lopez OL, Minoshima S, Mizuno Y, Molina JA, Mukaetova-Ladinska EB, Pasquier F, Perry RH, Schulz JB, Trojanowski JQ, Yamada M. Diagnosis and management of dementia with Lewy bodies: third report of the DLB Consortium. Neurology. 2005;65:1863–1872. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- Morgan K. The three new pathways leading to Alzheimer's disease. Neuropathol Appl Neurobiol. 2011;37:353–357. doi: 10.1111/j.1365-2990.2011.01181.x. [DOI] [PubMed] [Google Scholar]
- Mosher KI, Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer's disease. Biochem Pharmacol10. 2014 doi: 10.1016/j.bcp.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, Binder L, Counts SE, DeKosky ST, de Toledo-Morrell L, Ginsberg SD, Ikonomovic MD, Perez SE, Scheff SW. Mild cognitive impairment: pathology and mechanisms. Acta Neuropathol. 2012;123:13–30. doi: 10.1007/s00401-011-0884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell KL, Hyman BT, Growdon JH, Hedley-Whyte ET. Application of the National Institute on Aging (NIA)-Reagan Institute criteria for the neuropathological diagnosis of Alzheimer disease. J Neuropathol Exp Neurol. 1999;58:1147–1155. doi: 10.1097/00005072-199911000-00004. [DOI] [PubMed] [Google Scholar]
- O'Banion MK. It may take more than a shot: alternatives to immunotherapy for Alzheimer's disease. Biol Psychiatry. 2013;74:316–317. doi: 10.1016/j.biopsych.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Orr SJ, Morgan NM, Elliott J, Burrows JF, Scott CJ, McVicar DW, Johnston JA. CD33 responses are blocked by SOCS3 through accelerated proteasomal-mediated turnover. Blood. 2007;109:1061–1068. doi: 10.1182/blood-2006-05-023556. [DOI] [PubMed] [Google Scholar]
- Panza F, Logroscino G, Imbimbo BP, Solfrizzi V. Is there still any hope for amyloid-based immunotherapy for Alzheimer's disease? Curr Opin Psychiatry. 2014;27:128–137. doi: 10.1097/YCO.0000000000000041. [DOI] [PubMed] [Google Scholar]
- Piaceri I, Nacmias B, Sorbi S. Genetics of familial and sporadic Alzheimer's disease. Front Biosci (Elite Ed) 2013;5:167–177. doi: 10.2741/e605. 167–177. [DOI] [PubMed] [Google Scholar]
- Puli L, Pomeshchik Y, Olas K, Malm T, Koistinaho J, Tanila H. Effects of human intravenous immunoglobulin on amyloid pathology and neuroinflammation in a mouse model of Alzheimer's disease. J Neuroinflammation. 2012;9:105. doi: 10.1186/1742-2094-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM. Alzheimer's disease. N Engl J Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, Rothamel K, Stranger BE, Bennett DA, Evans DA, De Jager PL, Bradshaw EM. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer's disease susceptibility. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddt666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed-Geaghan EG, Savage JC, Hise AG, Landreth GE. CD14 and toll-like receptors 2 and 4 are required for fibrillar A{beta}-stimulated microglial activation. J Neurosci. 2009;29:11982–11992. doi: 10.1523/JNEUROSCI.3158-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Dols-Icardo O, Bullido MJ, Pastor P, Rodriguez-Rodriguez E, Lopez de MA, de Pancorbo MM, Perez-Tur J, Alvarez V, Antonell A, Lopez-Arrieta J, Hernandez I, Tarraga L, Boada M, Lleo A, Blesa R, Frank-Garcia A, Sastre I, Razquin C, Ortega-Cubero S, Lorenzo E, Sanchez-Juan P, Combarros O, Moreno F, Gorostidi A, Elcoroaristizabal X, Baquero M, Coto E, Sanchez-Valle R, Clarimon J. Assessing the role of the TREM2 p.R47H variant as a risk factor for Alzheimer's disease and frontotemporal dementia. Neurobiol Aging. 2014;35:444. doi: 10.1016/j.neurobiolaging.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Schellenberg GD, Montine TJ. The genetics and neuropathology of Alzheimer's disease. Acta Neuropathol. 2012;124:305–323. doi: 10.1007/s00401-012-0996-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely CA, Zandi PP. Non-steroidal anti-inflammatory drugs and Alzheimer's disease: the epidemiological evidence. CNS Neurol Disord Drug Targets. 2010;9:132–139. doi: 10.2174/187152710791012026. [DOI] [PubMed] [Google Scholar]
- Vlad SC, Miller DR, Kowall NW, Felson DT. Protective effects of NSAIDs on the development of Alzheimer disease. Neurology. 2008;70:1672–1677. doi: 10.1212/01.wnl.0000311269.57716.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DG, Dalsing-Hernandez JE, Campbell NA, Lue LF. Decreased expression of CD200 and CD200 receptor in Alzheimer's disease: a potential mechanism leading to chronic inflammation. Exp Neurol. 2009;215:5–19. doi: 10.1016/j.expneurol.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DG, Link J, Lue LF, Dalsing-Hernandez JE, Boyes BE. Gene expression changes by amyloid beta peptide-stimulated human postmortem brain microglia identify activation of multiple inflammatory processes. J Leukoc Biol. 2006;79:596–610. doi: 10.1189/jlb.0705377. [DOI] [PubMed] [Google Scholar]
- Walker DG, Lue LF, Adler CH, Shill HA, Caviness JN, Sabbagh MN, Akiyama H, Serrano GE, Sue LI, Beach TG. Changes in properties of serine 129 phosphorylated alpha-synuclein with progression of Lewy-type histopathology in human brains. Exp Neurol. 2013;240:190–204. doi: 10.1016/j.expneurol.2012.11.020. Epub@2012 Nov 28.:190-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter RB. The role of CD33 as therapeutic target in acute myeloid leukemia. Expert Opin Ther Targets. 2014;18:715–718. doi: 10.1517/14728222.2014.909413. [DOI] [PubMed] [Google Scholar]
- Walter RB, Hausermann P, Raden BW, Teckchandani AM, Kamikura DM, Bernstein ID, Cooper JA. Phosphorylated ITIMs enable ubiquitylation of an inhibitory cell surface receptor. Traffic. 2008;9:267–279. doi: 10.1111/j.1600-0854.2007.00682.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Neumann H. Alleviation of neurotoxicity by microglial human Siglec-11. J Neurosci. 2010;30:3482–3488. doi: 10.1523/JNEUROSCI.3940-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winblad B, Graf A, Riviere ME, Andreasen N, Ryan JM. Active immunotherapy options for Alzheimer's disease. Alzheimers Res Ther. 2014;6:7. doi: 10.1186/alzrt237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Chu C, Jia J. Association studies of 19 candidate SNPs with sporadic Alzheimer's disease in the North Chinese Han population. Neurol Sci. 2012;33:1021–1028. doi: 10.1007/s10072-011-0881-0. [DOI] [PubMed] [Google Scholar]
- Zotova E, Bharambe V, Cheaveau M, Morgan W, Holmes C, Harris S, Neal JW, Love S, Nicoll JA, Boche D. Inflammatory components in human Alzheimer's disease and after active amyloid-beta42 immunization. Brain. 2013;136:2677–2696. doi: 10.1093/brain/awt210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Identification of CD33 rs3865444 genotypes. The different pattern of DNA bands resulting from PCR amplification of DNA followed by restriction enzyme digestion with NlaIII used to identify the different CD33 rs3865444 genotypes (C/C, C/A, A/A). m- sizes of DNA marker bands (bp-base pairs).
Supplemental Figure 2. Effect of CD33 rs3865444 genotype and disease state on levels of synaptophysin in human brain samples. (A). Scatter plot showing relative levels of truncated synaptophysin with CD33 rs3865444 genotypes in temporal cortex samples (C/C: n=21; C/A: n=26; A/A: n=20). One way ANOVA showed no significant differences between genotypes. (B). Scatter plot showing significantly decreased levels of synaptophysin in AD cases (n= 25) compared to ND cases (n=42) (P<0.05, unpaired t test). (C). Scatter plot showing relative levels of synaptophysin in samples divided into CD33 rs3865444 genotype and disease state. Two-way ANOVA (with Sidak’s test for multiple comparisons) demonstrated no significant differences between genotypes for synaptophysin levels, or between genotype and disease groups, but a significant effect of disease state (P< 0.05). Lines indicate mean values ± SEM.