Abstract
Alzheimer’s disease (AD) is characterized by deficits in cerebral metabolic rates of glucose in the posterior cingulate (PC) and precuneus in AD subjects, and in APOEε4 carriers, decades prior to the onset of measureable cognitive deficits. However, the cellular and molecular basis of this phenotype remains to be clarified. Given the roles of astrocytes in energy storage and brain immunity, we sought to characterize the transcriptome of AD PC astrocytes. Cells were laser capture microdissected from AD (n=10) and healthy elderly control (n=10) subjects for RNA sequencing. We generated >5.22 billion reads and compared sequencing data between controls and AD patients. We identified differentially expressed mitochondria-related genes including TRMT61B, FASTKD2, and NDUFA4L2, and using pathway and weighted gene co-expression analyses, we identified differentially expressed immune response genes. A number of these genes, including CLU, C3, and CD74, have been implicated in Abeta generation or clearance. This data provides key insights into astrocyte-specific contributions to AD and we present this data set as a publicly available resource.
Keywords: Alzheimer’s disease, posterior cingulate, astrocytes, RNA sequencing, immune response, mitochondria
1. Introduction
With the rapidly growing prevalence of Alzheimer’s disease (AD), improved understanding of the molecular and cellular basis of the disease is needed in order to identify and develop improved diagnostics and treatments. Numerous imaging studies have set the stage for characterizing AD in live subjects. Using FDG-PET (fluorodeoxyglucose position emission tomography), researchers have characterized an association between AD and progressive reductions of CMRgI (cerebral metabolic rates of glucose) in both AD patients (1–4) and young adult APOEε4 carriers four decades before the expected onset of cognitive deficits (2). Affected brain regions include the posterior cingulate (PC), and parietal, temporal, and frontal cortices (1, 5–7), with the PC and precuneus demonstrating the earliest metabolic deficits in AD [1,2]. Such deficits may be associated with metabolic dysfunctions in neurons or glial cells (8, 9), reductions in the density or activity of terminal neuronal fields or perisynaptic glial cells (10, 11), or a combination of both, and additionally highlights a metabolic role of the PC in AD.
In previous work, we identified widespread down-regulated expression of electron transport and mitochondrial translocase genes in non-tangle bearing neurons microdissected from the PC of AD subjects (12). In a separate study, decreased cytochrome c oxidase activity was also identified across all six layers of the PC cortex in AD subjects (13). While these findings provide evidence that mitochondrial dysfunction may play a role in characteristic CMRgI deficits in AD, it is unclear as to what pathogenic contributions may be derived from astrocytes in the PC. Importantly, previous studies have shown that astrocytes demonstrate changes with respect to aging and AD. Examples include the occurrence of structural changes in astrocytes in the dentate gyrus of aged rats (14), distinct clustering of astrocytes and astroglial hypertrophy in the hippocampi of aged rats (15), release of proinflammatory factors from astrocytes in AD (16) and buildup of Abeta molecules within astrocytes (16) that may result from deficits in the ability of the cell to degrade Abeta, and recent work suggesting that activation of glial cells may have a primary role in impacting the health of neurons in AD (17). Given this previous research and astrocytes’ roles in energy storage and metabolism (18, 19), immunity (20), and the greater than two-fold increase in the number of astrocytes compared to neurons in the human brain (20–23), we hypothesized that PC astrocytes demonstrate significant expression changes in AD. With the role of astrocytes in energy storage and metabolism, we further hypothesize that genes involved in mitochondrial processes are also dysregulated in AD PC astrocytes.
A number of studies have evaluated human and murine astrocyte transcriptomes using microarray technology (24–27) or focused approaches such as Northern blotting (28) or real time PCR (polymerase chain reaction) (29). Simpson et al. (25) expression profiled GFAP (glial fibrillary acidic protein) positive astrocytes microdissected from the temporal cortex of elderly subjects and identified differentially expressed genes with respect to Braak stage and APOE (apolipoprotein) ε4 carrier status. In a separate study, cortical astrocytes were collected from young and old mice and expression profiled to reveal altered expression of key genes involved in neuronal signaling (30). In this study, we used laser capture microdissection (LCM) to collect ALDH1L1- (aldehyde dehydrogenase 1 family, member L1) positive astrocytes from PC cortex in both AD subjects (n=10) and healthy elderly controls (n=10). GFAP was not used in our study due to a number of caveats with this marker including: (1) GFAP demonstrates varying levels of expression across different cell types and regions (21, 31, 32), is expressed in other glial cells outside of astrocytes (21), is highly expressed in reactive astrocytes (21, 33–35), and is often not identifiable in healthy tissue (21). However, ALDH1L1 is more specific to astrocytes than GFAP, does not preferentially mark reactive astrocytes, is expressed in mature astrocytes, and demonstrates a wider pattern of expression compared to GFAP such that GFAP-positive cells make up a subset of ALDH1L1-positive cells (21, 33–35). It is also important to note that our previous analysis of PC neurons was limited by the use of microarrays that used pre-defined probes to target only nuclear-encoded, and not mitochondrially-encoded, genes. However, in our study here we capitalize on current genomic technologies to perform an unbiased evaluation of all transcripts present in each sample and to take advantage of the increased analytical sensitivity and wide dynamic range associated with next generation sequencing. We thus performed next generation RNA sequencing (RNAseq) of astrocyte total RNA and describe here the first reported sequencing data set of PC astrocytes in AD.
2. Materials and Methods
2.1 Sample acquisition
Post-mortem brain samples were collected at the Banner Sun Health Research Institute’s Brain and Body Donation Program from 10 clinically classified late-onset AD subjects (4 males and 6 females; 5 APOEε3/4 subjects and 5 APOEε4/4 subjects) with a mean age at death of 83.9 and 10 ND (no disease) healthy elderly control subjects (6 males and 4 females; 5 APOEε3/4 subjects and 5 APOEε4/4 subjects) with a mean age of death of 85.1. Clinically confirmed AD subjects had Braak stages ranging from IV to VI (36) and neuritic plaque densities of frequent (37). Control subjects had Braak stages ranging from I to IV, plaque densities ranging from zero to moderate, and were clinically confirmed to not demonstrate dementia. Samples were collected with a mean post-mortem interval (PMI) of 2.42 hours from the PC cortex (Brodmann’s areas 23 and 31). Following dissection, samples were flash frozen, sectioned (10 µm), and mounted on glass or PEN membrane slides (Carl Zeiss Microscopy; Thornwood, NY). Information for each subject is shown in Table 1. Total RNA was isolated from a single section from each subject using the Qiagen RNeasy kit to evaluate RNA integrity. Samples demonstrated Agilent Bioanalyzer RINs (RNA Integrity Number) between 7.4 and 9.4 such that all samples were used for downstream analyses.
Table 1.
Subject information
| Sample | Gender | APOE | PMIa | Expired age |
Plaque density |
Braak score |
NIA-Rb |
|---|---|---|---|---|---|---|---|
| control1 | M | 3/3 | 3.25 | 80 | sparse | II | criteria not met |
| control2 | F | 3/3 | 2.25 | 82 | zero | II | criteria not met |
| control3 | F | 3/3 | 2.50 | 95 | zero | III | criteria not met |
| control4 | M | 3/3 | 2.66 | 78 | zero | II | criteria not met |
| control5 | F | 3/4 | 1.50 | 85 | moderate | III | criteria not met |
| control6 | M | 3/4 | 2.00 | 91 | zero | IV | criteria not met |
| control7 | M | 3/4 | 2.33 | 76 | sparse | I | criteria not met |
| control8 | F | 3/4 | 2.50 | 85 | zero | III | criteria not met |
| control9 | M | 3/3 | 3.00 | 82 | zero | IV | criteria not met |
| control10 | M | 3/4 | 1.87 | 97 | zero | III | criteria not met |
| AD1 | F | 3/3 | 1.50 | 85 | frequent | VI | high |
| AD2 | M | 3/3 | 2.95 | 82 | frequent | V | high |
| AD3 | F | 3/3 | 2.15 | 84 | frequent | V | high |
| AD4 | M | 3/4 | 2.25 | 78 | frequent | VI | high |
| AD5 | F | 3/4 | 2.16 | 78 | frequent | VI | high |
| AD6 | F | 3/4 | 2.66 | 85 | frequent | V | high |
| AD7 | F | 3/4 | 2.70 | 89 | frequent | VI | high |
| AD8 | M | 3/4 | 3.92 | 79 | frequent | V | high |
| AD9 | M | 3/3 | 2.05 | 86 | frequent | VI | high |
| AD10 | F | 3/3 | 2.15 | 93 | frequent | IV | intermediate |
PMI = post mortem interval (hours)
NIA-R=National Institute on Aging and Reagan Institute neuropathological AD severity score
2.2 Staining and laser capture microdissection
PC brain sections were rapidly stained with Alexa Fluor 350 conjugated ALDH1L1 (anti-Aldehyde Dehydrogenase 1 Family, Member L1) rabbit polyclonal antibody (ABIN882166; antibodies-online; Atlanta, GA) to identify astrocytes. For each section, antibody was diluted 1:100 in PBS (phosphate buffered saline) with ProtectRNA RNase inhibitor (Sigma-Aldrich; St. Louis, MO). For staining, each section was first fixed in ice cold acetone for 3 minutes, washed 3X with PBS with RNase inhibitor, and diluted ALDH1L1 antibody was applied and incubated in the dark on ice for 10 minutes. The section was then washed 3X with PBS with RNase inhibitor, followed by consecutive washes with molecular water, 70% ethanol, 95% ethanol, and 100% ethanol. The section was then allowed to dry at room temperature. For each subject, approximately 300 astrocytes were LCMed from PC cortex onto the caps of AdhesiveCap opaque tubes (Carl Zeiss Microscopy) using the PALM LCM System (Carl Zeiss Microscopy; Thornwood, NY). Examples of ALDH1L1+ astrocytes, as well as a pre- and post-LCMed astrocyte, are shown in Supplementary Figure 1. 10 µL of Extraction buffer from the Arcturus PicoPure RNA Isolation Kit (Life Technologies; Grand Island, NY) was pipetted directly onto the cap and incubated at 42°C for 30 minutes to create cell lysate. Lysates were collected by centrifugation at 800×g for 2 minutes and stored at −80°C until all samples were microdissected.
2.3 RNA isolation
Total RNA was isolated from all cell lysates using the Arcturus PicoPure RNA Isolation Kit (Life Technologies; Grand Island, NY) following the manufacturer’s protocol. DNase treatment was additionally performed during RNA isolation using the RNase-free DNase set (Qiagen; Valencia, CA) following Appendix A of the Arcturus PicoPure manufacturer’s protocol.
2.4 Library preparation
All isolated total RNA from each LCM collection was used to generate and linearly amplify cDNA using the Ovation RNAseq System v2 (Nugen; San Carlos, CA) following the manufacturer’s protocol. This system uses a primer mix to initiate amplification from the 3’ end of genes as well as throughout the entire transcriptome. Amplified cDNA was input into the TruSeq DNA Sample Preparation Kit v2 (Illumina; San Diego, CA) and sequencing libraries were generated following the manufacturer’s protocol with the exception that 10 cycles of PCR was used for library enrichment. Final libraries were analyzed on a Bioanalyzer DNA 1000 chip (Agilent Technologies; Santa Clara, CA). Equimolar pools of libraries were created for sequencing and re-assessed on the Bioanalyzer prior to sequencing.
2.5 Paired end sequencing
Library pools were clustered onto Illumina v3 flowcells using the Illumina Truseq PE Cluster Kit v3 on the Illumina cBot. Clustered flowcells were sequenced by synthesis on the Illumina HiSeq2000 using Illumina’s Truseq PE Cluster Kit v3 and Illumina’s TruSeq SBS Kits v3 for paired 83 base pair read lengths.
2.6 Sequencing data analysis
Raw sequencing data in the form of BCL files were converted to FASTQ files using Illumina’s BCLConverter software. Data was aligned against the human reference genome (build 37) using TopHat1.2 (38, 39) and Cufflinks (38) was used to assemble aligned reads into transcripts. HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) was used to generate a counts table from Cufflinks output and DESeq2 v1.4.0 (40)(http://www.bioconductor.org/packages/devel/bioc/html/DESeq2.html) was used to calculate normalized read counts for each gene/transcript and to perform expression analysis. To evaluate inter-sample variability, we performed Pearson’s correlation analyses in R using normalized counts across all samples to identify any outliers. DESeq2 uses a generalized linear model to evaluate differential expression while accounting for biological variance and uses a Wald test statistic to evaluate significance. A DESeqDataSet object is created using HTSeq counts and the DESeq() wrapper function is called to perform differential analyses. The fold change is determined by dividing the average normalized read counts of AD samples over control samples for each transcript. Independent filtering and Cook cut-off parameters were set to ON to remove outliers and genes with low normalized read counts. P-values were corrected using the Benjamini and Hochberg False Discovery Rate and results were annotated using the Biomart online portal (http://www.ensembl.org/biomart/martview).
MetaCore from Thompson Reuters (v6.10 build 40284) was used for GeneGo pathway analysis. Weighted gene co-expression network analysis (WGCNA) (41, 42) was performed on differentially expressed genes (corrected P<0.05). Gene sets were loaded into the WGCNA R package and annotation of generated modules was performed using DAVID (43).
All data generated through this study is accessible through NCBI’s (National Center for Biotechnology Information) dbGaP (database of Genotypes and Phenotypes; accession phs000745.v1.p1).
2.7 qPCR (quantitative polymerase chain reaction) validation
qPCR was performed to experimentally validate differentially expressed transcripts in two phases: first, qPCR was performed on total RNA from astrocytes microdissected from 3 control subjects and 3 AD subjects and second, across total RNA isolated from whole PC sections from all 20 sequenced subjects (during initial sample quality control analyses described in section 2.1). For the first phase, validation was performed on cDNA synthesized and linearly amplified from LCMed astrocyte pools using the Nugen Ovation RNA-Seq System V2 kit. Additional remaining cDNA from original collections used for sequencing were used for validations for AD4, AD6, and ND9. Based on sample availability, newly microdissected astrocyte pools were collected for AD8, ND1, and ND2. Total RNA was isolated using the PicoPure RNA isolation kit and was used to generate cDNA for validation using the Nugen Ovation RNA-Seq System V2 kit. REV3L (REV3-like, polymerase (DNA directed), zeta, catalytic subunit) was selected as a reference gene because it demonstrated consistent high expression (greater than 1000 normalized read counts in each of all 20 samples) in RNAseq data and also demonstrated the lowest variance across all 20 samples. For the second phase, remaining total RNA from initial sample quality control analyses (section 2.1) was used and cDNA was generated using Qiagen’s QuantiTect Reverse Transcription Assay. Primers were designed using Primer3 and validated by performing a dilution series using Clontech Human RNA from Takara Bio Inc. (Kyoto, Japan) which was also reverse transcribed using the QuantiTect Reverse Transcription assay. Cq values were plotted against the log of the input to determine binding efficiency of each primer. QPCR reactions were performed in 10uL reactions using QuantiFast SYBR Green RT-PCR kit (Hilden, Germany) per the manufacturer’s instructions on the Roche Lightcycler 480 system (Basel, Switzerland), with the exception that the initial reverse transcription step was omitted. Reactions were performed in triplicate with one non-template control.
3. Result and Discussion
3.1 RNA sequencing
We sequenced 10 AD PC astrocyte pools and 10 ND (control) PC astrocyte pools to generate over 5.22 billion total reads and over 295 Gb of Q30 sequencing data. Both AD and ND groups were controlled for APOE genotype. We sequenced an average of 123,214,013 mapped reads per sample. Pearson’s correlation analyses on normalized read counts across all samples resulted in r values all greater than 0.88. All samples were thus included for differential expression analyses.
3.2 Differentially expressed genes in AD PC astrocytes
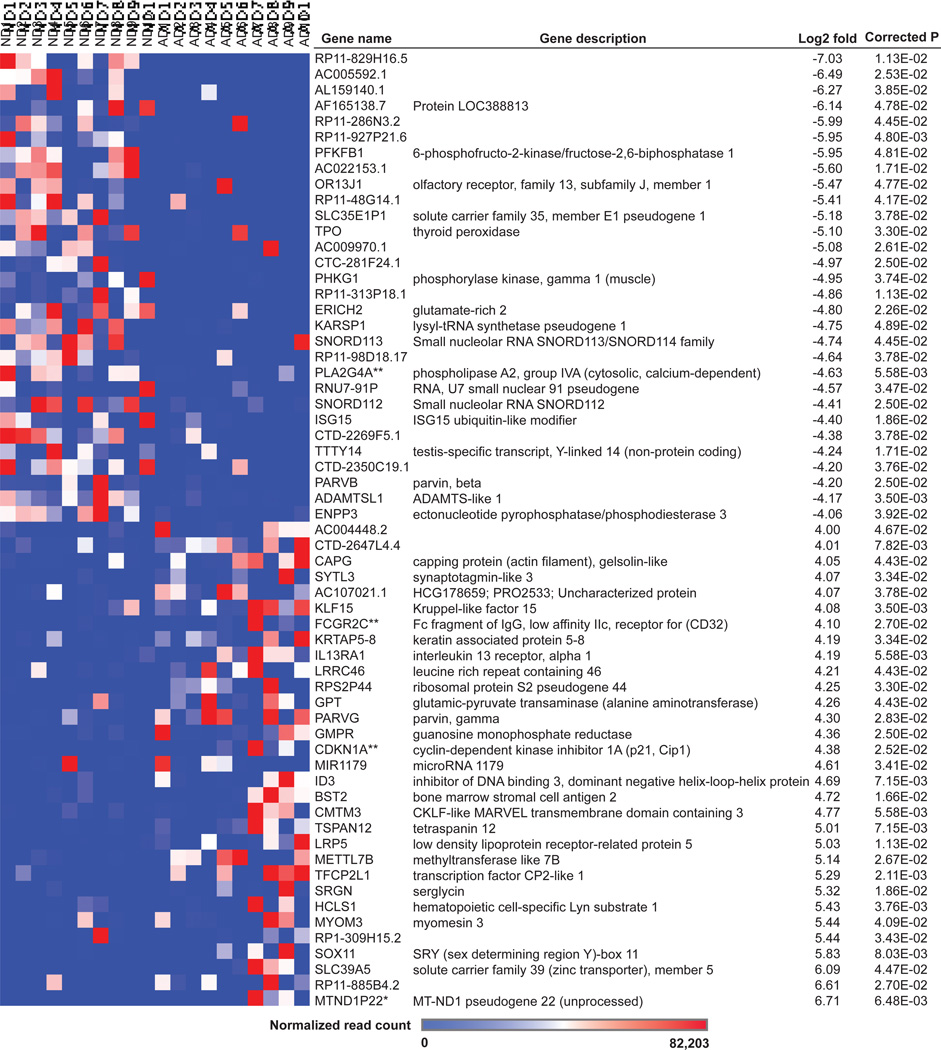
Comparison of AD samples to controls led to the identification of 226 differentially expressed genes (corrected P<0.05; Supplementary Table 1). Genes demonstrating the greatest log2 fold changes are shown in Figure 1. 55.8% of significant genes demonstrated up-regulated expression.
Figure 1. Differentially expressed genes in AD PC astrocytes.
Differentially expressed genes (corrected P<0.05) with the highest log2 fold changes are shown. *mitochondrial gene; **immune response gene
Differentially expressed genes include mitochondrial genes, defined as mitochondrially-encoded genes or MitoCarta genes (44) (Table 2). These genes include FASTKD2 (Fas-activated serine/threonine phosphoprotein kinase domains 2; log2 ratio=−3.50), TRMT61B (tRNA methyltransferase 61 homolog B; log2 ratio=−2.81), PITRM1-AS1 (PREP/MP1; pitrilysin metalloproteinase 1 antisense RNA 1; log2 ratio=−3.20), NDUFA4L2 (NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2; log2 ratio=3.23), and MTND1P22 (mitochondrially-encoded NADH dehydrogenase 1 pseudogene 22; log2 ratio=6.71). FASTKD2, which localizes to the mitochondrial matrix, has been predicted to be involved in regulating apoptosis in breast cancer such that knockdown of the gene resulted in inhibition of apoptosis (45). Homozygous nonsense mutations in FASTKD2 have also been previously reported in mitochondrial encephalomyopathy, a condition associated with decreased cytochrome c oxidase activity (46). Decreased expression of FASTKD2 in AD PC astrocytes thus suggests perturbation of apoptosis regulation in these cells. TRMT61B, which also localizes to mitochondria, methylates adenosine at position 58 of cytoplasmic mitochondrial transport RNAs (tRNAs) and is hypothesized to improve processing, stability, or function of tRNAs required for translation of respiratory factors (47).
Table 2.
Differentially expressed MitoCarta genes in AD PC astrocytes (corrected P<0.05)
| Ensembl ID | Gene name |
Description | Log2 fold |
P | Corrected P |
|---|---|---|---|---|---|
| ENSG00000118246 | FASTKD2 | FAST kinase domains 2 | −3.67 | 6.41E-08 | 1.37E-03 |
| ENSG00000237399 | PITRM1-AS1 | PITRM1 antisense RNA 1 | −3.61 | 4.67E-04 | 4.78E-02 |
| ENSG00000065911 | MTHFD2 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | −3.18 | 2.95E-04 | 3.93E-02 |
| ENSG00000171103 | TRMT61B | tRNA methyltransferase 61 homolog B (S. cerevisiae) | −3.01 | 3.19E-04 | 4.09E-02 |
| ENSG00000168394 | TAP1 | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 2.83 | 3.51E-04 | 4.25E-02 |
| ENSG00000122140 | MRPS2 | mitochondrial ribosomal protein S2 | 2.88 | 6.84E-05 | 2.40E-02 |
| ENSG00000047230 | CTPS2 | CTP synthase 2 | 3.15 | 9.63E-05 | 2.65E-02 |
| ENSG00000185633 | NDUFA4L2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 | 3.23 | 3.23E-04 | 4.09E-02 |
| ENSG00000251407 | MTND1P22 | MT-ND1 pseudogene 22 (unprocessed) | 6.71 | 5.14E-06 | 6.48E-03 |
PITRM1/PREP/MP1 encodes an enzyme that is localized to the mitochondrial matrix (48) and that has been shown to degrade Abeta (49). Activity of this enzyme has also been reported to be decreased in the temporal lobe of AD subjects and transgenic AD murine brains (50). Decreased expression of a transcript antisense to PITRM1 may be associated with changes in regulation of PITRM1 expression. NDUFA4L2 has been reported to inhibit activity of complex I under hypoxic conditions through HIF-1 (hypoxia-inducible transcription factor-1) (51) such that its increased expression in AD PC astrocytes may impact complex I activity. Lastly, we identified altered expression of a mitochondrially-encoded pseudogene, MTND1P22. Pseudogenes, represented by DNA sequences that are similar to protein-coding genes but do not generate functional proteins, have historically been assumed to be non-functional but more recent research suggests that pseudogene transcripts may represent regulatory long non-coding RNAs (52). The differentially expressed mitochondrial pseudogene identified here is an unprocessed pseudogene, resulting from gene duplication, whereas processed pseudogenes result from a retrotransposition event (52). To date, there have been no reports of biological evidence of mitochondrially-encoded pseudogenes, although nuclear-encoded mitochondrial pseudogenes are known (53). Our understanding of these pseudogenes remains limited. However, given our current understanding of pseudogenes and the sequence similarity of this transcript with the NADH dehydrogenase 1 gene, MTND1P22 may have a role in transcription regulation. Additional differentially expressed mitochondrial genes include CTPS2 (CTP synthase 2; log2 ratio=3.15), MRPS2 (mitochondrial ribosomal protein S2; log2 ratio=2.88), MTHFD2 (methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase; log2 ratio=−3.18), and TAP1 (transporter 1, ATP-binding cassette, subfamily B; log2 ratio=2.83).
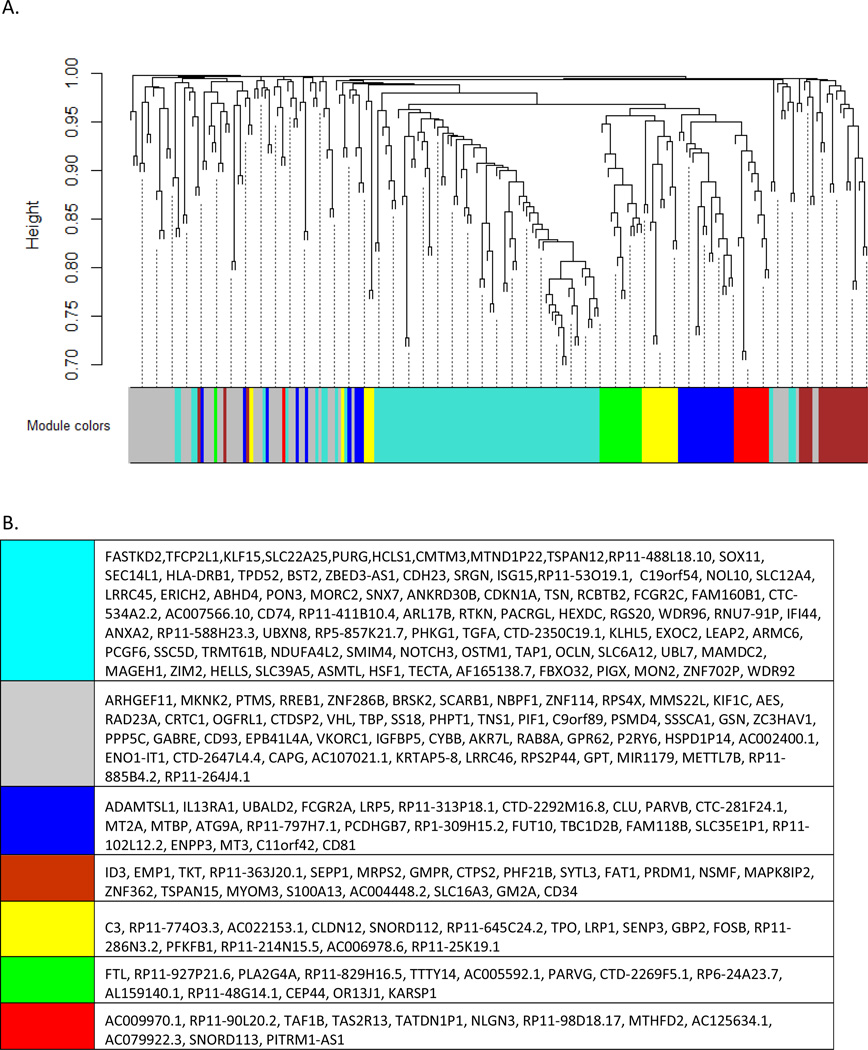
Pathway analysis of differentially expressed genes revealed that the most significantly impacted pathway in AD PC astrocytes is immune system response (Table 3) with 82.4% of significant genes in this pathway demonstrating up-regulated expression in AD PC astrocytes. Furthermore, WGCNA, which performs functional organization of genes based on co-expression, on differentially expressed genes in AD astrocytes generated 7 coexpression modules, whereby the largest module (Figure 2A; turquoise module) demonstrated the highest DAVID enrichment scores for immune response and immune processes, including antigen processing and presentation and lymphocyte activation. Genes in each module are shown in Figure 2B. Given the role of astrocytes in central nervous system immunity under circumstances when insults are present (20), identification that immune system response processes are the most heavily perturbed pathways in AD PC astrocytes correlates with known astrocyte functions. These changes may be associated with activation of astrocytes in response to one or all of the following: the presence of beta-amyloid, weakening of the blood brain barrier (BBB), and exposure of sequenced cells to inflammatory cytokines.
Table 3.
Pathway analysis of differentially expressed genes identified in AD PC astrocytes (corrected P<0.05)
| # | Pathway/process | P | Ratio | %Down | %Up | Genes |
|---|---|---|---|---|---|---|
| 1 | Immune system response | 4.21E-05 | 21/970 | 17.6 | 82.4 | ARHGEF11,C3,CD74,CD81,CD93,CDKN1A,CLU,CYBB,FCGR2A,GSN,HLA-DRB1,IL13RA1,ISG15,PLA2G4A,PSMD4,TAP1,TNS1 |
| 2 | Hematopoiesis | 3.10E-02 | 6/270 | 20.0 | 80.0 | CDKN1A,CYBB,FTL,TAP1,TPO |
| 3 | Transcription regulation | 1.48E-01 | 1/18 | 0.0 | 100.0 | TBP |
| 4 | Vasoconstriction | 2.95E-01 | 4/311 | 75.0 | 25.0 | ARHGEF11,OCLN,PHKG1,PLA2G4A |
| 5 | Calcium signaling | 3.31E-01 | 5/430 | 50.0 | 50.0 | CDKN1A,HLA-DRB1,IL13RA1,P2RY6,PHKG1,PLA2G4A |
| 6 | Oxidative stress regulation | 4.04E-01 | 6/578 | 14.3 | 85.7 | CYBB,FTL,HSF1,IL13RA1,PLA2G4A,RPS4X,SEPP1 |
| 7 | Blood clotting | 4.48E-01 | 3/278 | 33.3 | 66.7 | FCGR2A,PLA2G4A,VKORC1 |
| 8 | Tissue remodeling and wound repair | 4.98E-01 | 5/524 | 40.0 | 60.0 | CDKN1A,LRP5,OCLN,PLA2G4A,TGFA |
| 9 | Inflammatory response | 5.55E-01 | 6/673 | 25.0 | 75.0 | CYBB,IL13RA1,PLA2G4A,TNS1 |
| 10 | Mitogenic signaling | 5.59E-01 | 5/560 | 20.0 | 80.0 | CDKN1A,IGFBP5,IL13RA1,PLA2G4A,TGFA |
| 11 | Obesity | 5.64E-01 | 2/212 | 0.0 | 100.0 | CDKN1A,TNS1 |
| 12 | Myogenesis regulation | 5.72E-01 | 1/95 | 0.0 | 100.0 | CDKN1A |
| 13 | Vasodilation | 5.79E-01 | 3/337 | 66.7 | 33.3 | ARHGEF11,FOSB,PHKG1 |
| 14 | Vitamin and cofactor metabolism and its regulation | 5.91E-01 | 6/697 | 33.3 | 66.7 | ENPP3,FTL,MTHFD2,PHPT1,SCARB1,VKORC1 |
| 15 | Lipid biosynthesis and regulation | 6.43E-01 | 3/370 | 100.0 | 0.0 | PLA2G4A |
| 16 | Cystic fibrosis disease | 6.75E-01 | 5/636 | 14.3 | 85.7 | ANXA2,CYBB,FOSB,PSMD4,RAB8A,TAP1,TGFA |
| 17 | Vascular development (Angiogenesis) | 6.89E-01 | 4/522 | 25.0 | 75.0 | CDKN1A,MKNK2,PLA2G4A,VHL |
| 18 | Protein degradation | 6.94E-01 | 2/269 | 0.0 | 100.0 | CDKN1A,HSF1 |
| 19 | Nucleotide metabolism and its regulation | 6.97E-01 | 3/401 | 33.3 | 66.7 | CTPS2,ENPP3,GMPR |
| 20 | Cell differentiation | 7.25E-01 | 7/922 | 57.1 | 42.9 | CDKN1A,FOSB,IL13RA1,NOTCH3,OCLN,PHKG1,PLA2G4A |
Figure 2. WGCNA dendrogram of differentially expressed genes in AD PC astrocytes.
226 differentially expressed genes (corrected P<0.05) were used for WGCNA. (A) Seven coexpression modules were generated and the total genes for each module are as follows: blue=26, brown=22, green=14, gray=53, red=12, turquoise=83, yellow=16. Each open ended arm in the dendrogram represents a separate gene. (B) Genes falling within each module are listed.
Differentially expressed immune system response genes include CLU (clusterin, apolipoprotein J (APOJ); log2 ratio=1.97), C3 (complement component 3; log2 ratio=2.82), and CD74 (cluster of differentiation 74 molecule, major histocompatibility complex, class II invariant chain; log2 ratio=3.52). Two large genome wide association studies that evaluated over 23,000 individuals implicated a SNP, rs11136000, in CLU that is associated with late-onset AD (54, 55). CLU encodes a heterodimeric chaperone that has been found to inhibit Abeta oligomer uptake in human astrocytes (56) such that increased expression of this gene in AD PC astrocytes may affect Abeta clearance. C3 encodes a member of the complement system and this gene has been reported to demonstrate an age-related increase in expression in control (C57BL/6) mice with more significant increases in transgenic APP (amyloid precursor protein) mice during formation of Abeta (57). Lastly, we identified up-regulated expression of CD74 in AD PC astrocytes. CD74, which is involved in antigen presentation to T cells during an immune response, has also been found to interact with Abeta and obstruct Abeta production in HeLa cells (58). Up-regulated expression of CD74 in AD PC astrocytes may thus represent an effect resulting from multiple causes encompassing both immune system and Abeta responses. Notably, the immune system response genes described here have all been shown to be associated with APP/Abeta such that APP/Abeta may be a key initiating factor in immune response pathways. Of relevance, the proximity of LCMed astrocytes to Abeta plaques was not captured during microdissection but the findings reported here may be influenced by the higher plaque load in the AD subjects.
3.3 Experimental validation of differentially expressed genes
Based on sample availability, qPCR experiments were performed to validate expression changes. qPCR was first performed across 3 control and 3 AD subjects using cDNA generated from total RNA isolated from microdissected PC astrocytes to validate gene expression changes of 7 selected differentially expressed genes, or transcripts, identified through the AD versus ND comparison (Supplementary Figure 2A and 2B). These genes/transcripts include RP11-488L18.10, RP11-797H7.1, TRMT61B, PPP5C, C3, TBP, and CLU. Due to limited availability of these samples, we also performed qPCR validation on total RNA that was isolated from an entire PC section from each subject during initial sample quality control analyses (n=20; Supplementary Figure 2A and 2C). These analyses were performed on 6 selected differentially expressed genes including TRMT61B, CTPS2, NDUF4AL2, CD74, CLU, and C3. Overall, the directionality of expression changes was validated across all genes. Notably, CLU and C3, both of which were evaluated by qPCR in microdissected astrocytes and whole PC sections, demonstrated larger increases in the astrocyte-specific data and lower increases in whole PC sections to show that the astrocyte-specific signal is diluted in whole PC sections.
4. Conclusions
In this study, we identified altered expression of mitochondrial and immune system response genes in PC astrocytes in the context of AD. Based on WGCNA and pathway analyses, immune response processes appear to be most heavily impacted in PC astrocytes in AD subjects. These results provide evidence that brain immunity and mitochondrial functions in PC astrocytes are perturbed in AD.
Given the role of astrocytes as immune sensors in the brain (20), these cells may represent a first line of defense in response to insults, which may include inflammation, injury, or infection. Differentially expressed immune response genes in AD subjects include numerous genes that have been previously implicated as having key roles associated with Abeta production and clearance. However, it remains to be clarified if alterations in immunity pathways parallel, overlap, or potentially precede, Abeta formation. Deeper investigations into young pre-AD and MCI subjects will be needed to determine if immune response processes are similarly affected prior to onset of AD, and if perturbation of immune response pathways coincides with CMRgI deficits seen in young APOEε4 carriers. Likewise, additional regional analyses are needed to evaluate if immune response pathways are also heavily impacted in brain regions differentially affected by AD.
While this study lends new insight into astrocytic changes in AD, we are limited by a few caveats. Although microdissection was used to collect astrocytes for sequencing, it is possible that fragments of adjacent endothelial cells or neurons may have also been collected. To mitigate this possibility, astrocytes were individually collected, but it is possible that astrocytic end feet may have been missed. As a secondary confirmation, we evaluated GFAP expression and identified increased expression in AD subjects (uncorrected P=1.63E-03, log2 fold=2.49), as expected due to up-regulation of GFAP in reactive astrocytes in AD (21, 33, 59), although we did not specifically collect GFAP-positive cells. Another limitation is that we do not know the mitochondrial content of the microdissected cells, whereby differential expression of mitochondrial genes may reflect differential mitochondrial loads. Differences in mitochondrial content in both astrocytes and neurons in the context of AD have yet to be investigated. Thirdly, it is not known if increased expression of mitochondrial genes directly translates to activation of energy metabolism pathways.
This study is also prefaced by significant region-specific transcriptional analyses in AD that have demonstrated discrete alterations in different areas of the brain. Key upregulated and downregulated genes have been identified in the amygdala and cingulate cortex of post-mortem AD brains, correlating to processes including inflammation and energy metabolism, respectively (60). Although analyses were performed on whole tissue, upregulation of inflammation genes and downregulation of energy metabolism genes parallel our findings here, in addition to our previous report of decreased expression of electron transport genes in PC neurons (12). In a separate study, microarray analysis of LCMed CA1 hippocampal gray matter from FFPE (formalin fixed paraffin embedded) sections of AD brains suggests that increased expression of glial transcription factors genes, among others, may be concentrated in white, and not gray, matter (61). Although transcriptional analysis of FFPE samples may be associated with decreased specificity, this study provides evidence of divergent expression changes also at the level of gray and white matter. In neuron-specific transcriptomic studies in AD, key expression alterations were also identified across multiple brain regions, including the PC, hippocampal CA1, middle temporal gyrus, in layer III cortical neurons (62, 63).
Such findings emphasize the need to consider cell-specific responses, roles, and contributions to AD pathogenesis. The benefits of RNAseq support our ability to perform cell-specific analyses by widening the dynamic range for transcript detection and by supporting an unbiased evaluation of all transcripts in a sample. Furthermore, identification of differentially expressed mitochondrial and immune response genes, and the respective roles of these genes in Abeta generation and clearance, lends valuable insight into previously uncharacterized cells in the PC. As we continue to improve our understanding of the discrete molecular events that occur in the PC in AD, we will strengthen our ability to identify potential targets for slowing and arresting disease progression at earlier stages of AD.
Supplementary Material
Highlights.
We performed RNAseq on posterior cingulate astrocytes from controls and Alzheimer’s subjects.
Differentially expressed immune response and mitochondrial genes were identified.
Cell-specific analyses are needed to understand cellular roles in AD pathogenesis.
Acknowledgments
We thank Dr’s Carol Barnes and Matthew Huentelman, and the Arizona Alzheimer’s Consortium and the Arizona Alzheimer’s Disease Core Center for supporting this study. This study was funded by NIH P30AG019610 through the Arizona Alzheimer’s Disease Core Center pilot program. We additionally thank the participating subjects and families, Cynthia Lechuga, and Dr. Kendall Jensen for support and assistance, and St. Joseph’s Medical Center and Hospital for LCM access.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol. 1997;42(1):85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- 2.Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci U S A. 2004;101(1):284–289. doi: 10.1073/pnas.2635903100. Epub 2003 Dec 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiman EM, Caselli RJ, Chen K, Alexander GE, Bandy D, Frost J. Declining brain activity in cognitively normal apolipoprotein E epsilon 4 heterozygotes: A foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer's disease. Proc Natl Acad Sci U S A. 2001;98(6):3334–3339. doi: 10.1073/pnas.061509598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, et al. Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334(12):752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 5.Alexander GE, Chen K, Pietrini P, Rapoport SI, Reiman EM. Longitudinal PET Evaluation of Cerebral Metabolic Decline in Dementia: A Potential Outcome Measure in Alzheimer's Disease Treatment Studies. Am J Psychiatry. 2002;159(5):738–745. doi: 10.1176/appi.ajp.159.5.738. [DOI] [PubMed] [Google Scholar]
- 6.Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000;57(3):339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- 7.Thal LJ, Kantarci K, Reiman EM, Klunk WE, Weiner MW, Zetterberg H, et al. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20(1):6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mark RJ, Pang Z, Geddes JW, Uchida K, Mattson MP. Amyloid beta-peptide impairs glucose transport in hippocampal and cortical neurons: involvement of membrane lipid peroxidation. J Neurosci. 1997;17(3):1046–1054. doi: 10.1523/JNEUROSCI.17-03-01046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piert M, Koeppe RA, Giordani B, Berent S, Kuhl DE. Diminished glucose transport and phosphorylation in Alzheimer's disease determined by dynamic FDG-PET. J Nucl Med. 1996;37(2):201–208. [PubMed] [Google Scholar]
- 10.Magistretti PJ, Pellerin L. Cellular bases of brain energy metabolism and their relevance to functional brain imaging: evidence for a prominent role of astrocytes. Cereb Cortex. 1996;6(1):50–61. doi: 10.1093/cercor/6.1.50. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz WJ, Smith CB, Davidsen L, Savaki H, Sokoloff L, Mata M, et al. Metabolic mapping of functional activity in the hypothalamo-neurohypophysial system of the rat. Science. 1979;205(4407):723–725. doi: 10.1126/science.462184. [DOI] [PubMed] [Google Scholar]
- 12.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105(11):4441–4446. doi: 10.1073/pnas.0709259105. Epub 2008 Mar 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valla J, Berndt JD, Gonzalez-Lima F. Energy hypometabolism in posterior cingulate cortex of Alzheimer's patients: superficial laminar cytochrome oxidase associated with disease duration. J Neurosci. 2001;21(13):4923–4930. doi: 10.1523/JNEUROSCI.21-13-04923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geinisman Y, Bondareff W, Dodge JT. Hypertrophy of astroglial processes in the dentate gyrus of the senescent rat. Am J Anat. 1978;153(4):537–543. doi: 10.1002/aja.1001530405. [DOI] [PubMed] [Google Scholar]
- 15.Landfield PW, Rose G, Sandles L, Wohlstadter TC, Lynch G. Patterns of astroglial hypertrophy and neuronal degeneration in the hippocampus of ages, memory-deficient rats. J Gerontol. 1977;32(1):3–12. doi: 10.1093/geronj/32.1.3. [DOI] [PubMed] [Google Scholar]
- 16.Akiyama H, Arai T, Kondo H, Tanno E, Haga C, Ikeda K. Cell mediators of inflammation in the Alzheimer disease brain. Alzheimer Dis Assoc Disord. 2000;14(Suppl 1):S47–S53. doi: 10.1097/00002093-200000001-00008. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Nievas BG, Stein TD, Tai HC, Dols-Icardo O, Scotton TC, Barroeta-Espar I, et al. Dissecting phenotypic traits linked to human resilience to Alzheimer's pathology. Brain. 2013;136(Pt 8):2510–2526. doi: 10.1093/brain/awt171. Epub 2013 Jul 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci U S A. 1994;91(22):10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsacopoulos M, Magistretti PJ. Metabolic coupling between glia and neurons. J Neurosci. 1996;16(3):877–885. doi: 10.1523/JNEUROSCI.16-03-00877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen CJ, Massie A, De Keyser J. Immune players in the CNS: the astrocyte. J Neuroimmune Pharmacol. 2013;8(4):824–839. doi: 10.1007/s11481-013-9480-6. Epub 2013 Jul 4. [DOI] [PubMed] [Google Scholar]
- 21.Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119(1):7–35. doi: 10.1007/s00401-009-0619-8. Epub 2009 Dec 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nedergaard M, Ransom B, Goldman SA. New roles for astrocytes: redefining the functional architecture of the brain. Trends Neurosci. 2003;26(10):523–530. doi: 10.1016/j.tins.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Sherwood CC, Stimpson CD, Raghanti MA, Wildman DE, Uddin M, Grossman LI, et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci U S A. 2006;103(37):13606–13611. doi: 10.1073/pnas.0605843103. Epub 2006 Aug 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson JE, Ince PG, Shaw PJ, Heath PR, Raman R, Garwood CJ, et al. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer's pathology and APOE genotype. Neurobiol Aging. 2011;32(10):1795–1807. doi: 10.1016/j.neurobiolaging.2011.04.013. Epub Jun 25. [DOI] [PubMed] [Google Scholar]
- 26.Yasui DH, Xu H, Dunaway KW, Lasalle JM, Jin LW, Maezawa I. MeCP2 modulates gene expression pathways in astrocytes. Mol Autism. 2013;4(1):3. doi: 10.1186/2040-2392-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasternack JM, Abraham CR, Van Dyke BJ, Potter H, Younkin SG. Astrocytes in Alzheimer's disease gray matter express alpha 1-antichymotrypsin mRNA. Am J Pathol. 1989;135(5):827–834. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, O'Connor T, Vassar R. The contribution of activated astrocytes to Abeta production: implications for Alzheimer's disease pathogenesis. J Neuroinflammation. 2011;8:150. doi: 10.1186/1742-2094-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orre M, Kamphuis W, Osborn LM, Melief J, Kooijman L, Huitinga I, et al. Acute isolation and transcriptome characterization of cortical astrocytes and microglia from young and aged mice. Neurobiol Aging. 2014;35(1):1–14. doi: 10.1016/j.neurobiolaging.2013.07.008. Epub Aug 15. [DOI] [PubMed] [Google Scholar]
- 31.Roelofs RF, Fischer DF, Houtman SH, Sluijs JA, Van Haren W, Van Leeuwen FW, et al. Adult human subventricular, subgranular, and subpial zones contain astrocytes with a specialized intermediate filament cytoskeleton. Glia. 2005;52(4):289–300. doi: 10.1002/glia.20243. [DOI] [PubMed] [Google Scholar]
- 32.Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32(12):638–647. doi: 10.1016/j.tins.2009.08.002. Epub Sep 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969–2000) Neurochem Res. 2000;25(9–10):1439–1451. doi: 10.1023/a:1007677003387. [DOI] [PubMed] [Google Scholar]
- 34.Pekny M, Leveen P, Pekna M, Eliasson C, Berthold CH, Westermark B, et al. Mice lacking glial fibrillary acidic protein display astrocytes devoid of intermediate filaments but develop and reproduce normally. Embo J. 1995;14(8):1590–1598. doi: 10.1002/j.1460-2075.1995.tb07147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pekny M, Pekna M. Astrocyte intermediate filaments in CNS pathologies and regeneration. J Pathol. 2004;204(4):428–437. doi: 10.1002/path.1645. [DOI] [PubMed] [Google Scholar]
- 36.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 37.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 38.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. Epub 2009 Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. Epub Oct 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103(46):17402–17407. doi: 10.1073/pnas.0608396103. Epub 2006 Nov 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. Epub 2005 Aug 12. [DOI] [PubMed] [Google Scholar]
- 43.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. Epub 2003 Apr 3. [PubMed] [Google Scholar]
- 44.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeung KT, Das S, Zhang J, Lomniczi A, Ojeda SR, Xu CF, et al. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Mol Cell Biol. 2011;31(11):2287–2298. doi: 10.1128/MCB.01381-10. Epub 2011 Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghezzi D, Saada A, D'Adamo P, Fernandez-Vizarra E, Gasparini P, Tiranti V, et al. FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am J Hum Genet. 2008;83(3):415–423. doi: 10.1016/j.ajhg.2008.08.009. Epub Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chujo T, Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. Rna. 2012;18(12):2269–2276. doi: 10.1261/rna.035600.112. Epub 2012 Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chow KM, Gakh O, Payne IC, Juliano MA, Juliano L, Isaya G, et al. Mammalian pitrilysin: substrate specificity and mitochondrial targeting. Biochemistry. 2009;48(13):2868–2877. doi: 10.1021/bi8016125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, Johnson KA, et al. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J Biol Chem. 2006;281(39):29096–29104. doi: 10.1074/jbc.M602532200. Epub 2006 Jul 18. [DOI] [PubMed] [Google Scholar]
- 50.Alikhani N, Guo L, Yan S, Du H, Pinho CM, Chen JX, et al. Decreased proteolytic activity of the mitochondrial amyloid-beta degrading enzyme, PreP peptidasome, in Alzheimer's disease brain mitochondria. J Alzheimers Dis. 2011;27(1):75–87. doi: 10.3233/JAD-2011-101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tello D, Balsa E, Acosta-Iborra B, Fuertes-Yebra E, Elorza A, Ordonez A, et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting Complex I activity. Cell Metab. 2011;14(6):768–779. doi: 10.1016/j.cmet.2011.10.008. Epub Nov 17. [DOI] [PubMed] [Google Scholar]
- 52.Poliseno L. Pseudogenes: newly discovered players in human cancer. Sci Signal. 2012;5(242):re5. doi: 10.1126/scisignal.2002858. [DOI] [PubMed] [Google Scholar]
- 53.Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12(6):885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. Epub 2009 Sep 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. Epub 2009 Sep 6. [DOI] [PubMed] [Google Scholar]
- 56.Mulder SD, Nielsen HM, Blankenstein MA, Eikelenboom P, Veerhuis R. Apolipoproteins E and J interfere with amyloid-beta uptake by primary human astrocytes and microglia in vitro. Glia. 2014;62(4):493–503. doi: 10.1002/glia.22619. Epub 2014 Jan 20. [DOI] [PubMed] [Google Scholar]
- 57.Reichwald J, Danner S, Wiederhold KH, Staufenbiel M. Expression of complement system components during aging and amyloid deposition in APP transgenic mice. J Neuroinflammation. 2009;6:35. doi: 10.1186/1742-2094-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuda S, Matsuda Y, D'Adamio L. CD74 interacts with APP and suppresses the production of Abeta. Mol Neurodegener. 2009;4:41. doi: 10.1186/1750-1326-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panter SS, McSwigan JD, Sheppard JR, Emory CR, Frey WH., 2nd Glial fibrillary acidic protein and Alzheimer's disease. Neurochem Res. 1985;10(12):1567–1576. doi: 10.1007/BF00988599. [DOI] [PubMed] [Google Scholar]
- 60.Loring JF, Wen X, Lee JM, Seilhamer J, Somogyi R. A gene expression profile of Alzheimer's disease. DNA Cell Biol. 2001;20(11):683–695. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- 61.Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer's disease. J Chem Neuroanat. 2011;42(2):118–126. doi: 10.1016/j.jchemneu.2011.06.007. Epub Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Ramsey K, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol Genomics. 2008;33(2):240–256. doi: 10.1152/physiolgenomics.00242.2007. Epub 2008 Feb 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Ramsey K, et al. Neuronal gene expression in non-demented individuals with intermediate Alzheimer's Disease neuropathology. Neurobiol Aging. 2010;31(4):549–566. doi: 10.1016/j.neurobiolaging.2008.05.013. Epub Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.