Abstract
The budding yeast Saccharomyces cerevisiae has served as a remarkable model organism for numerous seminal discoveries in biology. This paradigm extends to the mitochondria, a central hub for cellular metabolism, where studies in yeast have helped reinvigorate the field and launch an exciting new era in mitochondrial biology. Here, we discuss few recent examples in which yeast research has laid a foundation for our understanding of evolutionarily conserved mitochondrial processes and functions; from the key factors and pathways involved in the assembly of the OXPHOS complexes, to metabolite transport, lipid metabolism, and interorganelle communication. We also highlight new areas of yeast mitochondrial biology that will likely aid in our understanding of mitochondrial etiology of disease in the years to come.
Keywords: Yeast, Mitochondria, oxidative phosphorylation, transporters, lipid metabolism, interorganelle communication
Yeast are a powerful resource for understanding mitochondrial biology
The yeast Saccharomyces cerevisiae is among the most powerful model organisms with which to approach an understanding of human biology. Although the examples are numerous, one area that has become particularly prominent recently is in our understanding of mitochondrial function. Mitochondria are well known as the powerhouse of the cell, providing the majority of ATP synthesis. These organelles have a much broader reach, however, with impact on metabolite synthesis and catabolism, intracellular signaling and organellar communication. As a result, impaired mitochondrial function is associated with a staggering variety of chronic human diseases [1].
While human genetics and biochemical analyses of mammalian mitochondria have been valuable tools in our efforts to understand the basic biology of mitochondria and the mitochondrial etiology of disease, the ability of S. cerevisiae to model mitochondrial biology and disease has arguably been even more powerful [2]. Most mitochondrial processes are conserved across eukaryotes, and researchers have taken full advantage of the power of yeast genetics to dissect a number of these pathways over the last several decades. These studies have been greatly enhanced by the recent development of many yeast genome-wide collections, including deletions of nonessential genes [3], collections containing titratable alleles of essential genes [4], sequence verified plasmid libraries [5, 6], and collections of epitope-tagged open reading frames [7]. The application of these resources, including in powerful high throughput genetic interaction studies, called epistasis miniarray profiles (E-MAPS) [8], has helped assign function to many uncharacterized genes involved in a number of mitochondrial pathways. Many conserved mitochondrial processes have been characterized in yeast, and extensively reviewed elsewhere, including, mitochondrial import [9], mitochondrial dynamics [10] and quality control [11], and mitochondrial-derived signaling [12].
Because the instances in which yeast have aided in the characterization of mitochondrial pathways are numerous, this review focuses on only a few recent examples where inquiries in the fundamental processes of yeast mitochondria have taught us how mitochondria work in higher organisms. The areas of focus include assembly of the oxidative phosphorylation machinery, mitochondrial metabolite transport and lipid metabolism, and inter-organelle communication pathways involving the mitochondria. Although these areas are quite diverse, crosstalk between these pathways is quite prominent, and ultimately critical for mitochondrial function as illustrated in Figure 1. As we hope will become obvious, dissecting these pathways in yeast has had great impact on our understanding of human diseases, including discovery of many causal genes.
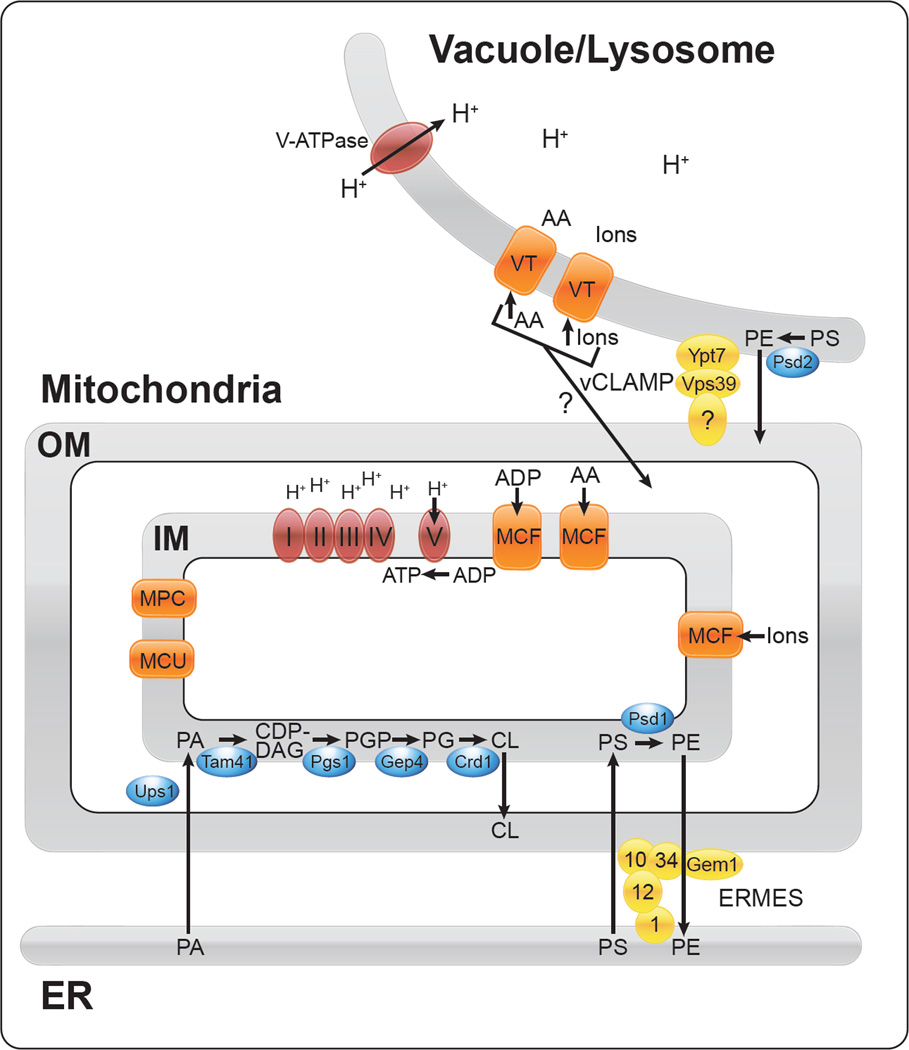
Figure 1.
An overview of the four areas of mitochondrial biology highlighted in this review: Assembly of the mitochondrial OXPHOS complexes (I–V) located in the inner mitochondrial membrane (red); import of metabolites into the mitochondrial matrix through transporters belonging to the mitochondrial carrier family (MCF), as well as additional transporters of interest to human disease (MPC, mitochondrial pyruvate carrier, and MCU, mitochondrial calcium uniporter) (orange); interorganelle lipid transport between the ER and mitochondria for synthesis of cardiolipin (CL) and phosphatidylethanolamine (PE) in the inner mitochondrial membrane (lipid enzymes in blue, organelle tethering complex ERMES in yellow); the physical and functional interaction between the vacuole/lysosome and mitochondria. The vCLAMP complex (yellow) tethers vacuole and mitochondria, and may be involved in exchange of metabolites between these organelles (amino acids, AA, ions, and PE). VT stands for vacuolar transporter.
Assembly of the mitochondrial oxidative phosphorylation complexes
Mitochondria are well known as the powerhouse of the cell, capturing energy in the form of ATP through oxidative phosphorylation (OXPHOS, see glossary), which couples electron transport in the inner mitochondrial membrane to the production of ATP. The five OXPHOS complexes include four large multi-protein complexes (complexes I – IV) that use the electrons from NADH and succinate to pump protons and establish the electrochemical potential across the mitochondrial inner membrane. The last complex, Complex V, harvests the energy of protons crossing back into the matrix for ATP production. These complexes contain between 4 and 46 different protein subunits, many of which are embedded in the membrane. Except Complex II, each complex contains proteins that are translated in both the mitochondrial matrix and the cytosol. Each of the complexes that transfer electrons (I–IV) also host a variety of redox active cofactors (flavin-adenine dinucleotide, heme, iron-sulfur clusters, etc.), which have the inherent propensity to generate reactive oxygen species if not handled properly. These factors combine to make OXPHOS complex assembly a very significant challenge for the cell [13, 14].
One of the evidences of the challenge presented by this assembly problem is the number of proteins that are specifically dedicated to ensuring that it happens appropriately. The vast majority of these assembly factors were discovered and/or functionally characterized in yeast. The assembly of Complex IV (cytochrome c oxidase) is perhaps the most comprehensively studied of any of the complexes, based largely on the power of yeast genetics. As this has been reviewed quite extensively elsewhere [15–18], it will not be addressed in this review. On the other hand, a molecular understanding of the assembly process of Complex I, has lagged behind for a variety of reasons, including that S. cerevisiae do not contain Complex I. Instead, other species have been used to study this assembly process, and this has resulted in the identification of a number of assembly factors and the beginning of an understanding of their mechanisms of action [19]. However, yeast has been instrumental in our understanding of the assembly of Complexes II and V.
Complex II
The last five years have witnessed a flurry of discoveries related to the assembly of Complex II, also called Succinate dehydrogenase complex (SDH), which to that point had largely been enigmatic. It is the simplest of the five OXPHOS complexes, containing only four proteins, all of which are encoded by the nuclear genome and translated in the cytosol. This relative simplicity might have led to the erroneous conclusion that Complex II assembles spontaneously, without the necessity for dedicated assembly factors. The discovery of four dedicated factors in the last few years has made it clear that even the simplest assembly problem in the electron transport chain is still quite complex [20]. Mutations in the genes encoding two of these four assembly factors of SDH, SDH5/SDHAF2 and SDHAF1, have been shown to cause human disease, either a familial tumor syndrome [21, 22] or a rapidly progressing infantile neurodegerative disease [21, 22], respectively. In one case, this factor was discovered using yeast genetics and biochemistry, while in the other, reconstitution studies in yeast demonstrated the role of the encoded protein in SDH assembly. Very recently, two additional factors have been discovered using yeast, both of which appear to have evolutionarily conserved functions in SDH assembly ([23, 24]; reviewed in [25]).
Complex V
Complex V or ATP synthase is a fascinating machine composed of two separable multi-protein sub-complexes, F0 and F1, and its structure, function and assembly has been recently reviewed [26]. F0 is embedded in the mitochondrial inner membrane and F1 is a soluble complex that attaches to the matrix face of F0. The relative rotation between F1 and F0 upon proton translocation provides the energy for ATP synthesis [27–29]. As with the other OXPHOS complexes, Complex V poses a complicated assembly problem [30], and cells use dedicated assembly factors to prevent mistakes. The first two Complex V assembly factors, Atp11 and Atp12, were discovered in a yeast genetic screen and later shown to be required for proper assembly of the F1 sector of ATP synthase [31–33]. These observations enabled the finding that human ATP11 and ATP12 also function in Complex V assembly [34] and that mutation of ATP12 causes mitochondrial disease in humans [35, 36]. A third factor, Fmc1, was discovered in yeast that appears to also be required for the assembly of F1, particularly at elevated temperature [37]. Two additional factors, Atp10 and Atp23, have been discovered that act on F0, both of which appear to be specifically required for the Atp6 protein to assembly into F0 normally [38–41]. Just recently, the discovery of the yeast INA complex containing Ina17 and Ina22, critical for the assembly of the F1F0 peripheral stalk, provided a new understanding of how the F0 and F1 sectors come together to form the intact ATP synthase [42]. Functional orthologs of these latter five factors are yet to be discovered in mammals.
Mitochondrial metabolite transport
Being the site of many chemical reactions of particular importance for cellular metabolism, including those in pathways with steps both inside and outside of the mitochondrial matrix, the transport of metabolites in and out of mitochondria is a critical feature of the physiology of this organelle. In general, the outer membrane does not provide a barrier to the transit of metabolites, although it does to most macromolecules. As a result, the regulated transport of metabolites across the mitochondrial inner membrane is of great importance and crucial in maintaining cellular bioenergetics and metabolic homeostasis. A number of human diseases are caused by defects in mitochondrial inner membrane transport. Although such diseases are rare, they are typically very severe [43]. As with many human diseases, the rare, severe, monogenic diseases affecting mitochondrial metabolite transport are likely to be instructive regarding important, but more complex, roles in common polygenic diseases, and those with a strong environmental component.
In many ways, metabolite transporters pose a more challenging set of problems to those intending to study them than do the enzymes that act on those metabolites. The genetics of metabolite transport is frequently complicated by complete or partial redundancy as well as by bypass mechanisms that obviate the necessity for a specific transport step [44]. Moreover, transporters do not perform a chemical modification and, therefore, in vitro assays of native function are only possible with a membrane-associated transporter embedded in an intact membrane. As a result, classical biochemistry discovered most of the enzymes of the core metabolic pathways, but our understanding of the transporters that perform equally important functions in these pathways lagged behind.
The Mitochondrial Carrier Family
While some molecules can pass through the mitochondrial inner membrane by passive diffusion, the majority, including charged and polar molecules, requires protein-provided assistance. In most cases, this facilitated transport is carried out by members of a distinct protein family that appears almost completely dedicated to this function [45]. Members of this family, known as the Mitochondrial Carrier Family (MCF), are characterized by three repeated homologous sequences of roughly 100 residues, each containing two transmembrane domains [46]. Based on genome analysis by sequence conservation, it appears that most eukaryotic genomes encode 35–55 different members of the MCF family [47], with 35 members present in yeast [48, 49].
A limited number of MCF transporters were purified and studied prior to the genomic era, but the vast majority was first identified based on the presence of their encoding gene in the yeast genome. Of course, this identification came with no information as to their potential functions, particularly the substrate(s) that they might transport. The Palmieri and Walker laboratories, their colleagues and others undertook the ambitious task of identifying transport substrates for many of these uncharacterized proteins [50]. This very successful effort directly led to identifying functions for dozens of yeast MCF transporters, which has facilitated the identification of substrates for homologous mammalian proteins. We highlight a few of these transporters here.
Mitochondrial ADP/ATP carrier
The archetypal MCF member in yeast is Aac2, which is the most abundant yeast ADP/ATP carrier. These proteins catalyze the exchange of ATP produced in the matrix for cytosolic ADP, thus enabling continuous ATP production and export to the cytosol [51]. Mutations in the human AAC2 homolog, ANT1, have been shown to cause mitochondrial DNA depletion syndrome and myopathy [52, 53]. In addition, both gene and protein has been a valuable model for deriving principles regarding the structure and function of these proteins.
The mitochondrial dicarboxylate carrier
The mitochondrial dicarboxylate carrier (DIC) is important for the anapleurotic filling of the TCA cycle via the mitochondrial import of succinate and malate. The DIC was one of the first to be identified by Palmieri and colleagues using the powerful combination of biochemical reconstitution of purified protein and yeast genetics [54]. Both the yeast and bovine protein had been previously purified and studied biochemically, but the sequence of the protein and the encoding gene was not known [55]. Based on the yeast protein sequence, the rat gene was cloned and it was shown to have similar transport characteristics [56]. The dicarboxylate carrier has subsequently been shown to be important for glucose-stimulated insulin secretion in pancreatic β-cells [57].
Mitochondrial coenzyme A importer
Mitochondrial coenzyme A is required for the entry of carbon from pyruvate into the TCA cycle via synthesis of acetyl-coA by pyruvate dehydrogenase. The first hints of the genetic basis of the mitochondrial coenzyme A importer Leu5p came when the leu5Δ mutant, which is unable to synthesize leucine, was shown to fail to accumulate coenzyme A in the mitochondria [58]. Two mammalian homologs were subsequently shown to perform a similar or identical function, one of which is the human Graves disease protein [58], which most likely is misnamed and has no role in Graves disease, also called solute carrier family 25 member 16 (SLC25A16). The second is SLC25A42 and was very convincingly shown, using in vitro reconstitution and transport assays as well as genetic complementation in yeast, to also function in mitochondrial coenzyme A import [59]. Orthologs were recently characterized in plants [60]. To our knowledge, this transporter or defects in it have not yet been implicated in human disease, but the fundamental knowledge of the proteins that enable this critical transport step in metabolism and the genes that encode them is likely to become valuable in our understanding of human disease in the future.
The mitoferrin proteins
Iron has essential roles in many cellular compartments, including mitochondria, which is the site of iron-sulfur cluster biogenesis. The mitoferrin proteins, which enable mitochondrial iron transport, were first discovered and characterized in yeast due to a relationship with the yeast homolog of the gene mutated in Friedreich’s ataxia. They were shown to be involved in mitochondrial iron metabolism [61, 62], but their function was unknown until work in S. cerevisiae demonstrated that Mrs3/Mrs4 function as mitochondrial iron transporters [63, 64]. Mitoferrin is an example where the genes were known in other systems, including in zebrafish [65], prior to elucidation of function in any species. The power of yeast genetics and biochemistry enabled functional discovery that may have been very difficult in other more complex systems.
Non-MCF Transporters
While this systematic approach of determining transport substrates of MCF proteins has proven to be of great value, it obviously is unable to assist in the discovery of transporters that are not members of this family. Two such examples have been discovered in the last few years. The mitochondrial calcium uniporter (MCU) is predominantly responsible for mitochondrial calcium uptake in mammalian cells. Although the S. cerevisiae model system lacks the MCU transporters and did not aid in the discovery of the transporter, yeast proved very useful for the comparative phylogenetics approach that enabled the discovery of the members of the MCU complex [66–72].
Like the MCU, the transporter that enables pyruvate, the product of cytosolic glycolysis, to enter the mitochondria and fuel the TCA cycle had been observed and studied for decades before its recent molecular identification. Two groups simultaneously discovered a complex containing at least two mitochondrial pyruvate carrier proteins (MPC1 and MPC2) that is necessary for mitochondrial pyruvate uptake [73, 74]. Mutations in MPC1 were shown to cause a severe neuromuscular phenotype in humans [73]. Based on reconstitution experiments in bacteria, MPC1 and MPC2 appear to also be sufficient for pyruvate transport [74]. In this case, genetic and biochemical approaches using yeast provided the key data that enabled this discovery. Since this initial discovery, four other groups have confirmed the role of the mitochondrial pyruvate carrier in mitochondrial pyruvate import, with major potential implications for human metabolism and metabolic disease [74–79]. While the structure of the MPC complex is unknown, the determination of the structure of distantly related sugar transporters suggests that it might function as a MPC1/MPC2 heterodimer [80].
Studying gene mutations
In addition to the utility of the yeast model system for discovering protein function, it also provides a facile mechanism for testing the consequences of mutations found in the genes encoding human transporters. The principle of human gene complementation of yeast deletion mutants has been used repeatedly in the study of mitochondrial transporters [50], including the discovery that mutations in the MPC1 gene encoding one subunit of the mitochondrial pyruvate carrier are loss-of-function mutations [73], as described above. The yeast system was also used to assess the functionality of mutations in the gene encoding the aspartate-glutamate carrier, which are tightly associated with a specific form of urea cycle deficiency [81]. A final example of this is the study of mutations in the gene encoding the ornithine transporter SLC25A15, found in patients with hyperornithinemia-hyperammonemia-homocitrullinuria syndrome [82]. This transporter was first found and characterized in yeast [83, 84], with the human ortholog being discovered and characterized later based on homology [85]. This powerful capability of the yeast system in allele characterization will become increasingly valuable as genome-sequencing efforts are increased and as remaining mitochondrial transporters are functionally annotated.
Mitochondrial Lipid Metabolism
Much like metabolite transport and OXPHOS assembly, our understanding of mitochondrial lipid biology has relied heavily on yeast biology and genetics. Proper mitochondrial lipid composition is key for most mitochondrial processes to function efficiently. In fact, a number of genes encoding proteins involved in mitochondrial lipid synthesis pathways were first identified for secondary affects on other mitochondrial processes such as import [86] and fusion [87]. The lipid composition of the mitochondrial membrane is distinct from other cellular membranes. It is highly enriched in phosphatidylethanolamine (PE), phosphatidylcholine (PC), and cardiolipin (CL), and deficient in sterols and sphingolipids [88]. Mitochondria import lipids such as phosphatidylserine (PS) and phosphatidic acid (PA) as precursors for the synthesis of PE and CL within the mitochondria. Sequential enzymes that function in the synthesis of these lipids often reside in separate mitochondrial subcomparments or in another organelle entirely. Thus, intra- and inter-organelle transport of lipids is required for both CL and PE synthesis [89]. The enzymes and transport steps involved in these pathways are highly conserved from yeast to man, and we will highlight several recent examples in which the power of yeast has greatly enhanced our understanding of mitochondrial lipid metabolism.
Cardiolipin
CL is the hallmark lipid of the mitochondria, and the majority of the genes involved in this pathway were first discovered in yeast, including two enzymes and one lipid transport protein in just the last few years alone. CL is a unique phospholipid in that contains two phosphatidyl groups linked to a glycerol backbone. It is localized almost exclusively in mitochondria, and associates with proteins in mitochondrial import, OXPHOS, and mitochondrial dynamics [90]. In addition to a core requirement for CL in basic mitochondrial processes, proper remodeling of CL’s acyl chains in the outer mitochondrial membrane is also a necessity for normal mitochondrial health. The last step of CL remodeling is carried out by the conserved acyltransferase Tafazzin/Taz1 [91]. Mutations in the human TAZ gene impair acyl chain remodeling and lead to Barth syndrome, an X-linked disease characterized by cyclic neutropenia and skeletal and cardiac myopathies [92, 93]. While the function of the TAZ gene was first identified in humans, studies on the yeast TAZ1 have greatly accelerated research in this area, including providing potential pathogenic mechanisms for loss of function mutations in TAZ.
CL is synthesized in the mitochondria, and starts with the formation of CDP-diacylglycerol (CDP-DAG) from phosphatidic acid (PA). CDP-DAG is then converted to phosphatidylglycerolphosphate (PGP) by phosphatidylglycerophosphate synthase 1 (Pgs1) [94], followed by dephosphorylation of PGP to phosphatidylglycerol (PG). Finally, cardiolipin synthase (Crd1) converts PG into CL through reaction with another CDP-DAG [95, 96], followed by remodeling by Cld1 and Taz1. The genes encoding PGS1 and CRD1 were discovered in yeast in 1998 [94–96], and paved the way for the identification of the orthologous mammalian genes several years later [97–99]. Remarkably, despite the fact that CL was first isolated in 1945 [100], the genes required for formation of CDP-DAG from PA in the mitochondria, as well as dephosphorylation of PGP to PG were only identified very recently.
PA is synthesized in the ER, and is transported to the mitochondria for CL synthesis. It was initially thought that a small subset of CDP-diacylglycerol synthase (Cds1), an ER-localized enzyme that converts PA to CDP-DAG in the ER, must also be present in mitochondria for conversion of PA to CDP-DAG [101, 102]. However, Tamura et al. recently demonstrated that Cds1 is exclusively ER localized, and showed that the highly conserved mitochondrial protein Tam41 directly catalyzes the formation of CDP-DAG from PA in the mitochondrial inner membrane [103]. Tam41 was originally isolated as a protein required for the activity of the Tim23 inner membrane translocase [86], and was first implicated in CL biosynthesis in 2008 [104]. The human sequence homolog of TAM41 is TAMM41, and its role in CL synthesis has yet to be confirmed.
Similar to the identification of Tam41, the yeast phosphatase GEP4, which is responsible for conversion of PGP to PG, was only identified recently by yeast genetics. Osman et al. first identified GEP4 in a genome-wide genetic array for genes required in the absence of mitochondrial prohibitins [105]. A number of CL-associated genes were present in the interaction set with prohibitins, and this subsequently helped the authors pin down Gep4 as the long sought after PGP phosphatase [106]. Subsequently, an unrelated phosphatase, protein tyrosine phosphatase, mitochondrial 1 (PTPMT1), was shown to catalyze the same reaction as GEP4 in mice [107].
Inter-and intraorganelle transport
In addition to the enzymatic steps of CL synthesis, yeast has also proven to be a resourceful system to understand both intra- and inter-organelle transport of lipids during the CL and PE synthesis pathways. The synthesis of CL starts from PA, which is generated in the ER and must make its way to the inner mitochondrial membrane for the first step of CL biosynthesis. Likewise, PE is generated from PS in the inner mitochondrial membrane by the conserved enzyme Psd1, and PS also must move from the ER to mitochondria [89]. The transfer of PA and PS to the mitochondria is predicted to occur at well-characterized contact sites between the ER and mitochondria called mitochondria-associated membranes (MAMs), which have been extensively reviewed elsewhere [108].
A recent study in yeast uncovered the first protein complex potentially involved in lipid transport at these contact sites [109]. The ER-mitochondrion encounter structure (ERMES) comprises four core protein subunits [maintenance of mitochondrial morphology (Mmm) protein 1; mitochondrial distribution and morphology (Mdm) protein 10, 12 and 34; and the Gem1 GTPase] that together function to tether the ER and mitochondrial membranes [110]. In support of a role for ERMES in lipid transport, several members of the complex contain conserved synaptotagmin-like-mitochondrial-lipid binding protein (SMP) domains, which are potential lipid binding motifs that may play a role in lipid transfer [111]. Additionally, Kornmann et al. showed that cells lacking ERMES components exhibited slower rates of PS to PC conversion, which requires ER-mitochondria lipid transfer. However, mammals contain no obvious homologs of the ERMES complex, and a recent study in yeast suggested that ERMES has no impact on PS to PE conversion rates [112]. Thus, the exact role of ERMES in mitochondrial lipid biosynthesis requires further clarification, and it is likely that other undiscovered systems work in conjunction with ERMES to mediate ER-mitochondrial lipid transfer.
After PA is moved to the mitochondria outer membrane from the ER, it must be transported to the mitochondrial inner membrane for conversion to CDP-DAG in CL biosynthesis. An impressive collection of recent studies in yeast has identified the conserved protein Ups1/PRELI as a mediator of intra-mitochondrial PA transport. Ups1 was initially described as a protein required for processing of the mitochondrial fusion protein Mgm1 [87]. The role of Ups1 in Mgm1 processing was subsequently linked to its direct function in CL metabolism by two separate groups[105, 113]. Both Osman et al. and Tamura et al. showed that loss of Ups1 severely compromised CL biosynthesis [105, 113], and Langer’s group went on to elegantly demonstrate that Ups1 functions as the long-sought after PA transfer protein, shuttling PA from the outer to the inner mitochondrial membrane in a CL-responsive manner [114]. The transfer activity of Ups1 also requires the conserved protein Mdm35 [115, 116], and Langer’s group has since demonstrated that the orthologs of Ups1 (PRELI) and Mdm35 (TRIAP1) function similarly in PA transport, in mammals [117].
Mitochondria-Vacuole Crosstalk
In classic textbook illustrations, mitochondria are depicted as lone-wolf organelles. This view of the mitochondria could not be further from reality, as it is now appreciated that they are part of an ever-expanding interconnected network of organelles within the cell. Recent studies have described both physical and functional interactions of mitochondria with a number of organelles, and these interactions are critical for mitochondrial function [118]. As with the other areas of mitochondrial biology we have discussed thus far, studies in yeast are leading the way in this area of mitochondrial biology as well.
Mitochondrial communication with other organelles
Many examples of mitochondrial communication with other organelles have been described in great detail elsewhere. The classic example is the yeast retrograde response, in which deficits in mitochondrial function are communicated to the nucleus through the action of the ReTroGrade regulation (RTG) transcription factors [119]. In this context, the functional status of the mitochondria is relayed to the cell’s transcriptional machinery, which in turn regulates the cellular metabolic state. Since the discovery of the retrograde response in yeast, similar retrograde pathways have also been described in other organisms which have been reviewed elsewhere [120]. As we discussed in the lipid metabolism section, mitochondria also form physical and functional contacts with the ER that are important for lipid metabolism, calcium homeostasis, and mitochondrial dynamics [121]. More recent studies in yeast have identified a physical tether between the mitochondria and plasma membrane [122], and outlined a close association between mitochondria and peroxisomes [123]. The mitochondria and peroxisome also share the same fission machinery [124], and recent studies in mammals have outlined a vesicle-mediated pathway for delivery of mitochondrial proteins to the peroxisomes [125].
The vacuole-mitochondria relationship
An emerging topic in the arena of inter-organelle crosstalk is the tight physical and functional association of mitochondria with the yeast lysosome-like vacuole. The vacuole is similar to the mammalian lysosome, and it is becoming increasingly clear that this organelle plays key role in cellular metabolism. The presence of a functional metabolic link between the mitochondria and vacuole is nothing new. During the initial characterization of genes important for vacuole function in the late 1980s and early 90s, it was discovered that loss of function mutations in the Vacuolar H+-ATPase (V-ATPase) prevents growth of yeast on nonfermentable carbon sources [126]. The V-ATPase is an evolutionarily conserved protein complex that pumps protons into the lumen of the yeast vacuole and mammalian lysosome, acidifying these organelles and generating a proton gradient that is used for uptake and storage of metabolites within the vacuole [127]. Several more recent studies have confirmed the requirement for vacuole function in mitochondrial respiratory metabolism, and the collection of V-ATPase protein-coding genes has been described as the largest class of non-mitochondrial localized proteins required for mitochondrial function [128].
Despite the longstanding functional interaction between the mitochondria and vacuole, the underlying mechanism for this connection is still unclear. Vacuolar acidity is required for two main vacuole functions: degradation of proteins and metabolites within the vacuole lumen, and storage of ions and amino acids [127]. It has been proposed that loss of V-ATPase activity compromises mitochondrial function through three potential avenues: 1) Failure to properly turnover mitochondrial proteins through autophagy; 2) increased production of oxidative stress; 3) and overload from failed storage of ions and metabolites [128]. Consistent with the second hypothesis, loss of V-ATPase function leads to a dramatic increase in oxidative stress, potentially through disruption of normal iron metabolism [129, 130]. An early study on V-ATPase function also demonstrated rescue of mitochondrial deficiencies in V-ATPase mutants with the addition of excess iron, supporting the idea that iron metabolism may play a role in vacuole-mitochondria crosstalk [131]. A more recent study examining mitochondrial function in the context of yeast aging, addressed the role of protein degradation and metabolite storage in vacuole-mitochondria crosstalk. In this study, Hughes and Gottschling showed that age-induced mitochondrial dysfunction is driven by loss of vacuolar acidity, and provided evidence against a role for protein degradation in this connection [132]. Instead, the authors demonstrated that the vacuole-mitochondria relationship is likely governed by compromised vacuolar storage of neutral amino acids in the vacuole lumen. It remains to be determined how imbalances in the cellular distribution of neutral amino acids impacts mitochondrial function, and whether this same metabolic connection exists in mammals as well. Vacuole-mitochondria crosstalk is clearly a complex process, and additional studies are required to separate out the role of oxidative stress and amino acids in this process.
A very recent and exciting development in the mitochondria-vacuole relationship was the discovery of a physical association between the vacuole and mitochondria mediated by a tether called vCLAMP, for vacuole and mitochondria patch [133, 134]. The discovery of vCLAMP was independently reported by the Ungermann and Schuldiner groups each using a separate approach. Ungermann and colleagues used a candidate gene approach to find genes that enhanced contact sites between vacuoles and mitochondria, upon overexpression, while the Schuldiner group identified vCLAMP in a microscopy-based screen looking for genes that impacted the amount of ERMES contacts per cell. Remarkably, both approaches led to the same gene, Vacuolar Protein Sorting 39 Homolog (VPS39), a well-characterized subunit of vacuole homotypic fusion and vacuole protein sorting (HOPS) machinery. The authors showed that Vps39, in conjunction with the Rab GTPase Ypt7, mediates tethering of vacuoles to mitochondria. How the two proteins associate with mitochondria is currently unclear.
Much like the ERMES complex at the ER-mitochondria interface, a comprehensive understanding of the function of vCLAMP remains to be worked out. vCLAMP shares some functional overlap with the ERMES complex, and it appears these two complexes are regulated in an interconnected manner [133, 134]. Elbaz-Alon et al. show that vCLAMP is important for lipid exchange between the vacuole and mitochondria, and that vacuole-mitochondria lipid exchange likely compensates for defects in ER-mitochondria lipid exchange in ERMES mutants, which may provide an explanation for the apparent discrepancy reported for the role of ERMES in lipid transport discussed earlier. Excitingly, Elbaz-Alon et al. also find that metabolite transporters are potentially enriched near vacuole-mitochondria contact sites [133, 134]. Thus, it is easy to envision a role for vCLAMP beyond lipid metabolism, and it will be interesting to see whether vCLAMP connections facilitate the amino acid-dependent vacuole-mitochondria crosstalk described by Hughes and Gottschling [132]. Unlike the other sections of this review, vacuolemitochondria communication has yet to be firmly established in mammals. However, we are highlighting this connection as an exciting area of recent discovery in yeast that we believe will ultimately bear fruit in the mammalian system as well. Homologs of Vps39 are apparent in mammals [135], and it has already been demonstrated that mouse mitochondria are physically connected to lysosome-related organelles of pigment cells [136].
Concluding remarks and future perspectives
The four areas that we have highlighted provide a small glimpse of the impact that fundamental studies in yeast have had on our understanding of mitochondrial biology. That impact has extended far beyond the knowledge of protein function and detailed mechanisms, to include many examples where the discovery of human disease genes was directly enabled by observations made in yeast. Many outstanding questions remain in the field of mitochondrial biology, and we have highlighted some of those that pertain to the four topics we discussed here (See Outstanding Questions Box). As we enter an era where the demand for applicability to human health and disease is being made of scientific inquiry like never before, we must not forget that frequently the most efficient way to understand the biology of the complex human is to interrogate fundamental mechanisms in yeast.
Box 1: Outstanding Questions.
What additional factors are important for OXPHOS assembly?
What are the causal genes in the remaining SDH assembly-related disease states without an assigned gene?
What are the substrates of the orphan mitochondrial carrier proteins?
Is there a high order structural organization of transporters and their related enzymes within the mitochondrial interior?
How are lipids distributed to the correct location within the mitochondria?
What is the mechanism of lipid transport by the ERMES and vCLAMP complexes?
What proteins play the role of ERMES at ER-mitochondrial contact sites in mammals?
Are there additional lipid transport proteins at ER-mitochondrial contact sites?
How does loss of vacuole/lysosome acidity compromise mitochondrial function?
Is the vCLAMP involved in amino acid related mitochondria-vacuole crosstalk?
What metabolites are exchanged between the mitochondrial and vacuole, and what is the role of vCLAMP in this process?
How is vCLAMP attached to the mitochondrial outer membrane?
Highlights.
Studies in yeast have identified key players in mitochondrial OXPHOS assembly
Yeast were instrumental for identifying mitochondrial metabolite transporters
Numerous players in mitochondrial lipid metabolism were first identified in yeast
Mitochondrial-vacuole crosstalk is an emerging area of mitochondrial biology
Acknowledgments
J. Rutter is supported by NIH grants GM094232, GM110755, and GM112057. A. Hughes is supported by NIH grant AG043095.
Glossary
- Cardiolipin
is a diphosphatidylglycerol lipid and an important component of the inner mitochondrial membrane, where it constitutes about 20% of the total lipid composition. Cardiolipin has multiple functions, from regulating structure, to serving as proton trap for oxidative phosphorylation, to apoptosis.
- Epistasis Miniarray Profiles (E-MAPs)
are genetic epistasis studies performed on a large-scale that are often used to assign function to uncharacterized genes.
- ER-mitochondrial encounter structure (ERMES)
is a protein complex composed of four core components (Mmm1, Mdm10, Mdm34, Mdm12) and one associated protein (Gem1) that physically tethers the ER and mitochondrial membranes at ER-mitochondria contact sites.
- Mitochondria-associated membranes (MAMs)
are reversibly tethered to mitochondria and part of the endoplasmic reticulum. They allow the import of some lipids into mitochondria and are involved in calcium homeostasis, mitochondrial function, autophagy and apoptosis.
- Mitochondrial calcium uniporter (MCU)
The multi-protein complex that enables the mitochondrial import of Ca2+ ions across the mitochondrial inner membrane into the matrix.
- Mitochondrial Carrier Family (MCF)
The family of transmembrane transporters that facilitate most of the solute transport across the mitochondrial inner membrane.
- Mitochondrial pyruvate carrier (MPC)
The protein complex that facilitates transport of pyruvate across the mitochondrial inner membrane. While known and characterized since the 1960s, the identity of the proteins in this complex was only discovered in 2012.
- Mitochondrial calcium uniporter (MCU)
The multi-protein complex that enables the mitochondrial import of Ca2+ ions across the mitochondrial inner membrane into the matrix.
- Mitoferrin
A member of the mitochondrial carrier family that is responsible for transport of iron ion across the mitochondrial inner membrane.
- Oxidative phosphorylation (OXPHOS)
a mitochondrial metabolic pathway that uses energy released by the oxidation of nutrients to produce (ATP). During oxidative phosphorylation, electrons are transferred from electron donors to electron acceptors such as oxygen, in redox reactions that eventually synthesize ATP.
- Phosphatidic acid (PA)
is a vital cell lipid that acts as a biosynthetic precursor for the formation of all acylglycerol lipids in the cell.
- Retrograde Pathway
is a signaling pathway that originates from dysfunctional mitochondria and ultimately activates a transcriptional program mediated by the transcription factors Rtg1 and Rtg3. The purpose of the pathway is to help replenish TCA cycle intermediates in low functioning mitochondria.
- Succinate dehydrogenase (SDH or Complex II)
A tetrameric complex in the mitochondrial inner membrane that catalyzes the oxidation of succinate to fumarate and reduction of quinone as an entry point to the electron transport chain as part of oxidative phosphorylation.
- Vacuole and mitochondria patch (vCLAMP)
is a protein complex consisting of subunits Vps39 and Ypt7 that functionally tethers the vacuole and mitochondria, and is thought to play a role in lipid and metabolite exchange between the organelles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baile MG, Claypool SM. The power of yeast to model diseases of the powerhouse of the cell. Front Biosci (Landmark Ed) 2013;18:241–278. doi: 10.2741/4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giaever G, Nislow C. The yeast deletion collection: a decade of functional genomics. Genetics. 2014;197(2):451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118(1):31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Jones GM, et al. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat Methods. 2008;5(3):239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17(4):536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 8.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 9.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta. 2013;1833(2):274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15(10):634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer F, Hamann A, Osiewacz HD. Mitochondrial quality control: an integrated network of pathways. Trends Biochem Sci. 2012;37(7):284–292. doi: 10.1016/j.tibs.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Chandel NS. Mitochondria as signaling organelles. BMC Biol. 2014;12:34. doi: 10.1186/1741-7007-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontanesi F, et al. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am J Physiol Cell Physiol. 2006;291(6):C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- 14.Ghezzi D, Zeviani M. Assembly factors of human mitochondrial respiratory chain complexes: physiology and pathophysiology. Adv Exp Med Biol. 2012;748:65–106. doi: 10.1007/978-1-4614-3573-0_4. [DOI] [PubMed] [Google Scholar]
- 15.DiMauro S, Tanji K, Schon EA. The many clinical faces of cytochrome c oxidase deficiency. Adv Exp Med Biol. 2012;748:341–357. doi: 10.1007/978-1-4614-3573-0_14. [DOI] [PubMed] [Google Scholar]
- 16.Fontanesi F, Soto IC, Barrientos A. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life. 2008;60(9):557–568. doi: 10.1002/iub.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalimonchuk O, Winge DR. Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim Biophys Acta. 2008;1783(4):618–628. doi: 10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soto IC, et al. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta. 2012;1817(6):883–897. doi: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mimaki M, et al. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817(6):851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion. 2010;10(4):393–401. doi: 10.1016/j.mito.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao HX, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325(5944):1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghezzi D, et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet. 2009;41(6):654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 23.Na U, et al. The LYR Factors SDHAF1 and SDHAF3 Mediate Maturation of the Iron- Sulfur Subunit of Succinate Dehydrogenase. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Vranken JG, et al. SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.05.012. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Vranken JG, et al. Protein-mediated assembly of succinate dehydrogenase and its cofactors. Crit Rev Biochem Mol Biol. 2014 doi: 10.3109/10409238.2014.990556. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonckheere AI, Smeitink JA, Rodenburg RJ. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis. 2012;35(2):211–225. doi: 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox GB, et al. Hypothesis. The mechanism of ATP synthase. Conformational change by rotation of the beta-subunit. Biochim Biophys Acta. 1984;768(3–4):201–208. doi: 10.1016/0304-4173(84)90016-8. [DOI] [PubMed] [Google Scholar]
- 28.Boyer PD. The ATP synthase--a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 29.Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286(5445):1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 30.Rak M, Gokova S, Tzagoloff A. Modular assembly of yeast mitochondrial ATP synthase. Embo j. 2011;30(5):920–930. doi: 10.1038/emboj.2010.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackerman SH. Atp11p and Atp12p are chaperones for F(1)-ATPase biogenesis in mitochondria. Biochim Biophys Acta. 2002;1555(1–3):101–105. doi: 10.1016/s0005-2728(02)00262-1. [DOI] [PubMed] [Google Scholar]
- 32.Ackerman SH, Tzagoloff A. Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc Natl Acad Sci U S A. 1990;87(13):4986–4990. doi: 10.1073/pnas.87.13.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman S, et al. Characterization of ATP12, a yeast nuclear gene required for the assembly of the mitochondrial F1-ATPase. J Biol Chem. 1991;266(12):7517–7523. [PubMed] [Google Scholar]
- 34.Wang ZG, White PS, Ackerman SH. Atp11p and Atp12p are assembly factors for the F(1)-ATPase in human mitochondria. J Biol Chem. 2001;276(33):30773–30778. doi: 10.1074/jbc.M104133200. [DOI] [PubMed] [Google Scholar]
- 35.De Meirleir L, et al. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J Med Genet. 2004;41(2):120–124. doi: 10.1136/jmg.2003.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meulemans A, et al. Defining the pathogenesis of the human Atp12p W94R mutation using a Saccharomyces cerevisiae yeast model. J Biol Chem. 2010;285(6):4099–4109. doi: 10.1074/jbc.M109.046920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre-Legendre L, et al. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J Biol Chem. 2001;276(9):6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- 38.Osman C, et al. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol Biol Cell. 2007;18(2):627–635. doi: 10.1091/mbc.E06-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng X, Neupert W, Tzagoloff A. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol Biol Cell. 2007;18(2):617–626. doi: 10.1091/mbc.E06-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ackerman SH, Tzagoloff A. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem. 1990;265(17):9952–9959. [PubMed] [Google Scholar]
- 41.Tzagoloff A, et al. Atp10p assists assembly of Atp6p into the F0 unit of the yeast mitochondrial ATPase. J Biol Chem. 2004;279(19):19775–19780. doi: 10.1074/jbc.M401506200. [DOI] [PubMed] [Google Scholar]
- 42.Lytovchenko O, et al. The INA complex facilitates assembly of the peripheral stalk of the mitochondrial F1Fo-ATP synthase. Embo j. 2014;33(15):1624–1638. doi: 10.15252/embj.201488076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmieri F. Diseases caused by defects of mitochondrial carriers: a review. Biochim Biophys Acta. 2008;1777(7–8):564–578. doi: 10.1016/j.bbabio.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Palmieri F, Pierri CL. Mitochondrial metabolite transport. Essays Biochem. 2010;47:37–52. doi: 10.1042/bse0470037. (doi): p. 10.1042/bse0470037. [DOI] [PubMed] [Google Scholar]
- 45.Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med. 2013;34(2–3):465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Saraste M, Walker JE. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144(2):250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- 47.Kunji ER. The role and structure of mitochondrial carriers. FEBS Lett. 2004;564(3):239–244. doi: 10.1016/S0014-5793(04)00242-X. [DOI] [PubMed] [Google Scholar]
- 48.Palmieri L, et al. Yeast mitochondrial carriers: bacterial expression, biochemical identification and metabolic significance. J Bioenerg Biomembr. 2000;32(1):67–77. doi: 10.1023/a:1005564429242. [DOI] [PubMed] [Google Scholar]
- 49.el Moualij B, et al. Phylogenetic classification of the mitochondrial carrier family of Saccharomyces cerevisiae. Yeast. 1997;13(6):573–581. doi: 10.1002/(SICI)1097-0061(199705)13:6<573::AID-YEA107>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 50.Palmieri F, et al. Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta. 2006;1757(9–10):1249–1262. doi: 10.1016/j.bbabio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 51.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta. 2008;1778(10):1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 52.Palmieri L, et al. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum Mol Genet. 2005;14(20):3079–3088. doi: 10.1093/hmg/ddi341. [DOI] [PubMed] [Google Scholar]
- 53.Echaniz-Laguna A, et al. Complete loss of expression of the ANT1 gene causing cardiomyopathy and myopathy. J Med Genet. 2012;49(2):146–150. doi: 10.1136/jmedgenet-2011-100504. [DOI] [PubMed] [Google Scholar]
- 54.Palmieri L, et al. The mitochondrial dicarboxylate carrier is essential for the growth of Saccharomyces cerevisiae on ethanol or acetate as the sole carbon source. Mol Microbiol. 1999;31(2):569–577. doi: 10.1046/j.1365-2958.1999.01197.x. [DOI] [PubMed] [Google Scholar]
- 55.Bisaccia F, Indiveri C, Palmieri F. Purification and reconstitution of two anion carriers from rat liver mitochondria: the dicarboxylate and the 2-oxoglutarate carrier. Biochim Biophys Acta. 1988;933(2):229–240. doi: 10.1016/0005-2728(88)90030-8. [DOI] [PubMed] [Google Scholar]
- 56.Fiermonte G, et al. The Sequence, Bacterial Expression, and Functional Reconstitution of the Rat Mitochondrial Dicarboxylate Transporter Cloned via Distant Homologs in Yeast and Caenorhabditis elegans. Journal of Biological Chemistry. 1998;273(38):24754–24759. doi: 10.1074/jbc.273.38.24754. [DOI] [PubMed] [Google Scholar]
- 57.Huypens P, et al. The dicarboxylate carrier plays a role in mitochondrial malate transport and in the regulation of glucose-stimulated insulin secretion from rat pancreatic beta cells. Diabetologia. 2011;54(1):135–145. doi: 10.1007/s00125-010-1923-5. [DOI] [PubMed] [Google Scholar]
- 58.Prohl C, et al. The yeast mitochondrial carrier Leu5p and its human homologue Graves' disease protein are required for accumulation of coenzyme A in the matrix. Mol Cell Biol. 2001;21(4):1089–1097. doi: 10.1128/MCB.21.4.1089-1097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiermonte G, et al. A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3',5'-diphosphate in human mitochondria. J Biol Chem. 2009;284(27):18152–18159. doi: 10.1074/jbc.M109.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zallot R, et al. Identification of mitochondrial coenzyme a transporters from maize and Arabidopsis. Plant Physiol. 2013;162(2):581–588. doi: 10.1104/pp.113.218081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foury F, Roganti T. Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J Biol Chem. 2002;277(27):24475–24483. doi: 10.1074/jbc.M111789200. [DOI] [PubMed] [Google Scholar]
- 62.Muhlenhoff U, et al. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J Biol Chem. 2003;278(42):40612–40620. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- 63.Froschauer EM, Schweyen RJ, Wiesenberger G. The yeast mitochondrial carrier proteins Mrs3p/Mrs4p mediate iron transport across the inner mitochondrial membrane. Biochim Biophys Acta. 2009;1788(5):1044–1050. doi: 10.1016/j.bbamem.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, et al. Mrs3p, Mrs4p, and frataxin provide iron for Fe-S cluster synthesis in mitochondria. J Biol Chem. 2006;281(32):22493–22502. doi: 10.1074/jbc.M604246200. [DOI] [PubMed] [Google Scholar]
- 65.Shaw GC, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440(7080):96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 66.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476(7360):341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336(6083):886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaudhuri D, et al. MCU encodes the pore conducting mitochondrial calcium currents. Elife. 2013;2:e00704. doi: 10.7554/eLife.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Stefani D, et al. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476(7360):336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mallilankaraman K, et al. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat Cell Biol. 2012;14(12):1336–1343. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patron M, et al. The mitochondrial calcium uniporter (MCU): molecular identity and physiological roles. J Biol Chem. 2013;288(15):10750–10758. doi: 10.1074/jbc.R112.420752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sancak Y, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342(6164):1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bricker DK, et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337(6090):96–100. doi: 10.1126/science.1218099. Epub 2012 May 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herzig S, et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337(6090):93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 75.Colca JR, et al. Identification of a mitochondrial target of thiazolidinedione insulin sensitizers (mTOT)--relationship to newly identified mitochondrial pyruvate carrier proteins. PLoS One. 2013;8(5):e61551. doi: 10.1371/journal.pone.0061551. Print 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Divakaruni AS, et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proc Natl Acad Sci U S A. 2013;110(14):5422–5427. doi: 10.1073/pnas.1303360110. Epub 2013 Mar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patterson JN, et al. Mitochondrial metabolism of pyruvate is essential for regulating glucose-stimulated insulin secretion. J Biol Chem. 2014 doi: 10.1074/jbc.M113.521666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rohatgi N, et al. Novel Insulin Sensitizer Modulates Nutrient Sensing Pathways and Maintains beta-Cell Phenotype in Human Islets. PLoS One. 2013;8(5):e62012. doi: 10.1371/journal.pone.0062012. Print 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Timon-Gomez A, Proft M, Pascual-Ahuir A. Differential regulation of mitochondrial pyruvate carrier genes modulates respiratory capacity and stress tolerance in yeast. PLoS One. 2013;8(11):e79405. doi: 10.1371/journal.pone.0079405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Y, et al. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature. 2014;515(7527):448–452. doi: 10.1038/nature13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wongkittichote P, et al. Prediction of the functional effect of novel SLC25A13 variants using a S. cerevisiae model of AGC2 deficiency. J Inherit Metab Dis. 2013;36(5):821–830. doi: 10.1007/s10545-012-9543-5. [DOI] [PubMed] [Google Scholar]
- 82.Ersoy Tunali N, et al. A novel mutation in the SLC25A15 gene in a Turkish patient with HHH syndrome: functional analysis of the mutant protein. Mol Genet Metab. 2014;112(1):25–29. doi: 10.1016/j.ymgme.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crabeel M, et al. The ARG11 gene of Saccharomyces cerevisiae encodes a mitochondrial integral membrane protein required for arginine biosynthesis. J Biol Chem. 1996;271(40):25011–25018. doi: 10.1074/jbc.271.40.25011. [DOI] [PubMed] [Google Scholar]
- 84.Palmieri L, et al. Identification of the yeast ARG-11 gene as a mitochondrial ornithine carrier involved in arginine biosynthesis. FEBS Lett. 1997;410(2–3):447–451. doi: 10.1016/s0014-5793(97)00630-3. [DOI] [PubMed] [Google Scholar]
- 85.Camacho JA, et al. Hyperornithinaemia-hyperammonaemia-homocitrullinuria syndrome is caused by mutations in a gene encoding a mitochondrial ornithine transporter. Nat Genet. 1999;22(2):151–158. doi: 10.1038/9658. [DOI] [PubMed] [Google Scholar]
- 86.Tamura Y, et al. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J Cell Biol. 2006;174(5):631–637. doi: 10.1083/jcb.200603087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sesaki H, et al. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. J Cell Biol. 2006;173(5):651–658. doi: 10.1083/jcb.200603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52(4):590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 89.Tamura Y, Sesaki H, Endo T. Phospholipid Transport via Mitochondria. Traffic. 2014;15(9):933–945. doi: 10.1111/tra.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Claypool SM, Koehler CM. The complexity of cardiolipin in health and disease. Trends Biochem Sci. 2012;37(1):32–41. doi: 10.1016/j.tibs.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu Z, et al. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol Microbiol. 2004;51(1):149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 92.Barth PG, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci. 1983;62(1–3):327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 93.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580(23):5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 94.Chang SC, et al. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J Biol Chem. 1998;273(16):9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 95.Chang SC, et al. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem. 1998;273(24):14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- 96.Tuller G, et al. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 1998;421(1):15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- 97.Chen D, Zhang XY, Shi Y. Identification and functional characterization of hCLS1, a human cardiolipin synthase localized in mitochondria. Biochem J. 2006;398(2):169–176. doi: 10.1042/BJ20060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Houtkooper RH, et al. Identification and characterization of human cardiolipin synthase. FEBS Lett. 2006;580(13):3059–3064. doi: 10.1016/j.febslet.2006.04.054. [DOI] [PubMed] [Google Scholar]
- 99.Kawasaki K, et al. Isolation of a chinese hamster ovary (CHO) cDNA encoding phosphatidylglycerophosphate (PGP) synthase, expression of which corrects the mitochondrial abnormalities of a PGP synthase-defective mutant of CHO-K1 cells. J Biol Chem. 1999;274(3):1828–1834. doi: 10.1074/jbc.274.3.1828. [DOI] [PubMed] [Google Scholar]
- 100.Pangborn MC. A simplified preparation of cardiolipin, with note on purification of lecithin for serologic use. J Biol Chem. 1945;161:71–82. [PubMed] [Google Scholar]
- 101.Kuchler K, Daum G, Paltauf F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J Bacteriol. 1986;165(3):901–910. doi: 10.1128/jb.165.3.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen H, et al. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J Biol Chem. 1996;271(2):789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- 103.Tamura Y, et al. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 2013;17(5):709–718. doi: 10.1016/j.cmet.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kutik S, et al. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol. 2008;183(7):1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Osman C, et al. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol. 2009;184(4):583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Osman C, et al. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 2010;29(12):1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J, et al. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 2011;13(6):690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Helle SC, et al. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833(11):2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 109.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(5939):477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci U S A. 2011;108(34):14151–14156. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26(16):1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nguyen TT, et al. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13(6):880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tamura Y, et al. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol. 2009;185(6):1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Connerth M, et al. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;338(6108):815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- 115.Potting C, et al. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 2010;29(17):2888–2898. doi: 10.1038/emboj.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tamura Y, Iijima M, Sesaki H. Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 2010;29(17):2875–2887. doi: 10.1038/emboj.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Potting C, et al. TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab. 2013;18(2):287–295. doi: 10.1016/j.cmet.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 118.Klecker T, Bockler S, Westermann B. Making connections: interorganelle contacts orchestrate mitochondrial behavior. Trends Cell Biol. 2014;24(9):537–545. doi: 10.1016/j.tcb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 119.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 120.Jazwinski SM. The retrograde response: a conserved compensatory reaction to damage from within and from without. Prog Mol Biol Transl Sci. 2014;127:133–154. doi: 10.1016/B978-0-12-394625-6.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kornmann B. The molecular hug between the ER and the mitochondria. Curr Opin Cell Biol. 2013;25(4):443–448. doi: 10.1016/j.ceb.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 122.Lackner LL, et al. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci U S A. 2013;110(6):E458–E467. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cohen Y, et al. Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol Biosyst. 2014;10(7):1742–1748. doi: 10.1039/c4mb00001c. [DOI] [PubMed] [Google Scholar]
- 124.Schrader M, Bonekamp NA, Islinger M. Fission and proliferation of peroxisomes. Biochim Biophys Acta. 2012;1822(9):1343–1357. doi: 10.1016/j.bbadis.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 125.Neuspiel M, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18(2):102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 126.Ohya Y, et al. Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a Pet- phenotype are ascribable to defects of vacuolar membrane H(+)-ATPase activity. J Biol Chem. 1991;266(21):13971–13977. [PubMed] [Google Scholar]
- 127.Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793(4):650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Merz S, Westermann B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009;10(9):R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Diab HI, Kane PM. Loss of vacuolar H+-ATPase (V-ATPase) activity in yeast generates an iron deprivation signal that is moderated by induction of the peroxiredoxin TSA2. J Biol Chem. 2013;288(16):11366–11377. doi: 10.1074/jbc.M112.419259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Milgrom E, et al. Loss of vacuolar proton-translocating ATPase activity in yeast results in chronic oxidative stress. J Biol Chem. 2007;282(10):7125–7136. doi: 10.1074/jbc.M608293200. [DOI] [PubMed] [Google Scholar]
- 131.Eide DJ, et al. The vacuolar H(+)-ATPase of Saccharomyces cerevisiae is required for efficient copper detoxification, mitochondrial function, and iron metabolism. Mol Gen Genet. 1993;241(3–4):447–456. doi: 10.1007/BF00284699. [DOI] [PubMed] [Google Scholar]
- 132.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492(7428):261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Elbaz-Alon Y, et al. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30(1):95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 134.Honscher C, et al. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30(1):86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 135.Caplan S, et al. Human Vam6p promotes lysosome clustering and fusion in vivo. J Cell Biol. 2001;154(1):109–122. doi: 10.1083/jcb.200102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Daniele T, et al. Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr Biol. 2014;24(4):393–403. doi: 10.1016/j.cub.2014.01.007. [DOI] [PubMed] [Google Scholar]