Abstract
The fields of immunology, microbiology, and nutrition converge in an astonishing way. Dietary ingredients have a profound effect on the composition of the gut microflora, which in turn regulates the physiology of metazoans. As such, nutritional components of the diet are of critical importance not only for meeting the nutrient requirements of the host, but also for the microbiome. During their coevolution, bacterial microbiota has established multiple mechanisms to influence the eukaryotic host, generally in a beneficial fashion. The microbiome encrypts a variety of metabolic functions that complements the physiology of their hosts. Over a century ago Eli Metchnikoff proposed the revolutionary idea to consume viable bacteria to promote health by modulating the intestinal microflora. The idea is more applicable now than ever, since bacterial antimicrobial resistance has become a serious worldwide problem both in medical and agricultural fields. The impending ban of antibiotics in animal feed due to the current concern over the spread of antibiotic resistance genes makes a compelling case for the development of alternative prophylactics. Nutritional approaches to counteract the debilitating effects of stress and infection may provide producers with useful alternatives to antibiotics. Improving the disease resistance of animals grown without antibiotics will benefit the animals’ health, welfare, and production efficiency, and is also a key strategy in the effort to improve the microbiological safe status of animal-derived food products (e.g. by poultry, rabbits, ruminants, or pigs). This review presents some of the alternatives currently used in food-producing animals to influence their health in relation to human health.
Keywords: intestinal microflora, probiotics, production efficiency, health
The gut is colonized with different types of microorganisms which have dynamic and diverse symbiotic relationships. The gut microbiome serves multiple functions including the conversion of several nutrients that the host cannot digest to end products, a process which has a direct impact on digestive physiology and immunology. Although age, diet, environment, and ethnicity of the subjects have crucial impacts on the microbial gut composition, the improvement of high-throughput technologies has made it possible to better understand the contribution of microbiota to the human health status (1).
The concept of prebiotics is relatively new; it was developed in response to the notion that non-digestible food ingredients (e.g. non-digestible oligosaccharides) are selectively fermented by one or more bacteria known to have positive effects on gut physiology. Bacteria fed with a preferential food substrate have a proliferative advantage over other bacteria (2). Some prebiotics have shown to selectively stimulate the growth of endogenous lactic acid bacteria (LAB) and Bifidobacteria in the gut to improve the health of the host (3, 4). Nevertheless, probiotics can only be effective if the requirements for their growth are present in the gastrointestinal tract (GIT). Probiotic numbers have been enhanced by prebiotics that selectively stimulate the growth and activity of one or a limited number of bacterial species already resident in the large intestine and, thus, improve host health (5–7). In this way, prebiotics selectively modify the colonic microflora and can potentially influence gut metabolism (4). However, the bacterial nutrient package will not be advantageous without the presence of the specific beneficial bacteria that use it, and similarly the live microbial product will not succeed if the environment into which it is introduced is unfavorable (6). The concept of symbiotic has been proposed recently to characterize foods with both prebiotic and probiotic properties as health-enhancing functional foods.
Probiotics and prebiotics to improve animal health, productivity, and food safety
The capacity to ferment complex polysaccharides to short-chain fatty acids (SCFAs) by intestinal microflora has a profound effect on energy homeostasis, providing as much as ~70% of energy in ruminants and 20–30% for several monogastric animals (8). In exchange, their hosts have organs that enable microbial fermentation of non-digestible food stuff (9), revealing a symbiotic evolution over time, as indicated by concurrent phylogenetic trees (10–14). Although the mechanisms by which the intestinal microflora assert these effects on the GIT remain essentially unknown, research in this area is focusing on elucidating these mechanisms as well as manipulating the bacteria and the gastrointestinal environment toward achieving optimal health through probiotics and prebiotics (15–18). Probiosis, although not a new concept, has only recently begun to receive an increasing level of scientific interest. In agriculture, probiotics and direct-fed microbials (DFMs) used in animal feed are becoming accepted as potential alternatives to antibiotics for use as growth promoters, and in selected cases, for control of specific enteric pathogens (19–24). A probiotic is defined as a live microbial food supplement which benefits the host by improving its intestinal microbial balance (25). The presence of normal gut microflora may improve the metabolism of the host animals in various ways, including absorptive capacity, protein metabolism, energy metabolism, fiber digestion, and gut maturation (5, 26). Balanced gastrointestinal microflora and immune-stimulation are major functional effects attributed to the consumption of probiotics (3, 6, 27). Many probiotic effects are mediated through immune regulation, particularly through balanced control of pro-inflammatory and anti-inflammatory cytokines (28, 29). For example, the treatment with the probiotic Enterococcus faecium (EF 55), a poultry-derived strain, resulted in a significantly higher number of lymphocytes in peripheral blood and a tendency to increase CD3, CD4, CD8, and IgM positive cells 3 days post-infection with Salmonella (30). Conversely, several animal and human studies have provided unequivocal evidence that specific strains of probiotics are able to stimulate multiple aspects of innate immunity (31–34) as well as to increase humoral immunity (27, 35–39). Alternatively, different studies have showed that the administration of a lactic acid based probiotic 2 hours after Salmonella challenge, had no effect during the first 12 hours on increasing cecal colonization by this pathogen, although marked and rapid decreases were observed between 12 and 24 hours post-challenge (40). Later, using the same challenge model and microarray analysis of gut mRNA expression, gene expression differences were found in birds treated with probiotic compared to saline treated birds, especially those associated with the NFκB complex and apoptosis, suggesting that increased apoptosis may be a mechanism by which this probiotic reduces Salmonella infection (41). In addition, Toll-like receptors (TLRs) represent evolutionary conserved pathogen recognition receptors which are necessary components in the protection against invading microorganisms (42). Certain probiotic strains may modulate TLR expression as a mechanism of their protective effect against pathogens.
Probiotics and DFMs for monogastrics
During the last 15 years, our laboratories have worked toward the identification of probiotic candidates for use in poultry as a defined LAB-based probiotic culture (FloraMax® B11). This has demonstrated an accelerated development of normal microflora in chickens and turkeys, providing increased resistance to Salmonella spp. infections (24, 33, 40, 43–47). Published experimental and commercial studies have shown that these selected probiotic organisms are able to reduce idiopathic diarrhea in commercial turkey brooding houses (48). Large-scale commercial trials indicated that appropriate administration of this probiotic mixture to turkeys and chickens increased performance and reduced costs of production (45, 49). Lactobacillus salivarius and Pediococcus parvulus of poultry gastrointestinal origin present in FloraMax® B11 have been identified by 16S rRNA sequence analyses (50). Tolerance and resistance to acidic pH, high osmotic concentration of NaCl, and bile salts of these isolates may have contributed to the efficacy of these isolates to survive and provide beneficial effects. Both strains also have in vitro antibacterial activity against Salmonella enterica serovar Enteritidis, Escherichia coli (O157:H7), and Campylobacter jejuni (50). In addition, previous research from our laboratory indicates very rapid induction of specific host-gene expression pathways, which are associated with reductions in enteric colonization with Salmonella (41). The aforementioned characteristics, may contribute to the efficacy previously reported in laboratory and field conditions (24). While many mechanisms of action have been proposed for the observed efficacy, precise modalities have not been completely described for this highly effective culture.
In spite of the success shown by the development of the LAB probiotic for use in commercial poultry (above), there is still an urgent need for commercial probiotics that are shelf-stable, cost-effective, and feed-stable (tolerance to the heat pelletization process) to increase compliance and widespread utilization. Among the large number of probiotic products in use today some are bacterial spore formers, mostly of the genus Bacillus. Used primarily in their spore form, some (though not all) have been shown to prevent selected gastrointestinal disorders and the diversity of species used and their applications are astonishing. While not all Bacillus spores are highly heat tolerant, some specific isolates are the toughest life form known on earth (51) and can be used under extreme heat conditions. Several studies have shown that either live vegetative cells or endospores of some isolates can prevent colon carcinogenesis (52) or discharge antimicrobial substances against Gram-positive bacteria, such as Staphylococcus aureus, E. faecium, and Clostridium difficile (53). These results provide evidence of colonization and antimicrobial activity of probiotic bacteria, thus, products containing Bacillus spores are used commercially as probiotics, and they offer potential advantages over the more common LAB products since they can be used as direct feed microbials. DFMs are widely fed in production animals and are typically composed of alive or dead yeast, fungal or bacterial cultures or also end products of fermentation (54–62). There is scientific evidence suggesting that some, but not all, isolates of ingested B. subtilis spores can, in fact, germinate in the small intestine (63–66). Together, these studies not only show that spores are not transient passengers in the gut, but that they have an intimate interaction with the host cells or microflora that can enhance their potential probiotic effect. Several commercial spore-forming Bacillus cultures have been shown to reduce food-borne pathogens (67). However, cost issues associated with achieving necessary concentrations of spores in feed have greatly limited commercial acceptance in the animal industry (57).
At the present time, our laboratory’s aim is to develop a novel, cost-effective, feed-stable probiotic with widespread utilization, simple delivery, and clinical efficacy for human and animal use. We have demonstrated that one Bacillus subtilis spores isolate is as effective as our LAB-based probiotic (FloraMax™) for Salmonella reduction (68, 69), and equal to bacitracin (an antibiotic) for the prevention of necrotic enteritis in experimental and commercial field trials.
Other isolates or combinations of isolates with increased potency and efficacy may be identified with continued research. Some of these environmental Bacillus isolates have been evaluated in vitro for antimicrobial activity against selected bacterial pathogens, heat stability, and the ability to grow to high numbers. Our preliminary data suggests that these isolates could be an effective alternative to antibiotic growth promoters for commercial poultry. Importantly, improved efficiency of amplification and sporulation is absolutely essential to gain widespread industry acceptance of a feed-based probiotic for ante mortem food-borne pathogen intervention. Recently, both vegetative growth and sporulation rate have been optimized in our laboratory, which may lead to new efficiencies for commercial amplification and manufacture of a cost-effective product at very high spore counts (68). In order to select even more effective isolates, we are currently focused on the mechanistic action of new Bacillus candidates. Preliminary studies conducted in our laboratory indicate a potential mechanistic action of these new Bacillus candidates at least partially involves the rapid activation of innate host immune mechanisms in chickens and turkeys (unpublished data). This data provides an exciting possibility for the identification of vastly superior and more potent probiotics in the near future.
Ruminant gastrointestinal fermentation and niches
The ruminant animal was noted for being ‘different’ than monogastric animals since ancient times, Leviticus (11, pp. 3–4) states ‘Whatsoever parteth the hoof, and is cloven-footed, and cheweth the cud, among the beasts, that shall ye eat’. Ruminants, such as cattle, exist in a unique symbiotic relationship with a pre-gastric microbial consortium of bacteria, protozoa, archaea, viruses, and fungi that ferment relatively low-quality feedstuffs and in turn supply the ruminant with nutrients to produce high-quality meat, milk, and fiber (70). The intestinal tract of ruminants is different from monogastrics because it contains a large (up to 100 liters) anaerobic fermentation chamber, the reticulo-rumen, located prior to feedstuff entry into the gastric stomach, the abomasum (Figs. 1 and 2). The mutualistic nature of the ruminant animal and the resident microbial population is critical to the success of ruminant animals in a variety of environmental niches around the world.
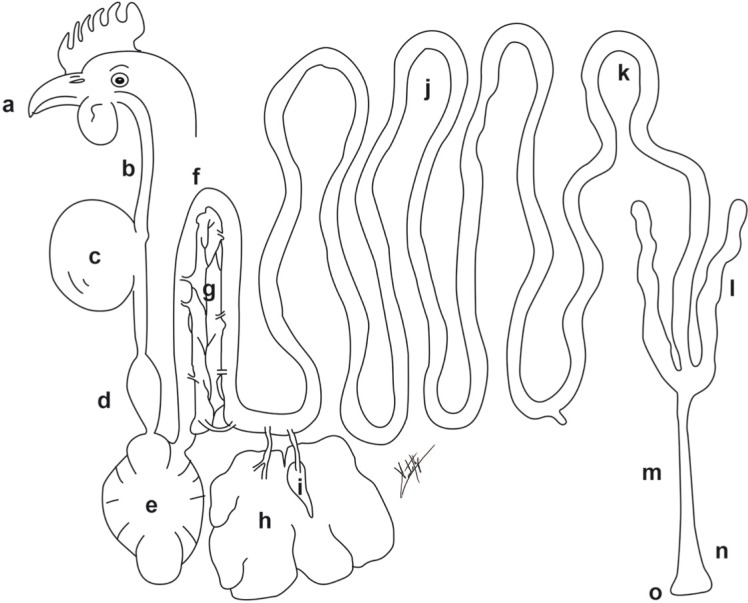
Fig. 1.
Digestive tract of a hen: (a) beak and mouth, (b) esophagus, (c) crop, (d) proventriculus, (e) gizzard, (f) duodenal loop, (g) pancreas, (h) liver, (i) gallbladder, (j) jejunum, (k) ileum, (l) ceca, (m) rectum, (n) cloaca, (o) vent.
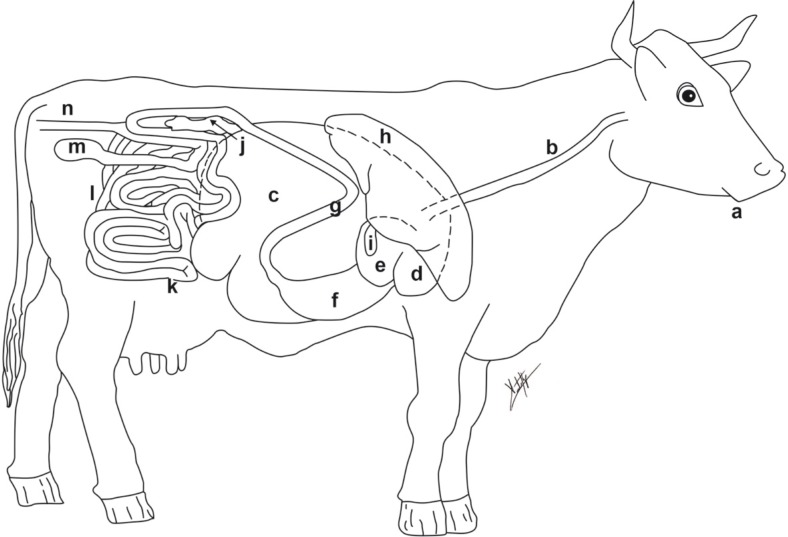
Fig. 2.
View from right of digestive tract of a cow: (a) mouth, (b) esophagus, (c) rumen, (d) reticulum, (e) omasum, (f) abomasum, (g) duodenum, (h) liver, (i) gallbladder, (j) pancreas, (k) small intestine, (l) large intestine, (m) ceca, (n) rectum.
The mutualism of the ruminant is dependent upon the pre-gastric ruminal fermentation which converts feedstuffs to SCFAs, and microbial cells (microbial crude protein), which are washed out of the rumen and are degraded by the host gastric stomach and absorbed by the intestinal tract (71). The primary SCFAs of interest in the ruminant are acetate, propionate, and butyrate, which when absorbed by the host, can provide up to 80% of the animals energy requirement (71). Although acetate is the primary SCFA produced in the ruminal fermentation, propionate is glucogenic and is more desirable in animals growing rapidly (e.g. beef cattle). The acetate:propionate ratio, an indicator of the energetic efficiency of the fermentation to the host animal, can be altered by simply increasing the amount of cereal grain in the ration. Grain feeding decreases the acetate:propionate ratio, meaning more energy is available to the host for growth. Further benefits to ruminants of this mutualistic relationship include the fact that B-complex vitamins are produced by the ruminal and gastrointestinal microbiome which eliminates the need for dietary B-vitamin supplementation.
Ruminant diets are comprised of many variable feedstuffs that contain complex carbohydrates (e.g. cellulose, hemicellulose, and lignin); therefore, many nutrient niches are available throughout the GIT (72). Thus, as dietary components change, the nutrient niches and bacterial species occupying them also change. Also, bacterial populations can be impacted by end products of the fermentation. For example, the primary driver of the shift in ruminal propionate proportions by cereal grain feeding is starch. Ruminal starch is fermented by Streptococci and Lactococci to produce SCFAs and lactic acid which results in a lowering of ruminal pH (73). As a result of this increase in lactic acid concentrations, populations of the lactate-utilizing ruminal bacteria Megasphaera elsdenii and Selenomonas ruminantium increase due to their ability to secondarily ferment lactic acid to form propionate (70).
In addition to the high-quality protein from bacterial cells (microbial crude protein), some feedstuffs are not degraded in the rumen and pass through the abomasum (often called ‘Undegraded Intake’ or ‘Rumen Escape’) and are degraded and absorbed in the small and large intestines. An additional microbial consortium occupies the cecum and colon of cattle that is distinct from the ruminal population (74). This microbial population performs a secondary fermentation in the lower gut that captures more dietary energy that bypasses ruminal fermentation. For instance, starch that reaches the hindgut is fermented, which causes higher fecal SCFA and lactate concentrations and a lower pH than that found in forage-based diets. As in humans, the fecal populations of cattle changes in response to diet (75).
Bacterial diversity of the ruminant GIT
The advent of next generation sequencing has produced a bloom of information available about the composition of the resident gastrointestinal microbiome under different conditions; and metagenomics, proteomics, and KEGG analyses (Kyoto Encyclopedia of Genes and Genomes) have led to an unprecedented understanding of the metabolic actions of and interactions between ruminal microorganisms and their host. However, our methods for understanding the biology and ecology of these consortia are still limiting. Approaching the ruminal and gastrointestinal population from a functional perspective by examining ‘guilds’ of bacteria occupying the same or related niches in a given environment offers promise for understanding the macro-biological impacts of the microbial population. A more subtle level of interaction within the microbiome also includes ‘crosstalk’ between members of the microbial ecosystem and the host, using compounds that mimic neuropeptides or other host signaling molecules, which has led to the description of the microbiome as a ‘microbial organ’ (76).
Most of our knowledge based on the microbial ecology of the rumen and GIT is derived from culture-based methodologies; however, over the past 5 years, more than 3,000 known bacterial species have been detected in cattle rumens and GITs, and estimates of the unidentified species in the rumen range from 10- to 100-fold higher. Based on our new insights into the composition of the ruminal microbiota, the ruminant GIT is dominated by two major bacterial phyla Firmicutes and Bacteroidetes, which comprise approximately 80–92% of the intestinal microbiota in the human and bovine gut (77, 78). The proportions and diversity of organisms representing this community are strongly influenced by diet as postulated in the nutrient-niche theory (72, 75, 78).
As an example of changes in the gastrointestinal microbiome caused by diet, starch can significantly alter the population composition of the lower GIT. Firmicutes populations decreased with increasing fecal starch concentrations, whereas Bacteroidetes increased as starch concentration increased (78). Prevotellaceae and Lachnospiraceae were the dominant members of Bacteroidetes and Firmicutes, respectively in grain-fed cattle; however in forage-fed cattle, the Ruminococcaceae and Bacteroidales became dominant. Grain feeding decreased Ruminococcaceae populations but increased Lachnospiraceae, Clostridiaceae, and Succinivibrionaceae populations (78). This shift in the ruminal population could be explained by the hypothesized role Ruminococci play in degradation of hemicellulose and xylan. Prevotellae and Bacteroides, on the other hand, utilize hemicellulose but also degrade starch and pectin rapidly, yet Bacteroides are more commonly found in cattle feces than Prevotella. Recent in press research has demonstrated that there are distinct ruminal microbial populations (or enterotypes) characterized from diets of: high grain, moderate grain and silage–forage; but that a small proportion of bacteria are common to all three diets (79). Feeding wet distiller’s grains to cattle changes the microbial community structure (most notably Prevotellae) in the feces (75) which significantly increased E. coli O157:H7 fecal shedding (80). Interestingly, while pens where cattle are housed are often covered with fresh feces, recent molecular studies have indicated that the bacterial communities of feedlot surfaces (and by extension other animal housing environments) are distinctive from fecal bacterial populations (81).
While much of our attention has been focused on the microbiome members that make up large proportions of the gastrointestinal population, members of other less known and/or less populous bacterial phyla have been isolated from gastrointestinal populations (82, 83). The Fusobacteria, Actinobacteria, Spirochetes, and Synergistes among others, have been found in ruminal and gastrointestinal samples from cattle but their ‘guild’ role has been unclear. These and other groups do contain a wide variety of fermentative capacities (impacting the metagenome), and may play a role of ‘utility player’ in this complex microbiome. Furthermore, some of these species may have a high specific activity, making their impact greater than their roles would suggest based on population alone, such as the obligate amino acid-fermenting/hyperammonia producing bacteria. Several ruminal Firmicutes (e.g. Clostridium, Enterococcus, Staphylococcus, and Peptostreptococcus) have displayed a marked ability to ferment peptides and amino acids, indicating that diets rich in available protein would provide a competitive advantage for these genera. Collectively, these population shifts illustrate the elasticity of the ruminant microbiome response to environmental (dietary) shifts.
Other members of the ruminal microbiome
While bacteria are the best understood members of the microbial consortium, protozoa (Eucarya) comprise as much as 50% of the biomass of the rumen, yet relatively little is known of the types of protozoa found in the rumen. Their role in starch sequestration and conversion of bacterial protein to high-quality protein has been noted but knowledge about the diversity of these important ruminal components has largely been limited to microscopy (84). Furthermore, while it has been long known that members of Kingdom Archaea, most notably methanogens, inhabit the rumen, only with the advent of molecular techniques have details of the Euryarchaeota population composition been reported (85, 86). While microscopy studies have previously demonstrated that protozoa and methanogens exist in symbiosis, this relationship can now be quantified and specific relationships between members of these understudied kingdoms can be determined. Fungi have also been commonly isolated from the rumen, yet the role of fungi is still unclear (87). Bacteriophages (bacterial viruses) have been frequently isolated from the rumen, and it has been hypothesized that they play a role in nutrient recycling in the rumen as well as diurnal variation of the ruminal populations (88).
Probiotics and DFMs for ruminants
A DFM is fed daily to improve the ruminal fermentation and production efficiency (89). Increasingly, companies claim some benefit to them in reducing E. coli O157:H7 shedding in cattle. E. coli O157:H7 is a food-borne pathogen capable of causing severe illness in humans. Different researchers have compared several of the commercially available growth enhancement probiotics and yeast products and found that feeding these probiotics provided no effect in regards to pathogen levels in cattle (90, 91). However, a probiotic culture comprised of Streptococcus bovis and Lactobacillus gallinarum from the rumen of cattle resulted in the reduction of E. coli O157 shedding when given to experimentally infected calves, and this decrease was attributed to an increase in SCFAs concentration in the gut (92). Probiotic products have been developed to specifically reduce E. coli O157:H7 shedding in cattle. A probiotic that contained S. faecium or a mixture of S. faecium, L. acidophilus, L. casei, L. fermentum and L. plantarum significantly reduced fecal shedding of E. coli O157:H7 in sheep, yet, a monoculture of Lactobacillus acidophilus was found to be ineffective (93). A DFM comprised of Bacillus subtilis did not affect the fecal prevalence or concentration of E. coli O157:H7 and did not impact average daily gain in feedlot cattle (94). Studies have also indicated that cultures of Lactobacillus acidilactici and Pediococcus could directly inhibit E. coli O157:H7, likely through the production of organic acids and low pH (95).
This Lactobacillus-based DFM is currently marketed as Bovamine™ and Bovamine Defend™ based on dosing levels, and both are widely used in the cattle industry because they have been reported to improve the growth efficiency of cattle, at least in a feedlot ration. There will likely not be a single DFM that can work effectively at reducing E. coli O157:H7 populations in cattle and improve production efficiency in all production systems (i.e. feedlots, cow–calf, stockers, and dairies). Therefore, alternative DFM cultures selected specifically for each production segment or situation need to be developed so that the food safety improvement can occur while economically balancing the cost of its inclusion in cattle rations thus ‘paying for’ the enhancement of food safety.
Bacteriocin-producing bacteria as probiotics
Bacteriocins have attracted great interest for their potential use as food preservatives, functional food ingredients (with both health and economic benefits), as well as antibacterial agents to prevent or reduce food-borne pathogens (96–99). In general, bacteriocins are ribosomally synthesized proteinaceous substances with antimicrobial activity against similar bacteria (100). One group of bacteriocins is enterocins; they are produced by representatives of the genus Enterococcus, the most frequent being the species E. faecium (101). Some enterococci are known to produce enterocins and to possess probiotic properties (102, 103), and the most studied enterocins are those produced by strains of E. faecium from different sources. E. faecium CCM4231 isolated from the rumen was the first bacteriocin producer strain of animal origin described in detail (104). Other bacteriocin-producing strains of animal origin are E. faecium EF55 of chicken origin, and rabbit feces-derived strain E. faecium 2019 (CCM7420). Moreover, animal waste isolates, E. faecium EK13 (CCM7419), and E. faecium AL41 have been found to produce bacteriocins and possess probiotic properties (104–108). Based on detailed studies (101), enterocins (Ents) are divided into four classes: 1) lantibiotic (lanthionine-containing peptide antibiotic) enterocins (e.g. cytolysin), 2) small non-lantibiotic peptides with three subgroups-enterocins of pediocin type (e.g. enterocin A, P), enterocins synthesized without a leader peptide (Ent Q or L50), and other linear, non-pediocin-type enterocins, (e.g. Ent B), 3) cyclic antibacterial peptides, and 4) large proteins.
Enterocin-producing enterococci and/or their enterocins have been studied in food animals (poultry and rabbits) and have been shown to maintain health due to their in vivo antimicrobial effect against Salmonella serotype Enteritidis PT4, coagulase-positive staphylococci, coliforms, clostridiae, and pseudomonas (109–114). The mechanisms of their antimicrobial activity can vary from bactericidal modulation of enzymatic activity, through to the inhibition of anionic transporters, or to form pores in the cytoplasmic membrane.
Although much work remains to be done to determine their exact mechanism of action, bacteriocins and bacteriocin-producing bacteria have the potential to improve animal health and by extension, improve the safety of animal products consumed by humans.
Conclusions
The interest in digestive physiology and the role of microorganisms has generated data whereby human and animal wellbeing can be enhanced and the risk of disease reduced. New molecular techniques allow an accurate assessment of the floral composition, resulting in improved strategies to elucidate the different mechanism of action that probiotics have to enhance performance of livestock animal species. Given the recent international legislation and domestic consumer pressures to withdraw growth-promoting antibiotics and limit antibiotics available for treatment of bacterial infections, probiotics can offer alternative options. New advances in the application of probiotics are directed to produce significant changes in gut physiology and provide even higher levels of health as well as increasing performance parameters in different animals.
Metchnikoff founded the research field of probiotics, aimed at modulating the intestinal microflora (115). However, other parts of the body containing endogenous microflora or problems relating to the immune system may also be candidates for probiotic therapy. Much research has been completed in an effort to understand and apply the natural benefits of non-pathogenic bacteria, but there is much still to do.
Acknowledgements
The authors acknowledge the support of the European Science Foundation (ESF), in the framework of the Research Networking Programe, The European Network for Gastrointestinal Health Research.
This paper is part of the Proceedings from the 2013 ENGIHR Conference in Valencia, Spain. More papers from this supplement can be found at http://www.microbecolhealthdis.net
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Tellez G, Higgins S, Donoghue A, Hargis B. Digestive physiology and the role of microorganisms. J Appl Poult Res. 2006;15:136–44. [Google Scholar]
- 2.Gibson GR, Roberfroit MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–12. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 3.Hammes WP, Hertel C. Research approaches for pre-and probiotics: challenges and outlook. Food Res Int. 2002;35:165–70. [Google Scholar]
- 4.Molinaro F, Paschetta E, Cassader M, Gambino R, Musso G. Probiotics, prebiotics, energy balance, and obesity: mechanistic insights and therapeutic implications. Gastroenterol Clin North Am. 2012;41:843–54. doi: 10.1016/j.gtc.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Teitelbaum JE, Walker WA. Nutritional impact of pre-and probiotics as protective gastrointestinal organisms. Annu Rev Nutr. 2002;22:107–38. doi: 10.1146/annurev.nutr.22.110901.145412. [DOI] [PubMed] [Google Scholar]
- 6.Parracho H, McCartney AL, Gibson GR. Probiotics and prebiotics in infant nutrition. Proc Nutr Soc. 2007;66:405–11. doi: 10.1017/S0029665107005678. [DOI] [PubMed] [Google Scholar]
- 7.Kerac M, Bunn J, Seal A, Thindwa M, Tomkins A, Sadler K, et al. Probiotics and prebiotics for severe acute malnutrition (PRONUT study): a double-blind efficacy randomised controlled trial in Malawi. Lancet. 2009;374:136–44. doi: 10.1016/S0140-6736(09)60884-9. [DOI] [PubMed] [Google Scholar]
- 8.Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm . Proc Nat Acad Sci USA. 2001;108:4645–52. doi: 10.1073/pnas.1000099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller R, Brooker B. Lactobacilli which attach to the crop epithelium of the fowl. Am J Clin Nutr. 1974;27:1305–12. doi: 10.1093/ajcn/27.11.1305. [DOI] [PubMed] [Google Scholar]
- 10.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–65. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Nat Acad Sci U S A. 2007;104:8627–33. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choct M. Managing gut health through nutrition. Br Poult Sci. 2009;50:9–15. doi: 10.1080/00071660802538632. [DOI] [PubMed] [Google Scholar]
- 13.Gill N, Wlodarska M, Finlay BB. The future of mucosal immunology: studying an integrated system-wide organ. Nat Immunol. 2010;11:558–60. doi: 10.1038/ni0710-558. [DOI] [PubMed] [Google Scholar]
- 14.Fraune S, Bosch TC. Why bacteria matter in animal development and evolution. Bioessays. 2010;32:571–80. doi: 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- 15.Yegani M, Korver D. Factors affecting intestinal health in poultry. Poult Sci. 2008;87:2052–63. doi: 10.3382/ps.2008-00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2010;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 17.Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota. The hygiene hypothesis expanded? Diabetes Care. 2010;33:2277–84. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathew AG, Cissell R, Liamthong S. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis. 2007;4:115–33. doi: 10.1089/fpd.2006.0066. [DOI] [PubMed] [Google Scholar]
- 20.Dominguez-Bello MG, Blaser MJ. Do you have a probiotic in your future? Microbes Infect. 2008;10:1072–6. doi: 10.1016/j.micinf.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter AJ, Adams MR, Woodward MJ, La Ragione RM. Control strategies for Salmonella colonization of poultry: the probiotic perspective. Food Sci Technol. 2009;5:103–15. [Google Scholar]
- 22.Sherman PM, Ossa JC, Johnson-Henry K. Unraveling mechanisms of action of probiotics. Nutr Clin Pract. 2009;24:10–14. doi: 10.1177/0884533608329231. [DOI] [PubMed] [Google Scholar]
- 23.MacDonald TT, Bell I. Probiotics and the immune response to vaccines. Proc Nutr Soc. 2010;69:442–6. doi: 10.1017/S0029665110001758. [DOI] [PubMed] [Google Scholar]
- 24.Tellez G, Pixley C, Wolfenden R, Layton S, Hargis B. Probiotics/direct fed microbials for Salmonella control in poultry. Food Res Int. 2012;45:628–33. [Google Scholar]
- 25.Isolauri E, Kirjavainen P, Salminen S. Probiotics: a role in the treatment of intestinal infection and inflammation? Gut. 2002;50:iii54–9. doi: 10.1136/gut.50.suppl_3.iii54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salminen S, Isolauri E. Intestinal colonization, microbiota, and probiotics. J Pediatr. 2006;149:S115–20. [Google Scholar]
- 27.Parvez S, Malik K, Ah Kang S, Kim HI. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100:1171–85. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 28.Braat H, van den Brande J, van Tol E, Hommes D, Peppelenbosch M, van Deventer S. Lactobacillus rhamnosus induces peripheral hyporesponsiveness in stimulated CD4+ T cells via modulation of dendritic cell function. Am J Clin Nutr. 2004;80:1618–25. doi: 10.1093/ajcn/80.6.1618. [DOI] [PubMed] [Google Scholar]
- 29.Borchers AT, Selmi C, Meyers FJ, Keen CL, Gershwin ME. Probiotics and immunity. J Gastroenterol. 2009;44:26–46. doi: 10.1007/s00535-008-2296-0. [DOI] [PubMed] [Google Scholar]
- 30.Levkut M, Revajová V, Lauková A, Ševcíková Z, Spišáková V, Faixová Z, et al. Leukocytic responses and intestinal mucin dynamics of broilers protected with Enterococcus faecium EF55 and challenged with Salmonella Enteritidis. Res Vet Sci. 2012;93:195–201. doi: 10.1016/j.rvsc.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 31. Alvarez-Olmos MI, Oberhelman RA. Probiotic agents and infectious diseases: a modern perspective on a traditional therapy. Clin Infect Dis. 2001;32:1567–76. doi: 10.1086/320518. [DOI] [PubMed] [Google Scholar]
- 32.Reveneau N, Geoffroy MC, Locht C, Chagnaud P, Mercenier A. Comparison of the immune responses induced by local immunizations with recombinant Lactobacillus plantarum producing tetanus toxin fragment C in different cellular locations. Vaccine. 2002;20:1769–77. doi: 10.1016/s0264-410x(02)00027-0. [DOI] [PubMed] [Google Scholar]
- 33.Farnell M, Donoghue A, De Los Santos FS, Blore P, Hargis B, Tellez G, et al. Upregulation of oxidative burst and degranulation in chicken heterophils stimulated with probiotic bacteria. Poult Sci. 2006;85:1900–6. doi: 10.1093/ps/85.11.1900. [DOI] [PubMed] [Google Scholar]
- 34.Jounai K, Ikado K, Sugimura T, Ano Y, Braun J, Fujiwara D. Spherical lactic acid bacteria activate plasmacytoid dendritic cells immunomodulatory function via TLR9-dependent crosstalk with myeloid dendritic cells. PLoS One. 2012;7:e32588. doi: 10.1371/journal.pone.0032588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arvola T, Laiho K, Torkkeli S, Mykkänen H, Salminen S, Maunula L, et al. Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: A randomized study. Pediatrics. 1999;104:e64. doi: 10.1542/peds.104.5.e64. [DOI] [PubMed] [Google Scholar]
- 36.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E, et al. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–9. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 37.FAO, WHO. Argentina: Córdoba; 2001. Report of a Joint FAO/WHO expert consultation on evaluation of health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Available from: http://www.mesanders.com/probio_report.pdf [cited 1–4 October 2001]. [Google Scholar]
- 38.Ouwehand A, Isolauri E, Salminen S. The role of the intestinal microflora for the development of the immune system in early childhood. Eur J Nutr. 2002;41(Suppl 1):i32–137. doi: 10.1007/s00394-002-1105-4. [DOI] [PubMed] [Google Scholar]
- 39.Salzman NH. Microbiota–immune system interaction: an uneasy alliance. Curr Opin Microbiol. 2011;14:99–105. doi: 10.1016/j.mib.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins J, Higgins S, Wolfenden A, Henderson S, Torres-Rodriguez A, Vicente J, et al. Effect of lactic acid bacteria probiotic culture treatment timing on Salmonella Enteritidis in neonatal broilers. Poult Sci. 2010;89:243–7. doi: 10.3382/ps.2009-00436. [DOI] [PubMed] [Google Scholar]
- 41.Higgins S, Wolfenden A, Tellez G, Hargis B, Porter T. Transcriptional profiling of cecal gene expression in probiotic-and Salmonella-challenged neonatal chicks. Poult Sci. 2011;90:901–13. doi: 10.3382/ps.2010-00907. [DOI] [PubMed] [Google Scholar]
- 42.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Higgins SE, Erf GF, Higgins JP, Henderson SN, Wolfenden AD, Gaona-Ramirez G, et al. Effect of probiotic treatment in broiler chicks on intestinal macrophage numbers and phagocytosis of Salmonella Enteritidis by abdominal exudate cells. Poult Sci. 2007;86:2315–21. doi: 10.3382/ps.2007-00123. [DOI] [PubMed] [Google Scholar]
- 44.Wolfenden A, Pixley C, Higgins J, Higgins S, Vicente J, Torres-Rodriguez A, et al. Evaluation of spray application of a Lactobacillus-based probiotic on Salmonella Enteritidis colonization in broiler chickens. Int J Poult Sci. 2007;6:493–6. [Google Scholar]
- 45.Vicente J, Higgins S, Hargis B, Tellez G. Effect of poultry guard litter amendment on horizontal transmission of Salmonella Enteritidis in broiler chicks. Int J Poult Sci. 2007;6:314–17. [Google Scholar]
- 46.Vicente JL, Torres-Rodriguez A, Higgins SE, Pixley C, Tellez G, Donoghue AM, et al. Effect of a selected Lactobacillus spp.-based probiotic on Salmonella enterica serovar Enteritidis-infected broiler chicks. Avian Dis. 2008;52:143–6. doi: 10.1637/7847-011107-ResNote. [DOI] [PubMed] [Google Scholar]
- 47.Menconi A, Wolfenden A, Shivaramaiah S, Terraes J, Urbano T, Kuttel J, et al. Effect of lactic acid bacteria probiotic culture for the treatment of Salmonella enterica serovar Heidelberg in neonatal broiler chickens and turkey poults. Poult Sci. 2011;90:561–5. doi: 10.3382/ps.2010-01220. [DOI] [PubMed] [Google Scholar]
- 48.Higgins S, Torres-Rodriguez A, Vicente J, Sartor C, Pixley C, Nava G, et al. Evaluation of intervention strategies for idiopathic diarrhea in commercial turkey brooding houses. J Appl Poult Res. 2005;14:345–8. [Google Scholar]
- 49.Torres-Rodriguez A, Higgins S, Vicente J, Wolfenden A, Gaona-Ramirez G, Barton J, et al. Effect of lactose as a prebiotic on turkey body weight under commercial conditions. J Appl Poult Res. 2007;16:635–41. [Google Scholar]
- 50.Menconi A, Kallapura G, Latorre JD, Morgan MJ, Pumford NR, Hargis BM, et al. Identification and characterization of lactic acid bacteria in a commercial probiotic culture. Biosci Microb Food Health. 2014;33:25–30. doi: 10.12938/bmfh.33.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vreeland RH, Rosenzweig WD, Powers DW. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature. 2000;407:897–900. doi: 10.1038/35038060. [DOI] [PubMed] [Google Scholar]
- 52.Park E, Jeon GI, Park JS, Paik HD. A probiotic strain of Bacillus polyfermenticus reduces DMH induced precancerous lesions in F344 male rat. Biol Pharm Bull. 2007;30:569–74. doi: 10.1248/bpb.30.569. [DOI] [PubMed] [Google Scholar]
- 53.O’Mahony L, Feeney M, O’Halloran S, Murphy L, Kiely B, Fitzgibbon J, et al. Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment Pharmacol Ther. 2001;15:1219–25. doi: 10.1046/j.1365-2036.2001.01027.x. [DOI] [PubMed] [Google Scholar]
- 54.Anadón A, Martínez-Larrañaga MR, Aranzazu Martínez M. Probiotics for animal nutrition in the European Union. Regulation and safety assessment. Regul Toxicol Pharmacol. 2006;45:91–5. doi: 10.1016/j.yrtph.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Barbosa TM, Serra CR, La Ragione RM, Woodward MJ, Henriques AO. Screening for bacillus isolates in the broiler gastrointestinal tract. Appl Environ Microbiol. 2005;71:968–78. doi: 10.1128/AEM.71.2.968-978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duc le H, Hong HA, Barbosa TM, Henriques AO, Cutting SM. Characterization of Bacillus probiotics available for human use. Appl Environ Microbiol. 2004;70:2161–71. doi: 10.1128/AEM.70.4.2161-2171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong HA, Duc le H, Cutting SM. The use of bacterial spore formers as probiotics. FEMS Microbiol Rev. 2005;29:813–35. doi: 10.1016/j.femsre.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Hong HA, Huang JM, Khaneja R, Hiep LV, Urdaci MC, Cutting SM. The safety of Bacillus subtilis and Bacillus indicus as food probiotics. J Appl Microbiol. 2008;105:510–20. doi: 10.1111/j.1365-2672.2008.03773.x. [DOI] [PubMed] [Google Scholar]
- 59.McNulty CA, Boyle P, Nichols T, Clappison P, Davey P. The public’s attitudes to and compliance with antibiotics. J Antimicrob Chemother. 2007;60(Suppl 1):i63–8. doi: 10.1093/jac/dkm161. [DOI] [PubMed] [Google Scholar]
- 60.Osipova IG, Makhaǐlova NA, Sorokulova IB, Vasil’eva EA, Gaǐderov AA. Spore probiotics. Zh Mikrobiol Epidemiol Immunobiol. 2003;3:113–19. [PubMed] [Google Scholar]
- 61.Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiol. 2007;153:3923–38. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 62.Wolken WA, Tramper J, van der Werf MJ. What can spores do for us? Trends Biotechnol. 2003;21:338–45. doi: 10.1016/S0167-7799(03)00170-7. [DOI] [PubMed] [Google Scholar]
- 63.Latorre JD, Hernandez-Velasco X, Kallapura G, Menconi A, Pumford NR, Morgan MJ, et al. Evaluation of germination, distribution and persistence of Bacillus subtilis spores through the gastrointestinal tract of chickens. Poult Sci. 2014;93:1–8.4. doi: 10.3382/ps.2013-03809. [DOI] [PubMed] [Google Scholar]
- 64.Casula G, Cutting SM. Bacillus probiotics: spore germination in the gastrointestinal tract. Appl Environ Microbiol. 2002;68:2344–52. doi: 10.1128/AEM.68.5.2344-2352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duc le H, Hong HA, Cutting SM. Germination of the spore in the gastrointestinal tract provides a novel route for heterologous antigen delivery. Vaccine. 2003;21:4215–24. doi: 10.1016/s0264-410x(03)00492-4. [DOI] [PubMed] [Google Scholar]
- 66.Hoa TT, Duc LH, Isticato R, Baccigalupi L, Ricca E, Van PH, et al. Fate and dissemination of Bacillus subtilis spores in a murine model. Appl Environ Microbiol. 2001;67:3819–23. doi: 10.1128/AEM.67.9.3819-3823.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aureli P, Fiore A, Scalfaro C, Casale M, Franciosa G. National survey outcomes on commercial probiotic food supplements in Italy. Int J Food Microbiol. 2010;137:265–73. doi: 10.1016/j.ijfoodmicro.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 68.Wolfenden RE, Pumford NR, Morgan MJ, Shivaramaiah S, Wolfenden AD, Tellez G. Evaluation of a screening and selection method for bacillus isolates for use as effective direct-fed microbials in commercial poultry. Int J Poult Sci. 2010;9:317–23. [Google Scholar]
- 69.Shivaramaiah S, Pumford NR, Morgan MJ, Wolfenden RE, Wolfenden AD, Torres-Rodríguez A, et al. Evaluation of Bacillus species as potential candidates for direct-fed microbials in commercial poultry. Poult Sci. 2011;90:1574–80. doi: 10.3382/ps.2010-00745. [DOI] [PubMed] [Google Scholar]
- 70.Hungate RE. New York, NY: Academic Press; 1966. The rumen and its microbes. [Google Scholar]
- 71.Yokoyama MG, Johnson KA. Microbiology of the rumen and intestine. In: Church DC, editor. The ruminant animal: digestive physiology and nutrition. Englewood Cliffs, NJ: Waveland Press; 1988. [Google Scholar]
- 72.Freter R. Control mechanisms of the large-intestinal microflora and its influence on the host. Acta Gastroenterol Latinoam. 1989;19:197–217. [PubMed] [Google Scholar]
- 73.Russell JB, Hino T. Regulation of lactate production in Streptococcus bovis: a spiraling effect that contributes to rumen acidosis. J Dairy Sci. 1985;68:1712–21. doi: 10.3168/jds.s0022-0302(85)81017-1. [DOI] [PubMed] [Google Scholar]
- 74.Frey JC, Pell AN, Berthiume R, Lapierre H, Lee S, Ha JK, et al. Comparative studies of microbial populations in the rumen, duodenum, ileum and faeces of lactating dairy cows. J Appl Microbiol. 2010;108:1982–93. doi: 10.1111/j.1365-2672.2009.04602.x. [DOI] [PubMed] [Google Scholar]
- 75.Durso LM, Wells JE, Harhay GP, Rice WC, Kuehn L, Bono JL, et al. Comparison of bacterial communities in faeces of beef cattle fed diets containing corn and wet distillers’ grain with solubles. Lett Appl Microbiol. 2012;55:109–14. doi: 10.1111/j.1472-765X.2012.03265.x. [DOI] [PubMed] [Google Scholar]
- 76.Lyte M. The microbial organ in the gut as a driver of homeostasis and disease. Med Hypoth. 2010;74:634–8. doi: 10.1016/j.mehy.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 77.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the veterbrate gut microbiota. Nat Rev Microbiol. 2008;6:766–88. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, et al. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol. 2011;77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim MS, Kim J, Kuehn LA, Bono JL, Berry ED, Kalchayanand N, et al. Investigation of bacterial diversity in the feces of cattle fed different diets. J Anim Sci. 2014;92:683–94. doi: 10.2527/jas.2013-6841. [DOI] [PubMed] [Google Scholar]
- 80.Wells JE, Shackelford SD, Berry ED, Kalchayanand N, Bosilevac JM, Wheeler TL. Impact of reducing the level of wet distillers grains fed to cattle prior to harvest on prevalence and levels of Escherichia coli O157:H7 in feces and on hides. J Food Prot. 2011;74:1611–17. doi: 10.4315/0362-028X.JFP-11-160. [DOI] [PubMed] [Google Scholar]
- 81.Durso LM, Harhay GP, Smith TP, Bono JL, DeSantis TZ, Clawson ML. Bacterial community analysis of beef cattle feedlots reveals pen surface is distinct from feces. Foodborne Pathog Dis. 2011;8:647–9. doi: 10.1089/fpd.2010.0774. [DOI] [PubMed] [Google Scholar]
- 82.Dowd SE, Callaway TR, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125–32. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Durso LM, Harhay GP, Smith TP, Bono JL, Desantis TZ, Harhay DM, et al. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl Environ Microbiol. 2010;76:4858–62. doi: 10.1128/AEM.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hristov AN, Callaway TR, Lee C, Dowd SE. Ruminal bacterial, archaeal, and fungal diversity of dairy cows with normal and reduced ruminal fauna. J Anim Sci. 2012;90:4449–57. doi: 10.2527/jas.2011-4624. [DOI] [PubMed] [Google Scholar]
- 85.Callaway TR, Dowd SE, Edrington TS, Anderson RC, Krueger N, Bauer N, et al. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J Anim Sci. 2010;88:3977–83. doi: 10.2527/jas.2010-2900. [DOI] [PubMed] [Google Scholar]
- 86.Hook SE, Steele MA, Northwood KS, Wright ADG, McBride BW. Impact of high-concentrate feeding and low ruminal pH on methanogens and protozoa in the rumen of dairy cows. Microb Ecol. 2011;62:94–105. doi: 10.1007/s00248-011-9881-0. [DOI] [PubMed] [Google Scholar]
- 87.Davies DR, Theodorou MK, Lawrence MIG, Trinci APJ. Distribution of anaerobic fungi in the digestive tract of cattle and their survival in faeces. J Gen Microbiol. 1993;139:1395–400. doi: 10.1099/00221287-139-6-1395. [DOI] [PubMed] [Google Scholar]
- 88.Klieve AV, Gregg K, Bauchop T. Isolation and characterization of lytic phages from Bacteroides ruminicola ss brevis . Curr Microbiol. 1991;23:183–7. [Google Scholar]
- 89.Martin SA, Nisbet DJ. Effect of direct-fed microbials on rumen microbial fermentation. J Dairy Sci. 1992;75:1736–44. doi: 10.3168/jds.S0022-0302(92)77932-6. [DOI] [PubMed] [Google Scholar]
- 90.Keen J, Elder R. Commercial probiotics are not effective for short-term control of enterohemorrhagic Escherichia coli O157 infection in beef cattle. In: 4th International Symposium and Workshop on Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infection, editor. Kyoto, Japan: VTEC; 2000. p. 92. [Google Scholar]
- 91.Swyers KL, Carlson BA, Nightingale KK, Belk KE, Archibeque SL. Naturally colonized beef cattle populations fed combinations of yeast culture and an ionophore in finishing diets containing dried distiller’s grains with solubles had similar fecal shedding of Escherichia coli O157:H7. J Food Prot. 2011;74:912–18. doi: 10.4315/0362-028X.JFP-10-484. [DOI] [PubMed] [Google Scholar]
- 92.Ohya T, Akiba M, Ito H. Use of a trial probiotic product in calves experimentally infected with Escherichia coli O157. Japan Agric Res Quart. 2001;35:189–94. [Google Scholar]
- 93.Lema M, Williams L, Rao DR. Reduction of fecal shedding of enterohemorrhagic Escherichia coli O157:H7 in lambs by feeding microbial feed supplement. Small Rum Res. 2001;39:31–9. doi: 10.1016/s0921-4488(00)00168-1. [DOI] [PubMed] [Google Scholar]
- 94.Arthur TM, Bosilevac JM, Kalchayanand N, Wells JE, Shackelford SD, Wheeler TL, et al. Evaluation of a direct-fed microbial product effect on the prevalence and load of Escherichia coli O157:H7 in feedlot cattle. J Food Prot. 2010;73:366–71. doi: 10.4315/0362-028x-73.2.366. [DOI] [PubMed] [Google Scholar]
- 95.Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. Isolation of bovine intestinal Lactobacillus plantarum and Pediococcus acidilactici with inhibitory activity against Escherichia coli O157 and F5. J Appl Microbiol. 2009;106:393–401. doi: 10.1111/j.1365-2672.2008.03959.x. [DOI] [PubMed] [Google Scholar]
- 96.Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol. 2001;71:1–20. doi: 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 97.Gallo RL, Murakami M, Ohtake T, Zaiou M. Biology and clinical relevance of naturally occurring antimicrobial peptides. J Allergy Clin Immunol. 2002;110:823–31. doi: 10.1067/mai.2002.129801. [DOI] [PubMed] [Google Scholar]
- 98.Welman AD, Maddox IS. Exopolysaccharides from lactic acid bacteria: perspectives and challenges. TRENDS Biotechnol. 2003;21:269–74. doi: 10.1016/S0167-7799(03)00107-0. [DOI] [PubMed] [Google Scholar]
- 99.Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. 2010;1:440–64. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 100.Nes IF, Holo H. Class II Antimicrobial peptides from lactic acid bacteria. Biopolymers. 2000;55:50–61. doi: 10.1002/1097-0282(2000)55:1<50::AID-BIP50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 101.Franz CM, Van Belkum MJ, Holzappfel HW, Abriouel H, Gálvéz A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev. 2007;31:293–310. doi: 10.1111/j.1574-6976.2007.00064.x. [DOI] [PubMed] [Google Scholar]
- 102. Franz CM, Stiles ME, Schleifer KH, Holzappfel WH. Enterococci in foods – a coriandrum for food safety. Int J Food Microbiol. 2003;88:105–22. doi: 10.1016/s0168-1605(03)00174-0. [DOI] [PubMed] [Google Scholar]
- 103.Franz CM, Huch M, Abriouel H, Holzappfel W, Gálvéz A. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol. 2011;151:125–40. doi: 10.1016/j.ijfoodmicro.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 104.Lauková A, Mareková M, Javorský P. Detection and antimicrobial spectrum of a bacteriocin-like substance produced by Enterococcus faecium CCM 4231. Lett Appl Microbiol. 1993;16:257–60. [Google Scholar]
- 105.Lauková A, Czikková S, Vasilková Z, Juriš P, Mareková M. Occurrence of bacteriocin production among environmental enterococci. Lett Appl Microbiol. 1998;27:178–82. doi: 10.1046/j.1472-765x.1998.00404.x. [DOI] [PubMed] [Google Scholar]
- 106.Lauková A, Mareková M, Štyriak I. Inhibitory effect of different enterocins against fecal bacterial isolates. Berl Muench Tierarztl Wochensschr. 2003;116:37–40. [PubMed] [Google Scholar]
- 107.Mareková M, Lauková A, De Vuyst L, Skaugen M, Nes IF. Partial characterization of bacteriocins produced by environmental strain Enterococcus faecium EK13. J Appl Microbiol. 2003;94:523–30. doi: 10.1046/j.1365-2672.2003.01861.x. [DOI] [PubMed] [Google Scholar]
- 108.Mareková M, Lauková A, Skaugen M, Nes IF. Isolation and characterization of a new bacteriocin, termed enterocin M, produced by environmental isolate Enterococcus faecium AL41. J Ind Microbiol Biotechnol. 2007;34:533–7. doi: 10.1007/s10295-007-0226-4. [DOI] [PubMed] [Google Scholar]
- 109.Simonová MP, Lauková A, Plachá I, Čobanová K, Strompfová V, Szabóová R, et al. Can enterocins affect phagocytosis and glutathione-peroxidase in rabbits? Cent Eur J Biol. 2013;8:730–4. [Google Scholar]
- 110.Szabóová R, Chrastinová L, Lauková A, Haviarová M, Simonová M, Strompfová V, et al. Bacteriocin producing strain E. faecium CCM4231 and its use in rabbits. Int J Probiotics Prebiotics. 2008;3:77–82. [Google Scholar]
- 111.Simonová MP, Lauková A, Chrastinová L, Strompfová V, Faix Š, Vasilková Z, et al. Enterococcus faecium CCM 7420, bacteriocin PPB CCM 7420 and their effect in the digestive tract of rabbits. Czech J Anim Sci. 2009;54:376–86. [Google Scholar]
- 112.Lauková A, Chrastinová L, Pogánysimonová M, Strompfová V, Plachá I, Čobanová K, et al. Enterococcus faecium AL41, its Enterocin M and their beneficial use in rabbits husbandry. Probiotics Antimicrob Proteins. 2012;4:243–9. doi: 10.1007/s12602-012-9118-7. [DOI] [PubMed] [Google Scholar]
- 113.Lauková A, Guba P, Nemcová R, Mareková M. Inhibition of Salmonella enterica serovar Duesseldorf by enterocin A in gnotobiotic Japanese quails. Vet Med Czech. 2004;49:47–51. [Google Scholar]
- 114.Lauková A, Strompfová V, Skřivanová V, Volek Z, Jindřichová E, Marounek M. Bacteriocin-producing strain of Enterococcus faecium EK13 with probiotic character and its application in the digestive tract of rabbits. Biol Bratislava. 2006;61:779–82. [Google Scholar]
- 115.Metchnikoff E. The prolongation of life: optimistic studies. Broadway, NY: Springer; 1907. [Google Scholar]