Abstract
The intestinal microbiota has become a relevant aspect of human health. Microbial colonization runs in parallel with immune system maturation and plays a role in intestinal physiology and regulation. Increasing evidence on early microbial contact suggest that human intestinal microbiota is seeded before birth. Maternal microbiota forms the first microbial inoculum, and from birth, the microbial diversity increases and converges toward an adult-like microbiota by the end of the first 3–5 years of life. Perinatal factors such as mode of delivery, diet, genetics, and intestinal mucin glycosylation all contribute to influence microbial colonization. Once established, the composition of the gut microbiota is relatively stable throughout adult life, but can be altered as a result of bacterial infections, antibiotic treatment, lifestyle, surgical, and a long-term change in diet. Shifts in this complex microbial system have been reported to increase the risk of disease. Therefore, an adequate establishment of microbiota and its maintenance throughout life would reduce the risk of disease in early and late life. This review discusses recent studies on the early colonization and factors influencing this process which impact on health.
Keywords: gut, microbiota, neonate, diet, maternal
The microbiota is composed of a significant number of different bacteria, approximately 160 species per person per fecal sample, and this ecosystem plays an important role in human health. The microbial colonization of the infant gut is known to play a key role in immunological and metabolic pathways impacting on human health. Disruptions during this complex process of microbial colonization have been shown to increase the disease susceptibility during life. Alteration in the compositional development of the gut microbiota of a newborn has been demonstrated in a few studies to predispose to diseases later in life. This review highlights the recent observations on the importance of microbial exposition during early life and the microbial development throughout life. This microbial colonization process promotes short- and long-term health benefits and different factors that modify the microbial composition.
Microbial colonization of the infant gut
Colonization of the infant gastrointestinal (GI) tract by microbes is an essential process in our life cycle since microbiota–host interactions have an important influence on human health and disease. Since the studies of Tissier (1) concerning the acquisition of the infant gut microbiota, the idea that fetuses are sterile in utero and that microbial colonization of the newborn starts during and after birth has been widely accepted. More than a century later, the hypothesis that the placenta barrier keeps fetuses sterile throughout a healthy pregnancy still remains a general dogma and, as a consequence, the presence of any bacteria in the uterus is generally considered as a potential danger for the fetus. This view arises from the fact that, during decades, microbiological analyses of pregnancy-related biological samples (chorioamnion, amniotic fluid, and meconium) were only performed in cases where an intrauterine infection was evident or suspected. Indeed, several studies have found a strong correlation between preterm deliveries and intrauterine infections (2, 3), the leading cause of infant mortality worldwide (4).
In contrast, relatively few studies have examined the uterine microbiota associated with healthy term pregnancies, partly because of the enduring influence of the sterile womb paradigm, and also due to the technical and ethical issues of collecting samples from healthy pregnancies before birth. However, investigations into the potential for bacterial transmission through the placental barrier have detected bacteria in placenta tissue (5), umbilical cord blood (6), amniotic fluid (7–9), and fetal membranes (9, 10) from healthy newborns without any indication of infection or inflammation.
Prenatal microbial contact: is there a fetal gut microbiota?
Meconium is not sterile, as was previously assumed, since it harbors a complex microbial community (8, 11–14). A recent study characterized the microbiota of meconium and fecal samples obtained from preterm babies during the first 3 weeks of life using culture-dependent and culture-independent techniques (12). Both approaches provided similar results and showed that spontaneously released meconium of such neonates contains a specific microbiota that differs from those observed in early fecal samples. Firmicutes was the main phylum detected in meconium while Proteobacteria was abundant in feces. Culture-based techniques showed that staphylococci predominated in meconium while enterococci and certain gram-negative bacteria, such as Escherichia coli, Klebsiella pneumoniae, or Serratia marcescens, were more abundant in fecal samples. In addition, 16S rRNA gene-based microarrays revealed the high prevalence of bacteria related to Streptococcus mitis and Lactobacillus plantarum in meconium, whereas those related to E. coli, Enterococcus, and K. pneumoniae predominated in the infant feces. In another study, the diversity of the meconium microbiome was assessed by multi-barcode 16S rRNA gene sequencing using samples collected from 23 newborns stratified by maternal diabetes status (13). All meconium samples contained a diversified microbiota, which was not affected by the mode of delivery; in comparison with adult feces, meconium samples showed a lower species diversity and a higher sample-to-sample variation. Taxonomic analyses suggested that the overall bacterial content in meconium significantly differed by maternal diabetes status. Specifically, the phyla Bacteroidetes and the genus Parabacteroides were enriched in those samples of the diabetes group (13). It has also reported that meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and later respiratory problems in infants (11).
While vaginal microbes associated with preterm birth can get access to the uterine environment through an ascending route, the mechanisms by which gut bacteria reach this human niche are not well understood. It has been suggested that bacteria travel to the placenta via the bloodstream after gut epithelium translocation. While the intestinal epithelial barrier generally prevents microbial entry into the circulatory system, dendritic cells can actively penetrate the gut epithelium, take up bacteria from the intestinal lumen, and transport the live bacteria throughout the body as they migrate to lymphoid organs (15, 16). To test whether maternal gut bacteria can be transferred to fetuses in utero, two pioneer studies investigated if oral administration of a genetically labeled Enterococcus faecium to pregnant mice resulted in its presence in amniotic fluid and meconium of term offspring after sterile cesarean section (C-section) (6, 8). Remarkably, E. faecium with the genetic label was cultured from the amniotic fluid and meconium of pups from inoculated mothers, but not from pups of control mice. Thus, these studies provided foundational evidence for maternal microbial transmission in mammals. Interestingly, physiological bacterial translocation is highly increased during pregnancy and lactation in rodents (17).
Globally, the dogma that the fetus resides in a sterile environment is being challenged by recent findings, and clearly, the question has arisen whether microbes that colonize the fetus may be related to a better or worse pregnancy outcome and to the future health status of the neonate. Thus, some microorganisms colonizing the placenta, such as Prevotella and Gardnerella, may provoke distinctive newborn inflammatory responses while others, such as Lactobacillus, may suppress these responses (18, 19). Moreover, a previous study carried out in pregnant women focused on the influence of oral microbiota composition in the pregnancy outcome and showed that some bacteria, such as Actinomyces naselundii, were associated with lower birth weight and earlier delivery, while others, such as lactobacilli, were linked with a higher birth weight and later delivery date (20). Thus, it is possible that oral bacteria can enter the uterine environment through the bloodstream and may influence the delivery process. These findings are also supported by recent extensive deep sequencing studies in normal healthy term pregnancies, identifying a low abundance, but metabolically rich placental microbiome, with a composition resembling the oral microbiome more than the vaginal, fecal, skin, or nasal microbiomes (5). In a recent review, maternal microbial transmission has been reported to occur in all animal kingdoms (21). ‘Heirloom’ microbes received from the mother may be uniquely evolved to the offspring’s genotype and vertical as compared with horizontal transmission may increase the chance for optimal mutualism (21).
We still know very little about the identity and number of microbes that traverse the placenta, whether they persist in the infant or whether their presence has short- or long-term health consequences. The advent of high-throughput system biology approaches will lead to a characterization of the ‘fetal microbiome’ in utero and its health connotations, including effects on immune imprinting.
Infant microbial colonization process
Microbial contact during prenatal life may imprint the offspring microbiota and immune system in preparation for the much larger inoculum transferred during vaginal delivery and breastfeeding. A remarkably wide diversity of bacteria can colonize the child when exposed to the postnatal environment – being reflected by a high inter-individual diversity in the gut microbiota composition of newborns (22, 23). Advances in metagenomic technologies have revealed the composition of the human gut microbiota from early infancy (22) through to old age (24).
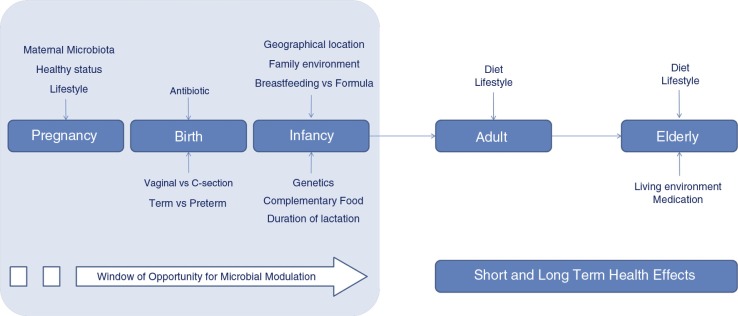
Following birth, the human intestine is rapidly colonized by an array of microbes and factors known to influence colonization include gestational age, mode of delivery (vaginal birth vs. assisted delivery), diet (breast milk vs. formula), sanitation, and antibiotic treatment (25, 26). First colonizers, facultative anaerobes, create a new environment that promotes the colonization of strict anaerobes as Bacteroides, Clostridium, and Bifidobacterium spp. The intestinal microbiota of neonates is characterized by low diversity and a relative dominance of the phyla Proteobacteria and Actinobacteria, with the microbiota becoming more diverse with the emergence and dominance of Firmicutes and Bacteroidetes as time after birth increases (27–29). By the end of the first year of life, infants possess an individually distinct microbial profile, converging toward the characteristic microbiota of an adult, such that by 2–5 years of age, the microbiota fully resembles that of an adult in terms of composition and diversity (22, 30, 31). Therefore, the first 3 years of life represent the most critical period for dietary interventions to improve child growth and development. This is the period when the intestinal microbiota, a vital asset for health and neurodevelopment (32) is established and its alteration during this period has the potential to profoundly affect host health and development. Gut microbiome development has been studied far more than that in other body habitats and oral, skin and respiratory infant microbial colonization processes are still being uncovered. Gut microbiota development is affected by different factors such as mode of delivery, diet, genetics, health status, gestational age, etc. (Fig. 1).
Fig. 1.
Factors influencing the infant gut microbiota development and the adult and elderly microbiota. The first 3 years of life represents the most critical period for dietary interventions aimed at microbiota modulation to improve child growth and development and positively affect health.
Factors influencing infant microbiota development
Mode of delivery
Initial postnatal microbial exposure occurs during and shortly after birth. Vaginally delivered infants are colonized by maternal vaginal and fecal bacteria, including Lactobacillus and Bifidobacterium spp. (33). Infants born via C-section are not directly exposed to maternal microbes and are instead colonized with microbes associated with the skin and the hospital environment. It is suggested that the microbiota composition in these infants may remain disturbed for months or even years (34, 35). During vaginal delivery there is an extensive transmission of vaginal bacteria from the mother to the child. The vaginal microbiota has a low diversity, being mainly composed of lactobacilli (36). During the first few days of life there is a relatively high load of lactobacilli in the neonatal gut, probably reflecting the vaginal microbiota (23). Human anatomy also supports that exposure to the mother’s fecal microbiota is an important transmission route during delivery (37). The facultative anaerobic Enterobacteriaceae represents one of the bacterial groups with the most probable direct transmission from mother to child through feces (38). It has also been suggested that Bacteroides fragilis is transmitted during, or just after, delivery (39). Vaginally delivered infants, but not infants born by C-section, shared a significantly higher proportion of gut microbiota 16S rRNA gene sequences with their own mother than with other mothers for up to 2 years of age, particularly within the Bacteroidetes and Firmicutes phyla (35). As these anaerobic phyla do not appear to grow outside the gut, it is probable that they are transmitted between human hosts (40).
It has also been argued that strictly anaerobic clostridia are unlikely to be transmitted directly during delivery due to the oxygen tension in the gut at birth. Although vegetative cells of human-derived clostridia are highly sensitive toward oxygen, the spore form survive harsh conditions – such as treatment with 3% chloroform – and are still able to colonize the gut of gnotobiotic mice (41). Spores would therefore provide an avenue for transmission at a later stage. Further support for lack of direct transmission of strictly anaerobic clostridia comes from the finding that clostridia were highly overrepresented in mothers, but not in their children up to the age of 4 months (23, 42).
It has been known from the mid-1980s that breaking the fecal and vaginal transmission route by C-section has a major impact on the infant gut microbiota (43). Despite major methodological changes and improvements, the main conclusion that C-section delays Bacteroidetes colonization still holds true (35). In recent reports, it has also been shown that microbiota diversity is decreased with C-section, compared to vaginal delivery (35).
Another important early source of colonizing microbiota is through the contact with the mother’s skin (44). Skin-associated bacteria belonging to the Staphylococcus genus, which is also widespread among different human mucosal surfaces, are one of the earliest colonizers of the human infant (45); they are rapidly outcompeted by other bacteria with age.
Infant diet
Another relevant and strong influence in the infant gut microbial development is the mode of feeding. In a single case study, the infant microbiota development was followed from birth to 2.5 years of age and results demonstrated a clear dietary influence on the microbiota composition (30). A major source for bacterial colonization of the infant gut is through bacteria in the mother’s milk, and it has been proposed that this mode of colonization plays a major role in the child’s health status (46). The mother’s milk microbiota mainly includes streptococci and staphylococci followed by other bacteria (47), which are among the earliest colonizers of the infant gut (22, 23). Breast-fed infants are exposed to the milk microbiota which has been reported to contain more than 700 species of bacteria (48). A higher maternal body mass index (BMI) was associated with lower microbial diversity in this study, as was emergency C-section compared with planned C-sections and vaginal birth. Breast milk also contains an abundance of complex oligosaccharides with prebiotic activity, stimulating the growth of specific bacterial groups such as staphylococci (47) and bifidobacteria (49). Gut microbiota diversity increases following weaning and introduction of solid food, with enhanced colonization of butyrate producers, including Bacteroides and certain Clostridium species (30, 50). The oral microbiota consists of a highly diverse assemblage of microorganisms (51). The child’s gut microbiota is also exposed to the mother’s and its own oral microbiota – either directly or indirectly through mother’s milk (48). Recent evidence suggests that oral bacteria can colonize the gut of infants, and as an example, oral Bifidobacterium dentium has been found to be both highly abundant and prevalent in the infant gut (52).
Antibiotics and preterm birth
Perturbation of optimal microbiota development, arising from preterm birth or antibiotics has likely long-term implications for microbial diversity and consequent health. In preterm infants, the microbiota is characterized by reduced diversity and higher levels of potentially pathogenic bacteria and lower numbers of Bifidobacterium and Bacteroides compared with full-term infants (53). Even short-term antibiotic treatment can significantly affect the evolution of the infant gut microbiota; in fact, the colonization pattern of Bifidobacterium seems to be particularly disturbed up to 8 weeks after treatment while Proteobacteria are increased (54). Preterm delivery is often confounded with both C-section and antibiotic usage since these three factors are often associated. Independent of these confounders, however, the microbiota of preterm infants is distinct to that of infants delivered at term. The main patterns are increased Enterococcus, together with Proteobacteria. In addition, there are lower levels of bifidobacteria compared to that expected for children delivered at term (12). Preterm birth is also associated with neonatal necrotizing enterocolitis (NEC), a severe inflammatory intestinal disorder. NEC has recently been linked to the microbiota, where high levels of Proteobacteria are predictive of the disease (55).
Environment and lifestyle
The family members and close relatives (siblings) have also been described as a relevant environmental factor influencing infant gut microbiota colonization. Thus, siblings have higher proportion of Bifidobacterium spp. than single infants (56). Geographical location also has an impact on the microbiota, as microbiota differences are related to dietary patterns and lifestyle in a specific area (city, town, country, religion, etc.). It has been reported that a ‘geographical gradient’ exists in the European infant microbiota where infants from Northern areas have higher levels of Bifidobacterium spp. and some Clostridium spp. and Atopobium spp., while Southern infants had a higher abundance of Eubacteria, Lactobacillus, and Bacteroides (50). Significant differences between the microbiota of Finnish and German infants (57) or between that of Estonian and Swedish ones (58) have been also reported. In the first study, the proportion of Bifidobacterium spp. was higher in the Finnish than in the German infants, who showed a higher abundance of the Bacteroides–Prevotella group and Akkermansia muciniphila. Another study reported higher levels of Bifidobacterium spp., Bacteroides–Prevotella, and Clostridium histolyticum in Malawi infants than Finnish infants at the age of 6 months (59). Furthermore, the gut microbiota of infants from Burkina Faso (rural village) was characterized by an enrichment of Bacteroides, while Enterobacteriaceae was predominant in Italian children (60). In a recent study, the healthy infant microbiota from the Amazonas of Venezuela, rural Malawi and US metropolitan was compared (31). Shared patterns of gut microbiome development were identified during the first year of life in all populations, and bifidobacteria was the most prevalent group, dominating the infant microbiota of all three groups during this period.
Host genetics
The relative contribution of the host genetics in shaping the gut microbial structure and function is not yet clearly defined and remains a subject of ongoing debate. Our understanding of the importance of host genetics in this regard is derived from studies in humans, animals, and comparative species analysis (61).
In humans, the most valuable information is derived from studies of twins and family relatedness. A DNA-based finger-print analysis of the microbiota of human adults with varying degree of relatedness ranging from parents and children, non-twin siblings, monozygotic twins, and unrelated subjects confirmed that host genotype has a significant effect on the composition of the dominant bacteria, with monozygotic twins demonstrating the highest similarity in their microbiota (62). These results were later substantiated by reanalysis using molecular profiling methods (63). A study with children under the age of 10 indicated that the degree of similarity in the bacterial community was higher in identical compared with non-identical twins and was lowest in the unrelated control group (64). In contrast, a metagenomics study using deep sequencing of samples from 31 monozygotic and 23 dizygotic twin pairs and their parents did not identify significant differences in bacterial diversity between the two types of twins, although the members of the same family were found to share a higher numbers of bacterial phylotypes (65, 66). It has been suggested that if the impact of host genotype on microbiota is less pronounced, detection of significant differences in healthy populations may require analysis of larger cohorts (67).
Defining host genotype–microbiota interactions in humans is complicated due to the well-established influence of diet and other environmental and maternal factors on the structure of the microbiota (see ‘Factors influencing infant microbiota development’ section). The use of genetically inbred lines of animals and germ-free and specific knock-out mice can overcome such confounding factors (68, 69). Eight different recombinant inbred lines, reflecting the genetic diversity found in humans, were profiled using ARISA-based fingerprinting methods (70). This study showed that genetic background significantly altered the composition of the microbiota but independently of the mouse gender, a somewhat surprising finding since differences in hormonal types and levels would be expected to have an influence on the gut microbiota. Using parental BALB/c and C57BL/6J mice and their reciprocal cross F1 hybrid, Buhnik-Rosenblau et al. (71) showed that host genetics had a major impact on the composition and level of colonization of the mouse gut by Lactobacillus johnsonii, a potential probiotic bacterium.
However, the molecular mechanisms by which specific host genes are responsible for shaping the gut microbiota are so far mainly derived from single gene studies although several genome-wide association studies are beginning to provide detailed insights into host genotype–microbiota interactions (67, 72). In another study, an Apoe-I knockout mouse with impaired glucose tolerance was used to assess the interaction between host genetics, diet, and the development of metabolic syndrome. Molecular profiling with pyrosequencing of 16S rRNA genes revealed that the Apoe-I genetic mutation contributed to 12% of the total structural variation in the gut microbiota, whereas dietary changes contributed 57% of the observed microbiota modulation (73). The effect of leptin deficiency resulting in obese phenotype has been extensively studied and shown to have a major impact on the gut microbiota composition, with the obese mice showing deceased levels of Bacteroides spp. (74). Understanding specific host traits associated with microbial interactions provides invaluable clues to identify the underlying mechanisms of microbiota modulation. Modulation via the immune system has already been suggested, but other host-genome polymorphisms leading to an alteration of the gut microbiota structure and function in a host-dependent manner also need to be considered. These include, among others, mucus production and mucin glycan modification (see next section ‘Mucin glycosylation’), bile metabolism (75), host-derived antimicrobial peptides (76), and mammalian hormones (77, 78).
There is now clear evidence that the host genotype influences the structure of the gut microbiota and even single gene mutations can lead to alterations of the microbiota composition. Detailed genome-wide association studies in humans are needed to provide a greater understanding of the mechanisms underlying gut microbiota and host genotype interactions and guide future microbiota-targeted intervention strategies.
Mucin glycosylation
The mucus layer overlies the intestinal epithelium and forms the anatomical site at which the microbiota first encounters the host. In addition to its association with the protection, lubrication and hydration of the intestinal epithelium, mucus plays a critical role in the maintenance of intestinal homeostasis by promoting microbial interactions with commensal bacteria, acting as decoys for pathogens and enhancing immune regulation (79). The thickness, composition, organization, and glycosylation pattern of the mucus layer varies along the GI tract and correlates with the association of distinct microbial communities with the mucosal surface (80). The major structural components of mucus are mucins, a family of high-molecular-mass glycoproteins (81, 82). To date, 22 human mucins have been identified and can be classified into two main categories, secreted and membrane-bound mucins (83). In the intestine, MUC2 is the predominant secretory mucin produced by goblet cells. Eight core structures of the mucin O-glycan chain have been identified (84), with core-1-4 glycans most commonly found in intestinal mucins (Fig. 2).
Fig. 2.
(a) Classification of human mucins into secreted (gel-forming and non-gel-forming) and membrane-bound mucins (MUCs). (b) The four common mucin type O-glycans (core-1-4) found in the GI.
The mucus-binding capacity of microbes increases their colonization ability at the mucosal interface and prolongs intestinal residency of beneficial microbes (85). Furthermore, bacterial adhesion to mucin glycans may be a mechanism by which the host selects bacterial species that present the complementary set of adhesins (86). The link between glycosylation of the intestinal mucus layer and microbiota composition has been highlighted in studies using mouse models deficient in specific mucin glycosyltransferase genes (87–93). Furthermore, recent work showed that host-derived fucose signaling, activated through the microbiota, can modulate pathogenic intestinal colonization (94, 95) indicating that mucin glycosylation has a selective capacity on microbial ecology. The loss of the α-1,2-fucosyltransferase FUT2, and therefore fucosylated host glycans, leads to a decreased diversity and differences in community composition in mice (87), whereas an association between the composition of the intestinal microbiota and the ABO blood group or FUT2 secretor status was reported in humans (96, 97). Host glycan-mediated alteration in the microbiota is also associated with increased susceptibility to infection and inflammatory bowel diseases (IBD) (98–100). These findings may explain the region-specific colonization of the GI tract and the likely influence of mucin glycosylation in selecting successive colonization of the intestine by gut bacteria throughout life. Earlier work investigating mucin gene expression in human embryonic and fetal intestine showed that mucin genes are expressed from the early mid-trimester of gestation and aberrations in expression may play a role in the development of intestinal disease (101–103).
More recently, MUC2 was implicated in the etiology of NEC, the most common GI cause of morbidity in premature infants (104). Goblet cells are significantly decreased in NEC, and the presence of functional MUC2 plays a critical role in the prevention of this disease (105). It is also important to note that abnormal gut colonization is a hallmark of NEC (106, 107), and that human milk (whose oligosaccharide structures are similar to mucin glycans) is associated with reduced NEC risk compared to formulas (108, 109), supporting a particular role for mucin-type O-glycans in microbial colonization.
Both human adult and fetal intestinal samples have been shown to contain the sialic acid Neu5AC as their most abundant component, with the level of expression of Neu5,8Ac2 in fetuses being higher compared to the adult intestine (110). In contrast to human adult intestinal mucins, an increasing gradient of sialic acid or decreasing gradient of fucose was not observed from ileum to distal colon in the fetal intestine (110–112). These findings suggest that region-specific glycosylation of the human intestine is acquired after birth, probably due to bacterial colonization and gut postnatal absorptive and digestive functions. Indeed, several studies have reported the influence of major commensal bacteria on the production of mucus, mucin glycans, and the development of goblet cells (113–115). Microbial profiling during the process of microbial colonization in germ free mice showed that the abrupt increase in the ratio of fucosylated to sialylated glycans during the initial stage of colonization correlates with the establishment of Bacteroidetes members (116) known to sense and regulate fucose (94, 95, 117). Further studies in germ free animals suggested that the simultaneous establishment of lactate-producing (e.g. Bifidobacterium adolescentis) and sulfate-liberating (e.g. Bacteroides spp.) bacteria during the initial stage of microbial colonization, as well as the availability of sulfated mucins, promoted a bloom in sulfate-reducing bacteria (118, 119), as reviewed recently by El Aidy et al. (120). Altogether these studies provide an insight into how bacterial communities adapt to changing mucin glycan profiles throughout life, and a possible mechanism of niche-development. However, a substantial amount of research is necessary to define the causal relationship between mucus development and bacterial succession at a mechanistic level. This link is of particular importance to increase our understanding of intestinal diseases where alterations in mucus secretion, mucin glycosylation, and microbial involvement are apparent, in particular in IBD and pre-malignant and malignant lesions.
Impact on development of child health
The world is experiencing a progressive increase of metabolic and immune mediated diseases, with a dramatically high increase in infant populations. This increase may be related to a parallel increase in the rates of C-section deliveries, which has exponentially increased far beyond the 15% recommended by the World Health Organization. Indeed, birth by C-section has been associated with the development of allergy and asthma, as well as type I diabetes, celiac disease and obesity (121–124). Allergic diseases comprise the most common chronic disease in childhood while obesity is the most prevalent nutritional problem in Western countries.
Allergic diseases
Allergic diseases have become a major public health problem in affluent societies (125). As changes in the genotype cannot explain such a rapid increase in the allergy prevalence, loss of protective factors or/and new risk environmental factors may contribute to the increased prevalence of these diseases since the middle of the last century (125). A reduced intensity and diversity of microbial stimulation may have resulted in an abnormal immune maturation in early childhood (126, 127). The developing neonatal immune system may depend on anticipated diverse inputs to mature normally, much like the central nervous system does (128). As other immune-mediated diseases such as multiple sclerosis, type I diabetes, and Crohn’s diseases also show rising incidences in economically developed countries (126, 128, 129), a theory of ‘microbial deprivation syndromes of affluence’ has been proposed (126, 127). A limited microbial pressure, resulting in insufficient T cell induction with regulatory and/or Th1-like properties to counteract allergy-inducing Th2 responses, may underlie this allergy epidemic (126, 130, 131).
The Th2-skewed state of the neonatal immune system (126, 132) is likely a consequence of the fetal immune environment during pregnancy (133). The fetal–maternal interface is characterized by high levels of Th2-like (134, 135) and anti-inflammatory (136) cytokines, as well as enrichment of T regulatory cells (137), most likely functioning to divert the maternal immune response away from damaging Th1-mediated immunity (138, 139). This neonatal Th2-skewing is even more marked in infants later developing allergic disease (132, 140, 141), supporting the notion that the prenatal immune environment can influence allergy development (133, 142). The neonatal Th2-bias should then develop toward a more balanced immune phenotype, including appropriate development of regulatory T cell responses (126) and maturation of Th1-like responses (131, 132). A failure of Th2-silencing during immune system maturation may underlie development of Th2-mediated allergic disease (126, 132, 143). Appropriate pre- and postnatal microbial stimulation may be needed to avoid this pathophysiological process (144, 145). In this respect, the gut microbiota is quantitatively the most important source of microbial stimulation and may provide a primary signal for the maturation of a balanced postnatal innate and adaptive immune system (126, 127, 146–150).
Early gut microbiota differences between infants who later do or do not develop allergic disease have been reported (151–160). There is considerable inconsistency between the results from the different studies and large differences in the microbiological techniques employed, however (151–161). It is also important to bear in mind that assessments of stool samples reflect luminal colonic microbiota and not necessarily the colonization of the small intestine, where the major part of the gut immune system is situated (162, 163). No specific microbes with consistently harmful or allergy protective roles have yet been identified; however, some studies have found associations between low rates of bifidobacteria (151, 152, 156, 164) or lactobacilli (156, 164) colonization during infancy and later allergy development. In addition, some studies have found evidence for early Clostridium difficile colonization as a risk factor for later allergy development (151, 153, 165, 166). As C. difficile is an opportunistic pathogen, expanding when gut microbiota niches are vacant (44), the increased detection rate of this bacterium in infants later developing allergy suggest that disruptions in infant gut microbial ecology precede disease development.
Early establishment of a diverse gut microbiota may be more important than the distribution of specific microbial species in shaping a normal immune maturation, with repeated exposure to new bacterial antigens enhancing the development of immune regulation (44, 126, 127, 167). In support of this theory, studies using cultivation-independent molecular techniques such as denaturing gradient gel electrophoresis (155, 157) or terminal restriction fragment length polymorphism (154, 159) of bacterial 16S rRNA phylotypes, demonstrated a low gut microbiota diversity during infancy in children developing eczema (154, 155, 159) or sensitization (157). The diversity within specific microbial phyla or genera cannot be identified with these types of molecular methods, however, and the sensitivity appears low as the median number of peaks/bands was much lower than the expected number of bacterial species (154, 155, 157, 159). Employing more sensitive and powerful next-generation high-throughput barcoded 16S rRNA gene 454-pyrosequencing, we could demonstrate that development of atopic eczema was preceded by a low diversity of the total microbiota at 1 month and a low diversity of the bacterial phylum Bacteroidetes and the genus Bacteroides at 1 month (158). Interestingly, a Western lifestyle with a low dietary fiber intake has been associated with a gut microbiota depleted of Bacteroidetes (60). Mice that fed a high-fiber diet had increased Bacteroidetes abundance, leading to enhanced circulating short-chain fatty acid levels and protection against allergic inflammation in the lung (168). Furthermore, delivery by C-section, a risk factor for allergy development (124, 127), was associated with decreased relative abundance and diversity within the Bacteroidetes phylum during the first year of life and less frequent detection of the major genus Bacteroides (35). We also demonstrated that the presence of the genus Bacteroides at 1 month was associated with high levels of the Th1-associated chemokines CXCL10 and CXCL11 during infancy (35). Further supporting the concept of the importance of early gut microbiota diversity in driving normal immune maturation, we recently found that children developing asthma at school age had a lower diversity of the total microbiota than non-asthmatic children at 1 week and 1 month of age (169). Speculatively, as viral lower respiratory tract infections (LRTIs) are particularly linked to asthma development among atopic children (125), a reduced mucosal barrier function could be a consequence of a less diverse microbial stimulation. This may be linked to high susceptibility of LRTIs, amplification of Th2 responses, and subsequent asthma development (125).
The continuously decreasing cost/throughput ratio of current sequencing platforms will allow analysis of the gut microbiota in larger pediatric cohorts with increased sensitivity and depth to further establish how the infant gut microbiota shapes immune and allergy development (127). Novel next-generation probiotic candidates may be identified in such studies, for example, indigenous gut bacteria-producing Treg-promoting short-chain fatty acids (170–172) and immunomodulatory Bacteroides strains (173). Probiotic interventions have so far failed to prevent asthma (174–177), although promising preventive effects have been observed for atopic eczema (127, 178–180). The preventive effects on eczema have primarily been observed in studies with combined pre- and postnatal administration with probiotic lactobacilli, although prenatal probiotic supplementation has not been given until the last trimester of pregnancy (127, 145, 178–181). If prenatal microbial exposure is vital for the preventative effect, the use of probiotics during the last trimesters of pregnancy, when circulating fetal T cells have developed, may have a relevant role in allergy and asthma prevention (145, 181). Such studies using traditional probiotics are now ongoing and similar interventions with next-generation probiotics may be tested in the future.
Obesity and related disorders
Obesity is among the major current health problems, which are increasing rapidly worldwide. Obesity is associated with a heightened risk of Western lifestyle diseases such as type 2 diabetes, cardiovascular diseases, sleep, and apnea, and also increases the mortality risk (182). The role of the gut microbiota in the development of these conditions (also known as metabolic syndrome) is discussed in a separate review in this supplement (dysbiosis of the gut microbiota in disease). However, there is evidence that the development of metabolic syndrome is affected by the gut microbiota early in life.
Excessive weight increments during the first month of life have been associated with a high risk of obesity development at 3 years of age (183). These results link with observations of early microbial differences in infants developing obesity at school age (184). This study reported lower Bifidobacterium levels and a higher prevalence of Staphylococcus during the first year of life in obese infants compared to those with a normal weight at 7 years of age.
Excessive weight gain may begin during the fetal period, as overweight mothers supply excessive energy to the fetus and problems with weight often persist from childhood to adulthood (185, 186). In obese pregnant women, a vicious circle of non-favorable metabolic development maybe generated if their altered gut microbiota composition is transferred to the infant (187–189). Furthermore, specific shifts in microbial composition were also associated with maternal factors such as BMI, weight, and weight gain over pregnancy (188–189). Infants from women with normal weight gain over pregnancy show higher levels of bifidobacteria than women with excessive weight gain, suggesting the potential role of Bifidobacterium group on infant microbiota and weight development (189). A relationship between the gut microbiome and maternal nutritional status during pregnancy has been reported (189, 190). Interestingly, both studies are supportive of the view that a gut microbiota profile favoring a higher number of Bifidobacterium spp. and a lower proportion of Staphylococcus spp. may provide protection against maternal excess weight development. Furthermore, a lower abundance of Bifidobacterium spp. in the milk microbiota of mothers with atopic disease compared with healthy mothers has been observed (191), and a similar difference is seen between healthy and obese mothers (188). The higher levels of bifidobacteria found in healthy infants are also in accordance with the protective role attributed to breast-feeding against developing obesity and other diseases later in life (192) and the predominance of bifidobacteria in the gut of breast-fed babies (193). The administration of probiotics during pregnancy is under consideration because of the positive effects that some strains exert on certain clinical conditions. It has been reported that probiotic consumption reduced the risk of central adiposity during the following 6 months after delivery (194). Other studies have provided clinical evidence of improved plasma glucose concentrations and insulin sensitivity during pregnancy and also 12 months after delivery when advantageous dietary intake is combined with probiotics (195). Maternal administration of Lactobacillus rhamnosus GG before and after delivery has been reported to induce specific changes in the initial neonatal colonization of bifidobacteria and also influences the breast milk microbiota compared with those receiving placebo (196). Interestingly, nutrition counseling and probiotic intervention were shown to reduce the risk of fetal overgrowth associated with gestational diabetes (197). Perinatal probiotic intervention was able to moderate the increased weight gain mainly in infants who later became overweight during the first years of life (198).
Therefore, microbiota modulation by probiotics early in life is receiving great interest. There is a ‘window of opportunity’ during the first months of life where the microbial colonization and immune system maturation are still in progress. Taken together, long-term health benefits for mothers and children may be conferred by balanced maternal nutrition during pregnancy influencing the infant microbiota and immune system development which impact on early and late health.
The composition and stability of the adult gut microbiota
The specific microbial diversity present in healthy adult subjects plays an important role in maintaining immune homeostasis. This links with the fact that microbiota alterations are related to gut-related diseases (199). Due to the important role of the microbiota in health, studies using next-generation sequencing methodologies are aimed at identifying and characterizing microbial diversity and functionality. While different projects have attempted to identify and describe the microbial composition in healthy subjects, this has been really difficult due to the high variation between individuals, the studied samples, and also, the methodology applied for that purpose. Furthermore, the composition that is typically measured is that which can be isolated from fecal samples, and this does not truly reflect the full diversity of the GI tract. This disconnects between fecal sample analysis, and the true composition of the different niches in the GI tract will need to be bridged if we are to better understand the role of the gut microbiota in human biology. Until we have technologies to bridge this gap, fecal samples and limited biopsy samples will still be used as proxies for the large intestinal microbiota. So as we enter the second decade of research into the gut microbiota, we are still faced with the reality that the descriptions that we provide of the composition and the functions may be misleading, but are currently our best guesses.
The current census, which is collated from numerous studies mainly based in developed countries, is that the gut microbiota in the large intestine is a two-phylum coalition comprising members of the Firmicutes and Bacteroidetes (24, 65). The next most abundant phylum is the Actinobacteria, which is mainly comprised of the genus Bifidobacterium. This consensus has been derived from thousands of fecal samples using inventories of the 16S rRNA gene, generated mainly on next-generation sequencing platforms such as the Roche 454 and Illumina systems. Hence, they represent snapshots in time of the composition of a stool sample, which may or may not have been residing in the rectum prior to sampling. Additionally, this approach can only really robustly identify bacteria down to the genus level and in some instances the species level, and does not capture fully the complete species or strain diversity within a sample or an individual. However, the striking conclusion is that an individual’s large intestinal gut microbiota sits on a continuum, which at one end is predominantly composed of members from the phylum Bacteroidetes, and at the other end is predominantly composed of Firmicutes. Additionally, when an individual’s large intestinal gut microbiota has reached its stable climax community, it appears to be stable for a significant period of time as Faith and colleagues showed (200). This study followed 37 health adults and reported that after 5 years 60% of the original strains were still present. One can speculate on the value of such stability as it benefits the host to provide a niche which ensures that important functions of the microbiome are always present. Moreover, this stability may also be of benefit to the innate immune system, as it will also be sensing the same microbiota and thus not respond to it.
While there are many studies on the diversity of the large intestine, few studies have attempted to investigate the diversity of the small intestine. These studies show that the small intestine contains mainly members of the streptococci and variable numbers of bacteria belonging to the genera Clostridium and Veillonella (201, 202). Additionally, unlike the large intestine, the bacterial composition shows temporal fluctuations, with the morning and afternoon profiles being significantly different. This dynamic nature will be driven in part by the frequent input of dietary components and will vary between individuals depending on the diet (203).
The gut microbiota in the elderly
The gut microbiota in the aging population has been the focus of much attention in recent years. Several studies (e.g. ELDERMET in Ireland and CENIT in Spain) are aimed at characterizing the microbiota of individuals as they progress into old age so that changes in microbial populations that are associated with aging can be determined. Such changes may be related to a decline in general health and well-being in the elderly and as such, the results of these studies may shed light on how we can manipulate the gut microbiota (using better nutrition and/or probiotics) to promote improved health.
The microbiota of elderly people (>65 years old) has been reported to show greater inter-individual variation than adults (204). This study also reported a link between gut microbiota composition, diet, and living in institution or community. Furthermore, centenarians had a different and less diverse microbiota than adults and younger elderly people (205). A large group of volunteers over 100 years old had increased facultative anaerobes, such as Proteobacteria and Bacilli, and a decrease in specific bacteria, such as Faecalibacterium prauznitzii and Clostridium cluster XIVa bacteria. It was also reported that centenarian’s microbiota show a decreased levels of Bacteroides, Bifidobacterium and Enterobacteriaceae, while Clostridium spp. levels were increased compared with younger adults (206). The relevance of these observed changes during aging are not yet fully understood and future studies (particularly dietary-intervention studies) are required to investigate whether changing the dietary pattern of elderly individuals can alter their gut microbiota in a way that is beneficial to their general health.
Strategies for microbiota modulation
During infancy the intestinal microbiota is less stable and more variable in its composition than in older children and adults. As diet plays a major role in the development of microbiota during this period, there is an opportunity for its manipulation. Generally the microbiota composition of the large intestine is considered to be quite stable (200, 207), although long-term metabolic effects can be observed even after transient perturbations of the microbiota in early life (208), while the composition of the small intestine is very dynamic and this reflects the daily input of dietary components into the system. In both cases, it is relatively easy to perturb the composition of the gut microbiota, for example, by diet, antibiotics, and GI surgery. Since it is difficult to investigate the small intestine, we are still unsure as to what impact dietary supplements, such as probiotics, have on the composition in this area of the intestinal tract. In the area of antibiotics, there is well-documented evidence that non-specific antibiotics, when taken orally, will cause collateral damage and kill off many of the organisms within the large intestine (209), but the gut microbiota shows remarkable resilience and can re-attain its original composition within a short period of time once the antibiotic intervention has been removed. However, subtle and long-term changes in the gut microbiota have been observed in response to antibiotic treatment (210, 211), although the effect on health of such changes is so far unclear.
Two of the most common approaches are interventional and involve an individual taking live organisms, probiotics, or a non-digestible carbohydrate, a prebiotic. The probiotic approach has been shown to be efficacious only in a few diseased cases, for example, in reducing the rates of NEC in preterm infants (212) and diarrhea (213). There is little evidence that they make any impact on the large intestinal composition, however. But one must not lose sight of the fact that these organisms may be having a significant impact in the small intestine, where they will be a significant proportion of the total community. The other dietary intervention, known as prebiotics, can have a major impact on the large intestine, when taken in significant quantities. However, many of these studies are in animal models and fail to translate through to real-world instances, for example, the equivalent dose of the prebiotic in humans equates to 50 g a day, which can result in many unpleasant side effects. Modification of the diet without a direct intervention, for example, changing the proportions of protein, carbohydrates, and fat, also changes the composition of the gut microbiota in the large intestine (200, 214). As we understand more about the establishment of the gut microbiota in healthy infants, it may be possible to determine which core bacterial species are required for the establishment of a ‘healthy’ microbiota. In future, we may be able to identify infants at risk of developing microbial dysbiosis (e.g. infants delivered by C-section, or infants with a history of antibiotic treatment) and manipulate their microbiotas during a window of opportunity (e.g. with cocktails of probiotics) to help establish a stable, healthy microbiota throughout life.
Conclusions
In the past 10 years, our understanding of the composition of the adult gut microbiota has undergone a significant change. We have moved from a position in which the predominant organisms in the gut microbiota were thought to be those that were grown easily on laboratory media, to one in which we now know that the gut microbiota is complex and host-specific (214). Furthermore, we now understand that while the majority of the most abundant bacteria can be grown in the laboratory, the bulk of the organisms cannot (214). Thus, if we are to understand the contribution of these organisms to host biology and move medicine into the 21st century, we need to be able to not only understand ‘who is there’, but also be able to determine ‘what they are doing’ and how these functions interact with the host. Therefore, the ecological and functional properties of a healthy gut microbiome in infants, children and adults still need to be identified.
While the infant gut microbiota seems to influence immune development and metabolic pathways, further studies on the appropriate timing of interventions and the complex interactions between the infant immune system and the gut microbiota are required to translate these findings into preventive strategies required to reduce the risk of disease. Strategies to treat conditions associated with a disrupted gut microbial ecology and inflammation in adults also represent important areas of future research. While the area of gut microbiology is not new and has received serious interest for over 100 years, technological advancements in the last decade have really allowed us to interrogate it in a manner which was unforeseen 20 years ago.
Acknowledgements
The authors acknowledge the support of the European Science Foundation, in the framework of the Research Networking Program, The European Network for GI Health Research. The main positions of this review were presented during the 2014 ENGIHR Conference: The Gut Microbiota Throughout Life (September 2014, Max Rubner-Insitut, Karlsruhe, Germany).
This paper is part of the Proceedings from the 2013 ENGIHR Conference in Valencia, Spain. More papers from this supplement can be found at http://www.microbecolhealthdis.net
Footnotes
Present address: Chair of Nutrition and Immunology, ZIEL – Research Center for Nutrition and Food Sciences, Technical University of Munich, Freising-Weihenstephan, Germany
Conflict of interest and funding
The authors have declared that they have no interests that might be perceived as posing a conflict or bias.
References
- 1.Tissier H. Paris: G. Carre and C. Naud; 1900. Recherches sur la flore intestinale des nourrissons (état normal et pathologique) [Google Scholar]
- 2.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blencowe H, Cousens S, Chou D, Oestergaard M, Say L, Moller AB, et al. The global epidemiology of 15 million preterm births. Reprod Health. 2013;10:S2. doi: 10.1186/1742-4755-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. 2005;51:270–4. doi: 10.1007/s00284-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 7.Bearfield C, Davenport ES, Sivapathasundaram V, Allaker RP. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG. 2002;109:527–33. doi: 10.1111/j.1471-0528.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–93. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Rautava S, Collado MC, Salminen S, Isolauri E. Probiotics modulate host–microbe interaction in the placenta and fetal gut: a randomized, double-blind, placebo-controlled trial. Neonatology. 2012;102:178–84. doi: 10.1159/000339182. [DOI] [PubMed] [Google Scholar]
- 10.Steel JH, Malatos S, Kennea N, Edwards AD, Miles L, Duggan P, et al. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr Res. 2005;57:404–11. doi: 10.1203/01.PDR.0000153869.96337.90. [DOI] [PubMed] [Google Scholar]
- 11.Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy. 2013;43:198–211. doi: 10.1111/cea.12063. [DOI] [PubMed] [Google Scholar]
- 12.Moles L, Gómez M, Heilig H, Bustos G, Fuentes S, de Vos W, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One. 2013;8:e66986. doi: 10.1371/journal.pone.0066986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J, Nomura Y, Bashir A, Fernandez-Hernandez H, Itzkowitz S, Pei Z, et al. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One. 2013;8:e78257. doi: 10.1371/journal.pone.0078257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One. 2014;9:e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–8. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 16.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–7. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 17.Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, Serrant P, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–32. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 18.Fichorova RN, Onderdonk AB, Yamamoto H, Delaney ML, DuBois AM, Allred E, et al. Maternal microbe-specific modulation of inflammatory response in extremely low-gestational-age newborns. MBio. 2011;2:e00280–10. doi: 10.1128/mBio.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stout MJ, Conlon B, Landeau M, Lee I, Bower C, Zhao Q, et al. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol. 2013;208:226.e1–7. doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dasanayake AP, Li Y, Wiener H, Ruby JD, Lee MJ. Salivary Actinomyces naeslundii genospecies 2 and Lactobacillus casei levels predict pregnancy outcomes. J Periodontol. 2005;76:171–7. doi: 10.1902/jop.2005.76.2.171. [DOI] [PubMed] [Google Scholar]
- 21.Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avershina E, Storrø O, Øien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol. 2014;87:280–90. doi: 10.1111/1574-6941.12223. [DOI] [PubMed] [Google Scholar]
- 24.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. PNAS. 2011;108:4586–91. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adlerberth I, Wold AE. Establishment of the gut microbiota in western infants. Acta Paediatr. 2009;98:229–38. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 26.Marques TM, Wall R, Ross RP, Fitzgerald G, Ryan CA, Stanton C. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol. 2010;21:149–56. doi: 10.1016/j.copbio.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Backhed F. Programming of host metabolism by the gut microbiota. Ann Nutr Metab. 2011;58:44–52. doi: 10.1159/000328042. [DOI] [PubMed] [Google Scholar]
- 30.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108:4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borre YE, Moloney RD, Clarke G, Dinan TG, Cryan JF. The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. Adv Exp Med Biol. 2014;817:373–403. doi: 10.1007/978-1-4939-0897-4_17. [DOI] [PubMed] [Google Scholar]
- 33.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salminen S, Gibson GR, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53:1388–9. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–66. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 36.Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7:e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Muinck EJ, Øien T, Storrø O, Johnsen R, Stenseth NC, Rønningen KS, et al. Diversity, transmission and persistence of Escherichia coli in a cohort of mothers and their infants. Environ Microbiol Rep. 2011;3:352–9. doi: 10.1111/j.1758-2229.2010.00231.x. [DOI] [PubMed] [Google Scholar]
- 39.Rudi K, Storro O, Oien T, Johnsen R. Modelling bacterial transmission in human allergen-specific IgE sensitization. Lett Appl Microbiol. 2012;54:447–54. doi: 10.1111/j.1472-765X.2012.03229.x. [DOI] [PubMed] [Google Scholar]
- 40.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 42.Sekelja M, Berget I, Naes T, Rudi K. Unveiling an abundant core microbiota in the human adult colon by a phylogroup-independent searching approach. ISME J. 2011;5:519–31. doi: 10.1038/ismej.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennet R, Nord CE. Development of the faecal anaerobic microflora after caesarean section and treatment with antibiotics in newborn infants. Infection. 1987;15:332–6. doi: 10.1007/BF01647733. [DOI] [PubMed] [Google Scholar]
- 44.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eggesbo M, Moen B, Peddada S, Baird D, Rugtveit J, Midtvedt T, et al. Development of gut microbiota in infants not exposed to medical interventions. APMIS. 2011;119:17–35. doi: 10.1111/j.1600-0463.2010.02688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69:1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6:e21313. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–51. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- 49.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108:4653–8. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 51.Cephas KD, Kim J, Mathai RA, Barry KA, Dowd SE, Meline BS, et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS One. 2011;6:e23503. doi: 10.1371/journal.pone.0023503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avershina E, Storrø O, Øien T, Johnsen R, Wilson R, Egeland T, et al. Succession and correlation-networks of bifidobacteria in a large unselected cohort of mothers and their children. Appl Environ Microbiol. 2013;79:497–507. doi: 10.1128/AEM.02359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrett E, Kerr C, Murphy K, O’Sullivan O, Ryan CA, Dempsey EM, et al. The individual-specific and diverse nature of the preterm infant microbiota. Arch Dis Child Fetal Neonatal Ed. 2013;98:F334–40. doi: 10.1136/archdischild-2012-303035. [DOI] [PubMed] [Google Scholar]
- 54.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56:5811–20. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–54. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 57.Grześkowiak Ł, Grönlund MM, Beckmann C, Salminen S, von Berg A, Isolauri E. The impact of perinatal probiotic intervention on gut microbiota: double-blind placebo-controlled trials in Finland and Germany. Anaerobe. 2012;18:7–13. doi: 10.1016/j.anaerobe.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 58.Sepp E, Julge K, Vasar M, Naaber P, Björksten B, Mikelsaar M. Intestinal microflora of Estonian and Swedish infants. Acta Paediatr. 1997;86:956–61. doi: 10.1111/j.1651-2227.1997.tb15178.x. [DOI] [PubMed] [Google Scholar]
- 59.Grzeskowiak L, Collado MC, Mangani C, Maleta K, Laitinen K, Ashorn P, et al. Distinct gut microbiota in southeastern African and northern European infants. J Pediatr Gastroenterol Nutr. 2012;54:812–16. doi: 10.1097/MPG.0b013e318249039c. [DOI] [PubMed] [Google Scholar]
- 60.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hansen J, Gulati A, Sartor RB. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr Opin Gastroenterol. 2010;26:564–71. doi: 10.1097/MOG.0b013e32833f1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JAGM, de Vos WM. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis. 2001;13:129–13. [Google Scholar]
- 63.Tims S, Zoetendal EG, de Vos WM, Kleerebezem M. Host genotype and the effect on microbial communities. In: Nelson KE, editor. Metagenomics of the human body. Springer, New York: Springer Science+Business Media, LLC; 2011. pp. 15–41. [Google Scholar]
- 64.Stewart JA. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54:1239–42. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- 65.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107:7503–8. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 68.Deloris Alexander A, Orcutt RP, Henry JC, Baker J, Jr, Bissahoyo AC, Threadgill DW. Quantitative pcr assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome. 2006;17:1093–104. doi: 10.1007/s00335-006-0063-1. [DOI] [PubMed] [Google Scholar]
- 69.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–33. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb Ecol. 2010;61:423–8. doi: 10.1007/s00248-010-9787-2. [DOI] [PubMed] [Google Scholar]
- 71.Buhnik-Rosenblau K, Danin-Poleg Y, Kashi Y. Predominant effect of host genetics on levels of Lactobacillus johnsonii bacteria in the mouse gut. Appl Environ Microbiol. 2011;77:6531–8. doi: 10.1128/AEM.00324-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nones K, Knoch B, Dommels YE, Paturi G, Butts C, McNabb WC, et al. Multidrug resistance gene deficient (mdr1a−/−) mice have an altered caecal microbiota that precedes the onset of intestinal inflammation. J Appl Microbiol. 2009;107:557–66. doi: 10.1111/j.1365-2672.2009.04225.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–41. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 74.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, et al. A top-down systems biology view of microbiome–mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–6. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 77.Lyte M, Bailey MT. Neuroendocrine–bacterial interactions in a neurotoxin-induced model of trauma. J Surg Res. 1997;70:195–201. doi: 10.1006/jsre.1997.5130. [DOI] [PubMed] [Google Scholar]
- 78.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 79.Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–53. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kleerebezem M, Vaughan EE. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu Rev Microbiol. 2009;63:269–90. doi: 10.1146/annurev.micro.091208.073341. [DOI] [PubMed] [Google Scholar]
- 81.Moniaux N, Escande F, Porchet N, Aubert JP, Batra SK. Structural organization and classification of the human mucin genes. Front Biosci. 2001;6:D1192–206. doi: 10.2741/moniaux. [DOI] [PubMed] [Google Scholar]
- 82.Porchet N, Buisine MP, Desseyn JL, Moniaux N, Nollet S, Degand P, et al. MUC genes: a superfamily of genes? Towards a functional classification of human apomucins. J Soc Biol. 1999;193:85–99. [PubMed] [Google Scholar]
- 83.Andrianifahanana M, Moniaux N, Batra SK. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochim Biophys Acta. 2006;1765:189–222. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 84.Jensen PH, Kolarich D, Packer NH. Mucin-type O-glycosylation–putting the pieces together. FEBS J. 2010;277:81–94. doi: 10.1111/j.1742-4658.2009.07429.x. [DOI] [PubMed] [Google Scholar]
- 85.van Tassell ML, Miller MJ. Lactobacillus adhesion to mucus. Nutrients. 2011;3:613–36. doi: 10.3390/nu3050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012;20:30–9. doi: 10.1016/j.tim.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 87.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013;110:17059–64. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sommer F, Adam N, Johansson ME, Xia L, Hansson GC, Bäckhed F. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One. 2014;9:e85254. doi: 10.1371/journal.pone.0085254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fu J, Wei B, Wen T, Johansson ME, Liu X, Bradford E, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–66. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johansson ME, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–91. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–29. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stone EL, Ismail MN, Lee SH, Luu Y, Ramirez K, Haslam SM, et al. Glycosyltransferase function in core 2-type protein O glycosylation. Mol Cell Biol. 2009;29:3770–82. doi: 10.1128/MCB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vanhooren V, Vandenbroucke RE, Dewaele S, van Hamme E, Haigh JJ, Hochepied T, et al. Mice overexpressing beta-1,4-Galactosyltransferase I are resistant to TNF-induced inflammation and DSS-induced colitis. PLoS One. 2013;8:e79883. doi: 10.1371/journal.pone.0079883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–17. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–9. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mäkivuokko H, Lahtinen SJ, Wacklin P, Tuovinen E, Tenkanen H, Nikkilä J, et al. Association between the ABO blood group and the human intestinal microbiota composition. BMC Microbiol. 2012;12:94. doi: 10.1186/1471-2180-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wacklin P, Tuimala J, Nikkilä J, Tims S, Mäkivuokko H, Alakulppi N, et al. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS One. 2014;9:e94863. doi: 10.1371/journal.pone.0094863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rausch P, Rehman A, Künzel S, Häsler R, Ott SJ, Schreiber S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–5. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tong M, McHardy I, Ruegger P, Goudarzi M, Kashyap PC, Haritunians T, et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn’s disease risk polymorphism. ISME J. 2014 doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larsson JM, Karlsson H, Crespo JG, Johansson ME, Eklund L, Sjövall H, et al. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- 101.Buisine MP, Devisme L, Savidge TC, Gespach C, Gosselin B, Porchet N, et al. Mucin gene expression in human embryonic and fetal intestine. Gut. 1998;43:519–24. doi: 10.1136/gut.43.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chambers JA, Hollingsworth MA, Trezise AE, Harris A. Developmental expression of mucin genes MUC1 and MUC2. J Cell Sci. 1994;107:413–24. doi: 10.1242/jcs.107.2.413. [DOI] [PubMed] [Google Scholar]
- 103.Reid CJ, Harris A. Developmental expression of mucin genes in the human gastrointestinal system. Gut. 1998;42:220–6. doi: 10.1136/gut.42.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clark JA, Doelle SM, Halpern MD, Saunders TA, Holubec H, Dvorak K, et al. Intestinal barrier failure during experimental necrotizing enterocolitis: protective effect of EGF treatment. Am J Physiol Gastrointest Liver Physiol. 2006;291:G938–49. doi: 10.1152/ajpgi.00090.2006. [DOI] [PubMed] [Google Scholar]
- 105.Martin NA, Mount Patrick SK, Estrada TE, Frisk HA, Rogan DT, Dvorak B, et al. Active transport of bile acids decreases mucin 2 in neonatal ileum: implications for development of necrotizing enterocolitis. PLoS One. 2011;6:e27191. doi: 10.1371/journal.pone.0027191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Caplan MS, Jilling T. New concepts in necrotizing enterocolitis. Curr Opin Pediatr. 2001;13:111–15. doi: 10.1097/00008480-200104000-00004. [DOI] [PubMed] [Google Scholar]
- 107.Claud EC, Walker WA. Hypothesis: inappropriate colonization of the premature intestine can cause neonatal necrotizing enterocolitis. FASEB J. 2001;15:1398–403. doi: 10.1096/fj.00-0833hyp. [DOI] [PubMed] [Google Scholar]
- 108.McGuire W, Anthony MY. Donor human milk versus formula for preventing necrotizing enterocolitis in preterm infants: systematic review. Arch Dis Child Fetal Neonatal Ed. 2003;88:F11–14. doi: 10.1136/fn.88.1.F11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lucas A, Cole TJ. Breast milk and neonatal necrotising enterocolitis. Lancet. 1990;336:1519–23. doi: 10.1016/0140-6736(90)93304-8. [DOI] [PubMed] [Google Scholar]
- 110.Robbe-Masselot C, Maes E, Rousset M, Michalski JC, Capon C. Glycosylation of human fetal mucins: a similar repertoire of Oglycansalong the intestinal tract. Glycoconj J. 2009;26:397–413. doi: 10.1007/s10719-008-9186-9. [DOI] [PubMed] [Google Scholar]
- 111.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of 0-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–16. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta JP, et al. Evidence of regio-specific glycosylation in human intestinal mucins – presence of an acidic gradient along the intestinal tract. J Biol Chem. 2003;278:46337–48. doi: 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- 113.Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Khailova L, Dvorak K, Arganbright KM, Halpern MD, Kinouchi T, Yajima M, et al. Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2009;297:G940–9. doi: 10.1152/ajpgi.00141.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bergström A, Kristensen MB, Bahl MI, Metzdorff SB, Fink LN, Frøkiaer H, et al. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res Notes. 2012;5:402. doi: 10.1186/1756-0500-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.El Aidy S, Derrien M, Merrifield CA, Levenez F, Doré J, Boekschoten MV, et al. Gut bacteria-host metabolic interplay during conventionalisation of the mouse germfree colon. ISME J. 2013;7:743–55. doi: 10.1038/ismej.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meng D, Newburg DS, Young C, Baker A, Tonkonogy SL, Sartor RB, et al. Bacterial symbionts induce a FUT2-dependent fucosylated niche on colonic epithelium via ERK and JNK signaling. Am J Physiol Gastrointest Liver Physiol. 2007;293:G780–7. doi: 10.1152/ajpgi.00010.2007. [DOI] [PubMed] [Google Scholar]
- 118.El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–79. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 119.Deplancke B, Hristova KR, Oakley HA, McCracken VJ, Aminov R, Mackie RI, et al. Molecular ecological analysis of the succession and diversity of sulfatereducing bacteria in the mouse gastrointestinal tract. Appl Environ Microbiol. 2000;66:2166–74. doi: 10.1128/aem.66.5.2166-2174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.El Aidy S, van den Abbeele P, van de Wiele T, Louis P, Kleerebezem M. Intestinal colonization: how key microbial players become established in this dynamic process: microbial metabolic activities and the interplay between the host and microbes. Bioessays. 2013;35:913–23. doi: 10.1002/bies.201300073. [DOI] [PubMed] [Google Scholar]
- 121.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between caesarean section and childhood asthma. Clin Exp Allergy. 2008;38:629–33. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 122.Decker E, Engelmann G, Findeisen A, Gerner P, Laass M, Ney D, et al. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatrics. 2010;125:e1433–40. doi: 10.1542/peds.2009-2260. [DOI] [PubMed] [Google Scholar]
- 123.Barros FC, Matijasevich A, Hallal PC, Horta BL, Barros AJ, Menezes AB, et al. Cesarean section and risk of obesity in childhood, adolescence, and early adulthood: evidence from 3 Brazilian birth cohorts. Am J Clin Nutr. 2012;95:465–70. doi: 10.3945/ajcn.111.026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cho CE, Norman M. Cesarean section and development of the immune system in the offspring. Am J Obstet Gynecol. 2013;208:249–54. doi: 10.1016/j.ajog.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 125.Hansel TT, Johnston SL, Openshaw PJ. Microbes and mucosal immune responses in asthma. Lancet. 2013;381:861–73. doi: 10.1016/S0140-6736(12)62202-8. [DOI] [PubMed] [Google Scholar]
- 126.Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2012;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- 127.West CE, Jenmalm MC, Prescott SL. The gut microbiota and its role in the development of allergic disease: a wider perspective. Clin Exp Allergy. 2014 doi: 10.1111/cea.12332. [DOI] [PubMed] [Google Scholar]
- 128.McDade TW. Early environments and the ecology of inflammation. Proc Natl Acad Sci U S A. 2012;109:17281–8. doi: 10.1073/pnas.1202244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kondrashova A, Seiskari T, Ilonen J, Knip M, Hyoty H. The ‘Hygiene hypothesis’ and the sharp gradient in the incidence of autoimmune and allergic diseases between Russian Karelia and Finland. APMIS. 2013;121:478–93. doi: 10.1111/apm.12023. [DOI] [PubMed] [Google Scholar]
- 130.Schaub B, Liu J, Hoppler S, Schleich I, Huehn J, Olek S, et al. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009;123:774–82. doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 131.Vuillermin PJ, Ponsonby AL, Saffery R, Tang ML, Ellis JA, Sly P, et al. Microbial exposure, interferon gamma gene demethylation in naive T-cells, and the risk of allergic disease. Allergy. 2009;64:348–53. doi: 10.1111/j.1398-9995.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- 132.Abrahamsson TR, Sandberg Abelius M, Forsberg A, Björkstén B, Jenmalm MC. A Th1/Th2-associated chemokine imbalance during infancy in children developing eczema, wheeze and sensitization. Clin Exp Allergy. 2011;41:1729–39. doi: 10.1111/j.1365-2222.2011.03827.x. [DOI] [PubMed] [Google Scholar]
- 133.Jenmalm MC. Childhood immune maturation and allergy development: regulation by maternal immunity and microbial exposure. Am J Reprod Immunol. 2011;66:75–80. doi: 10.1111/j.1600-0897.2011.01036.x. [DOI] [PubMed] [Google Scholar]
- 134.Tsuda H, Michimata T, Hayakawa S, Tanebe K, Sakai M, Fujimura M, et al. A Th2 chemokine, TARC, produced by trophoblasts and endometrial gland cells, regulates the infiltration of CCR4+ T lymphocytes into human decidua at early pregnancy. Am J Reprod Immunol. 2002;48:1–8. doi: 10.1034/j.1600-0897.2002.01117.x. [DOI] [PubMed] [Google Scholar]
- 135.Guo PF, Du MR, Wu HX, Lin Y, Jin LP, Li DJ. Thymic stromal lymphopoietin from trophoblasts induces dendritic cell-mediated regulatory TH2 bias in the decidua during early gestation in humans. Blood. 2010;116:2061–9. doi: 10.1182/blood-2009-11-252940. [DOI] [PubMed] [Google Scholar]
- 136.Svensson J, Jenmalm MC, Matussek A, Geffers R, Berg G, Ernerudh J. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J Immunol. 2011;187:3671–82. doi: 10.4049/jimmunol.1100130. [DOI] [PubMed] [Google Scholar]
- 137.Mjösberg J, Berg G, Jenmalm MC, Ernerudh J. FOXP3+ regulatory T cells and T helper 1, T helper 2, and T helper 17 cells in human early pregnancy decidua. Biol Reprod. 2010;82:698–705. doi: 10.1095/biolreprod.109.081208. [DOI] [PubMed] [Google Scholar]
- 138.Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- 139.Boij R, Svensson J, Nilsson-Ekdahl K, Sandholm K, Lindahl TL, Palonek E, et al. Biomarkers of coagulation, inflammation, and angiogenesis are independently associated with preeclampsia. Am J Reprod Immunol. 2012;68:258–70. doi: 10.1111/j.1600-0897.2012.01158.x. [DOI] [PubMed] [Google Scholar]
- 140.Sandberg M, Frykman A, Ernerudh J, Berg G, Matthiesen L, Ekerfelt C, et al. Cord blood cytokines and chemokines and development of allergic disease. Pediatr Allergy Immunol. 2009;20:519–27. doi: 10.1111/j.1399-3038.2008.00794.x. [DOI] [PubMed] [Google Scholar]
- 141.Abelius MS, Ernerudh J, Berg G, Matthiesen L, Nilsson LJ, Jenmalm MC. High cord blood levels of the T-helper 2-associated chemokines CCL17 and CCL22 precede allergy development during the first 6 years of life. Pediatr Res. 2011;70:495–500. doi: 10.1203/PDR.0b013e31822f2411. [DOI] [PubMed] [Google Scholar]
- 142.Abelius MS, Lempinen E, Lindblad K, Ernerudh J, Berg G, Matthiesen L, et al. Th2-like chemokine levels are elevated in allergic children and influenced by maternal immunity during pregnancy. Pediatr Allergy Immunol. 2014;25:387–93. doi: 10.1111/pai.12235. [DOI] [PubMed] [Google Scholar]
- 143.Böttcher MF, Jenmalm MC, Björkstén B. Immune responses to birch in young children during their first 7 years of life. Clin Exp Allergy. 2002;32:1690–8. doi: 10.1046/j.1365-2222.2002.01463.x. [DOI] [PubMed] [Google Scholar]
- 144.Douwes J, Cheng S, Travier N, Cohet C, Niesink A, McKenzie J, et al. Farm exposure in utero may protect against asthma, hay fever and eczema. Eur Respir J. 2008;32:603–11. doi: 10.1183/09031936.00033707. [DOI] [PubMed] [Google Scholar]
- 145.Jenmalm MC, Duchén K. Timing of allergy-preventive and immunomodulatory dietary interventions – are prenatal, perinatal or postnatal strategies optimal? Clin Exp Allergy. 2013;43:273–8. doi: 10.1111/cea.12003. [DOI] [PubMed] [Google Scholar]
- 146.Sjögren YM, Tomicic S, Lundberg A, Böttcher MF, Björkstén B, Sverremark-Ekstrom E, et al. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy. 2009;39:1842–51. doi: 10.1111/j.1365-2222.2009.03326.x. [DOI] [PubMed] [Google Scholar]
- 147.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sommer F, Bäckhed F. The gut microbiota – masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–38. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 149.Weng M, Walker WA. The role of gut microbiota in programming the immune phenotype. J Dev Orig Health Dis. 2013;4:203–14. doi: 10.1017/S2040174412000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Forsberg A, Abrahamsson TR, Björkstén B, Jenmalm MC. Pre- and postnatal Lactobacillus reuteri supplementation decreases allergen responsiveness in infancy. Clin Exp Allergy. 2013;43:434–42. doi: 10.1111/cea.12082. [DOI] [PubMed] [Google Scholar]
- 151.Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108:516–20. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 152.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–34. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 153.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–7. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129–34. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 155.Forno E, Onderdonk AB, McCracken J, Litonjua AA, Laskey D, Delaney ML, et al. Diversity of the gut microbiota and eczema in early life. Clin Mol Allergy. 2008;6:11. doi: 10.1186/1476-7961-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009;39:518–26. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- 157.Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–52.e1–5. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 158.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434–40.e2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 159.Ismail IH, Oppedisano F, Joseph SJ, Boyle RJ, Licciardi PV, Robins-Browne RM, et al. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr Allergy Immunol. 2012;23:674–81. doi: 10.1111/j.1399-3038.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 160.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013;132:601–7. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 161.Adlerberth I, Strachan DP, Matricardi PM, Ahrne S, Orfei L, Aberg N, et al. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol. 2007;120:343–50. doi: 10.1016/j.jaci.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 162.Travis J. On the origin of the immune system. Science. 2009;324:580–2. doi: 10.1126/science.324_580. [DOI] [PubMed] [Google Scholar]
- 163.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–41. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Johansson MA, Sjögren YM, Persson JO, Nilsson C, Sverremark-Ekström E. Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PLoS One. 2011;6:e23031. doi: 10.1371/journal.pone.0023031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Böttcher MF, Nordin EK, Sandin A, Midtvedt T, Björkstén B. Microflora-associated characteristics in faeces from allergic and nonallergic infants. Clin Exp Allergy. 2000;30:1590–6. doi: 10.1046/j.1365-2222.2000.00982.x. [DOI] [PubMed] [Google Scholar]
- 166.van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128:948–55.e1–3. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 167.Wold AE. The hygiene hypothesis revised: is the rising frequency of allergy due to changes in the intestinal flora? Allergy. 1998;53:20–5. doi: 10.1111/j.1398-9995.1998.tb04953.x. [DOI] [PubMed] [Google Scholar]
- 168.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–66. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 169.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–50. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 170.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 173.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kalliomäki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet. 2003;361:1869–71. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- 175.Kuitunen M, Kukkonen K, Juntunen-Backman K, Korpela R, Poussa T, Tuure T, et al. Probiotics prevent IgE-associated allergy until age 5 years in cesarean-delivered children but not in the total cohort. J Allergy Clin Immunol. 2009;123:335–41. doi: 10.1016/j.jaci.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 176.Abrahamsson TR, Jakobsson T, Björkstén B, Oldaeus G, Jenmalm MC. No effect of probiotics on respiratory allergies: a seven-year follow-up of a randomized controlled trial in infancy. Pediatr Allergy Immunol. 2013;24:556–61. doi: 10.1111/pai.12104. [DOI] [PubMed] [Google Scholar]
- 177.Wickens K, Stanley TV, Mitchell EA, Barthow C, Fitzharris P, Purdie G, et al. Early supplementation with Lactobacillus rhamnosus HN001 reduces eczema prevalence to 6 years: does it also reduce atopic sensitization? Clin Exp Allergy. 2013;43:1048–57. doi: 10.1111/cea.12154. [DOI] [PubMed] [Google Scholar]
- 178.Abrahamsson TR, Jakobsson T, Böttcher MF, Fredrikson M, Jenmalm MC, Björkstén B, et al. Probiotics in prevention of IgE-associated eczema: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:1174–80. doi: 10.1016/j.jaci.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 179.Kukkonen K, Savilahti E, Haahtela T, Juntunen-Backman K, Korpela R, Poussa T, et al. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:192–8. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 180.Rautava S, Kainonen E, Salminen S, Isolauri E. Maternal probiotic supplementation during pregnancy and breast-feeding reduces the risk of eczema in the infant. J Allergy Clin Immunol. 2012;130:1355–60. doi: 10.1016/j.jaci.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 181.Jenmalm MC. Should more be done during pregnancy to reduce allergies in children? Clin Pract. 2012;9:227–31. [Google Scholar]
- 182.Ogden CL, Yanovski SZ, Carroll MD, Flegal KM. The epidemiology of obesity. Gastroenterology. 2007;132:2087–10. doi: 10.1053/j.gastro.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 183.Taveras EM, Rifas-Shiman SL, Belfort MB, Kleinman KP, Oken E, Gillman MW. Weight status in the first 6 months of life and obesity at 3 years of age. Pediatrics. 2009;123:1177–83. doi: 10.1542/peds.2008-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kalliomäki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. 2008;87:534–8. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- 185.Baker JL, Michaelsen KF, Rasmussen KM, Sørensen TI. Maternal prepregnant body mass index, duration of breastfeeding, and timing of complementary food introduction are associated with infant weight gain. Am J Clin Nutr. 2004;80:1579–88. doi: 10.1093/ajcn/80.6.1579. [DOI] [PubMed] [Google Scholar]
- 186.Chu SY, Callaghan WM, Kim SY, Schmid CH, Lau J, England LJ, et al. Maternal obesity and risk of gestational diabetes mellitus. Diabetes Care. 2007;30:2070–6. doi: 10.2337/dc06-2559a. [DOI] [PubMed] [Google Scholar]
- 187.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010;92:1023–30. doi: 10.3945/ajcn.2010.29877. [DOI] [PubMed] [Google Scholar]
- 188.Collado MC, Laitinen K, Salminen S, Isolauri E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res. 2012;72:77–85. doi: 10.1038/pr.2012.42. [DOI] [PubMed] [Google Scholar]
- 189.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88:894–9. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 190.Santacruz A, Collado MC, García-Valdés L, Segura MT, Martín-Lagos JA, Anjos T, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 191.Grönlund MM, Gueimonde M, Laitinen K, Kociubinski G, Grönroos T, Salminen S, et al. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy. 2007;37:1764–72. doi: 10.1111/j.1365-2222.2007.02849.x. [DOI] [PubMed] [Google Scholar]
- 192.Le Huërou-Luron I, Blat S, Boudry G. Breast vs. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23:23–36. doi: 10.1017/S0954422410000065. [DOI] [PubMed] [Google Scholar]
- 193.Gueimonde M, Laitinen K, Salminen S, Isolauri E. Breast milk: a source of bifidobacteria for infant gut development and maturation? Neonatology. 2007;92:64–6. doi: 10.1159/000100088. [DOI] [PubMed] [Google Scholar]
- 194.Ilmonen J, Isolauri E, Poussa T, Laitinen K. Impact of dietary counselling and probiotic intervention on maternal anthropometric measurements during and after pregnancy: a randomized placebo-controlled trial. Clin Nutr. 2011;30:156–64. doi: 10.1016/j.clnu.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 195.Piirainen T, Isolauri E, Lagström H, Laitinen K. Impact of dietary counselling on nutrient intake during pregnancy: a prospective cohort study. Br J Nutr. 2006;96:1095–104. doi: 10.1017/bjn20061952. [DOI] [PubMed] [Google Scholar]
- 196.Gueimonde M, Sakata S, Kalliomäki M, Isolauri E, Benno Y, Salminen S. Effect of maternal consumption of Lactobacillus GG on transfer and establishment of fecal bifidobacterial microbiota in neonates. J Pediatr Gastroenterol Nutr. 2006;42:166–70. doi: 10.1097/01.mpg.0000189346.25172.fd. [DOI] [PubMed] [Google Scholar]
- 197.Luoto R, Laitinen K, Nermes M, Isolauri E. Impact of maternal probiotic-supplemented dietary counselling on pregnancy outcome and prenatal and postnatal growth: a double-blind, placebo-controlled study. Br J Nutr. 2010;103:1792–9. doi: 10.1017/S0007114509993898. [DOI] [PubMed] [Google Scholar]
- 198.Luoto R, Kalliomäki M, Laitinen K, Isolauri E. The impact of perinatal probiotic intervention on the development of overweight and obesity: follow-up study from birth to 10 years. Int J Obes (Lond) 2010;34:1531–7. doi: 10.1038/ijo.2010.50. [DOI] [PubMed] [Google Scholar]
- 199.Kayama H, Takeda K. Regulation of intestinal homeostasis by innate and adaptive immunity. Int Immunol. 2012;24:673–80. doi: 10.1093/intimm/dxs094. [DOI] [PubMed] [Google Scholar]
- 200.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Leimena MM, Ramiro-Garcia J, Davids M, van den Bogert B, Smidt H, Smid EJ, et al. A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genomics. 2013;14:530. doi: 10.1186/1471-2164-14-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–26. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Booijink CC, El-Aidy S, Rajilic-Stojanovic M, Heilig HG, Troost FJ, Smidt H, et al. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- 204.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 205.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Drago L, Toscano M, Rodighiero V, de Vecchi E, Mogna G. Cultivable and pyrosequenced fecal microflora in centenarians and young subjects. J Clin Gastroenterol. 2012;46:S81–4. doi: 10.1097/MCG.0b013e3182693982. [DOI] [PubMed] [Google Scholar]
- 207.Rajilic-Stojanovic M, Heilig HG, Tims S, Zoetendal EG, de Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2013;15:1146–1159. doi: 10.1111/1462-2920.12023. [DOI] [PubMed] [Google Scholar]
- 208.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.de La Cochetière MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Doré J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–92. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–23. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 212.AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev. 2014;4:CD005496. doi: 10.1002/14651858.CD005496.pub4. [DOI] [PubMed] [Google Scholar]
- 213.Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R, et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA. 2012;307:1959–69. doi: 10.1001/jama.2012.3507. [DOI] [PubMed] [Google Scholar]
- 214.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–30. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]