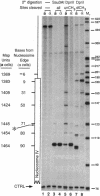
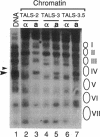
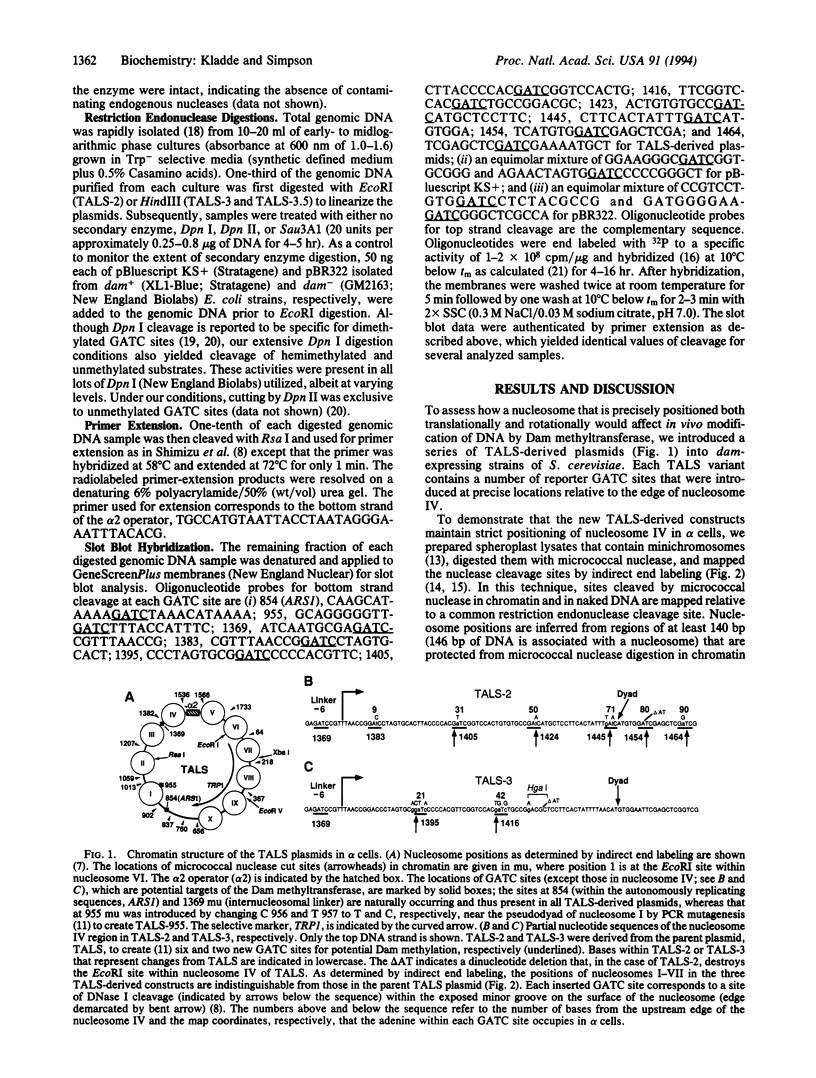
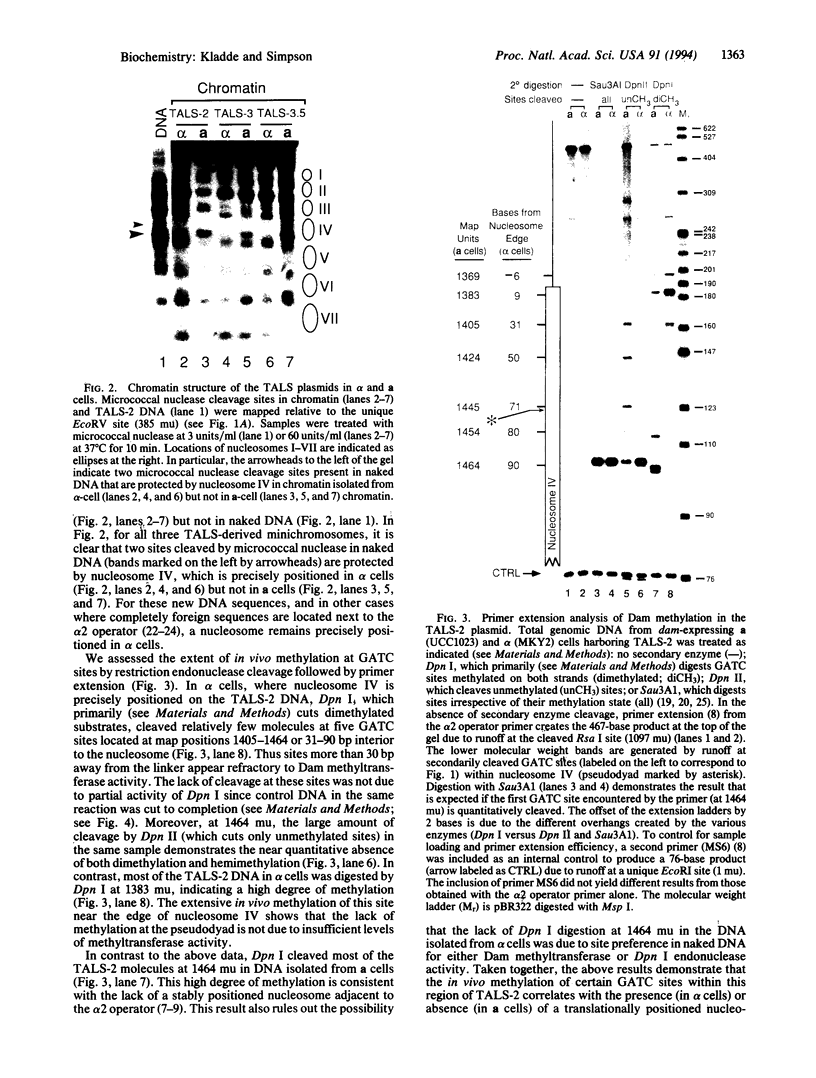
Abstract
Escherichia coli Dam DNA methyltransferase can methylate genomic GATC sites when expressed in Saccharomyces cerevisiae. Others have observed changes in the level of methylation at specific sites and suggested that these changes are related to transcriptional state or chromosomal context. To test directly the influence of nucleosome location on the ability of Dam methyltransferase to modify GATC sites in chromatin, we analyzed minichromosomes containing precisely positioned nucleosomes in dam-expressing yeast strains. Levels of methylation at individual GATC sites were rigorously quantified by an oligonucleotide-probing procedure. Within the linker and adjacent 21 bp of nucleosome-associated DNA, GATC sites were highly methylated, whereas methylation was severely inhibited by histone-DNA contacts nearer to the nucleosomal pseudodyad. Other DNA-protein complexes also interfere with Dam methylation. These data are consistent with a model in which nucleosomes exert a repressive influence on the biological functions of DNA by restricting access of trans-acting factors to DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hattman S., Kenny C., Berger L., Pratt K. Comparative study of DNA methylation in three unicellular eucaryotes. J Bacteriol. 1978 Sep;135(3):1156–1157. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J., Donald K. A., Griffiths D. E., Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991 Oct 25;19(20):5791–5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra M. F., Malone R. E. Expression of the Escherichia coli dam methylase in Saccharomyces cerevisiae: effect of in vivo adenine methylation on genetic recombination and mutation. Mol Cell Biol. 1985 Apr;5(4):610–618. doi: 10.1128/mcb.5.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J Biol Chem. 1975 Jun 10;250(11):4060–4066. [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lee K. P., Baxter H. J., Guillemette J. G., Lawford H. G., Lewis P. N. Structural studies on yeast nucleosomes. Can J Biochem. 1982 Mar;60(3):379–388. doi: 10.1139/o82-045. [DOI] [PubMed] [Google Scholar]
- Morse R. H., Pederson D. S., Dean A., Simpson R. T. Yeast nucleosomes allow thermal untwisting of DNA. Nucleic Acids Res. 1987 Dec 23;15(24):10311–10330. doi: 10.1093/nar/15.24.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Roth S. Y., Simpson R. T. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol Cell Biol. 1992 Sep;12(9):4015–4025. doi: 10.1128/mcb.12.9.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Nelson R. M., Long G. L. A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal Biochem. 1989 Jul;180(1):147–151. doi: 10.1016/0003-2697(89)90103-6. [DOI] [PubMed] [Google Scholar]
- Proffitt J. H., Davie J. R., Swinton D., Hattman S. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol Cell Biol. 1984 May;4(5):985–988. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Dean A., Simpson R. T. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990 May;10(5):2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Shimizu M., Johnson L., Grunstein M., Simpson R. T. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992 Mar;6(3):411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J Biol Chem. 1979 Oct 25;254(20):10123–10127. [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990 Jan 25;343(6256):387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Singh J., Klar A. J. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992 Feb;6(2):186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- Sussenbach J. S., Monfoort C. H., Schiphof R., Stobberingh E. E. A restriction endonuclease from Staphylococcus aureus. Nucleic Acids Res. 1976 Nov;3(11):3193–3202. doi: 10.1093/nar/3.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. H., Gottschling D. E., Zakian V. A. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992 Feb;6(2):197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]