Abstract
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm that, if not treated, will progress into blast crisis (BC) of either myeloid or B lymphoid phenotype. The BCR-ABL1 fusion gene, encoding a constitutively active tyrosine kinase, is thought to be sufficient to cause chronic phase (CP) CML, whereas additional genetic lesions are needed for progression into CML BC. To generate a humanized CML model, we retrovirally expressed BCR-ABL1 in the cord blood CD34+ cells and transplanted these into NOD-SCID (non-obese diabetic/severe-combined immunodeficient) interleukin-2-receptor γ-deficient mice. In primary mice, BCR-ABL1 expression induced an inflammatory-like state in the bone marrow and spleen, and mast cells were the only myeloid lineage specifically expanded by BCR-ABL1. Upon secondary transplantation, the pronounced inflammatory phenotype was lost and mainly human mast cells and macrophages were found in the bone marrow. Moreover, a striking block at the pre-B-cell stage was observed in primary mice, resulting in an accumulation of pre-B cells. A similar block in B-cell differentiation could be confirmed in primary cells from CML patients. Hence, this humanized mouse model of CML reveals previously unexplored features of CP CML and should be useful for further studies to understand the disease pathogenesis of CML.
Introduction
Chronic myeloid leukemia (CML) is characterized by the presence of the fusion protein BCR-ABL1, converting the ABL1 kinase into a constitutively active protein, resulting in an expansion of myeloid cells. Tyrosine kinase inhibitors (TKIs) have revolutionized the treatment of CML with the great majority of patients entering hematologic, cytogenetic and also molecular remission of varying depth.1, 2 However, CML leukemic stem cells appear not to be dependent on BCR-ABL1 kinase activity and some patients still fail tyrosine kinase inhibitor therapy.3 As a result, the chronic phase (CP) may transform into a more aggressive form of the disease termed blast crisis (BC), resembling either acute myeloid or B-cell leukemia, a state associated with a high mortality rate.
To ultimately achieve a cure for CML, there is a need to establish improved disease models allowing increased understanding of disease pathogenesis and to evaluate novel therapeutics. In this context, in vitro models using both cell lines and primary cells have been valuable, but lack the more complex physiological interaction that in vivo models provide. Syngeneic mouse models are by far the most common strategy to model CML,4, 5, 6, 7 but as mouse and man differ in several aspects, such as the number of mutations needed for cellular transformation,8 important insights into the disease pathogenesis may be overlooked unless also human cells also are studied in an in vivo setting.
So far, two main strategies have been explored to create humanized CML models in immunodeficient mice. The first involves transplantation of primary cells from CML patients, resulting in a general low engraftment and only few mice developing a myeloproliferative disorder associated with increased levels of myeloid cells in the bone marrow (BM) and spleen.9, 10 The second approach has been to retrovirally express BCR-ABL1 in cord blood (CB) hematopoietic progenitor cells followed by transplantation into NOD/SCID (non-obese diabetic/severe-combined immunodeficient) or NOD/SCID-β2M mice.11, 12 Such mice display an increase of myeloid cells, mainly of the erythroid and megakaryocytic lineage, and only rarely this is accompanied by increased peripheral white blood cell counts and splenomegaly.11, 12 Recently, it was demonstrated that coexpression of BCR-ABL1 and a dominant-negative isoform of IKZF1 (Ikaros) in lineage-negative human CB, followed by transplantation into NOD/SCID interelukin-2-receptor γ-deficient (NSG) mice, results in a rapid development of aggressive myeloid leukemia with myeloid sarcomas.13 In addition to the myeloid phenotypes that have been described previously by Chalandon et al.,11 control mice (BCR-ABL1 only) exhibit large clusters of histiocytes/macrophages in the BM and spleen, something also seen in other mouse models of CML.13, 14
In this study, we investigated if BCR-ABL1 expression alone in CB CD34+ cells transplanted into NSG mice would reveal novel features of CP CML. We describe that such mice display a myeloid cell expansion accompanied by an increase in macrophages/histiocytes and T-cell numbers, indicative of an inflammatory response. In primary mice, mast cells were the only myeloid lineage expressing BCR-ABL1 to be specifically expanded by the transgene. Following secondary transplantation, phenotypically aberrant CD25+ mast cells dominated the graft. Interestingly, we also found that BCR-ABL1 induced a differentiation block at the pre-B-cell stage, a finding that also was observed in BM samples obtained from patients with CML in CP.
Materials and methods
Isolation and retroviral transduction of CD34+ cells from CB
The collection and use of CB was approved by the Lund/Malmö Ethical Committee and performed after informed consent in accordance with the Declaration of Helsinki. Mononuclear cells (MNCs) were isolated by centrifugation over Lymphoprep (Axis-Shield PoC A/S, Oslo, Norway), pooled and CD34+ cells were enriched by the use of MACS (magnetic-activated cell sorting) separation columns and isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions.
Professor R Bhatia kindly provided the retroviral vectors MIGR1 (MSCV-IRES-GFP) and MIGR1P210 (MSCV-BCR-ABL1-IRES-GFP). CB CD34+ cells were either used fresh or after cryopreservation, and prestimulated (5 × 105 cells per /ml) for 48 h in Dulbecco's modified Eagle's media with GlutaMAX (Life Technologies, Grand Island, NY, USA), containing 10% fetal calf serum (Thermo Scientific, Waltham, MA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Thermo Scientific). The medium was supplemented with the following cytokines (Peprotech, Rocky Hill, NJ, USA): 50 ng/ml thrombopoietin, 100 ng/ml stem cell factor and 100 ng/ml Flt-3 ligand. Viral vector containing the supernatant, produced as described previously,15 was preloaded onto a 24-well non-tissue culture dish using Retronectin (Takara Bio, Otsu, Japan). The prestimulated CD34+ cells were resuspended in prestimulation medium, with the addition of 4 μg/ml protamine sulfate, and added to the wells at a density of 3 × 105 cells per ml. Forty-eight hours after transduction, cells were analyzed for green fluorescent protein (GFP) expression using a FACSCanto (BD Biosciences, San Jose, CA, USA).
In vivo mouse xenotransplantation assay
For the in vivo mouse xenotransplantation assay, we used 8–12-week-old male or female NSG mice that were subjected to 200 cGy total body irradiation 18–20 h before transplantation. After irradiation and for the duration of the experiment, mice were given antibiotics (ciprofloxacin) in drinking water and powder food. Mice were transplanted via tail vein with 1.8–2.6 × 105 unsorted cells per mouse 24 h after transduction. Counting of white blood cells, red blood cells and platelets in peripheral blood was performed at week 4, 8 and 12 after transplantation on an ABX Micros 60 cell counter (HORIBA ABX Corporate, Edison, NJ, USA). Mice were monitored daily and killed at signs of illness (anemia, weight loss and reduced motility). The control (MIG) mice in every experiment were kept alive until the last of the BCR-ABL1-transplanted mice (BA) mice were killed. Thus, the control mice were analyzed between 91 and 176 days after transplantation. For secondary transplantations, BM cells from primary mice were either transplanted unsorted (1–10 × 106 cells per mouse) or sorted for GFP (1–2 × 106 per mouse) and transplanted into NSG mice irradiated with 200 cGy.
Histopathology and immunohistochemistry
At the time of killing, long bones and spleen were collected for histopathology analysis. Spleens were weighed and cut in half. One part was fixed in 4% formaldehyde together with either tibia or femur. Subsequently, bones were decalcified for 2–3 weeks in 4% ethylenediaminetetracetic acid before paraffin embedding. The spleen was also embedded in paraffin. Bone and spleen sections were stained using hematoxylin and eosin for visualization of tissue structure, or used for immunohistochemistry with antibodies to visualize specific cells: CD20, CD68 (Dako, Glostrup, Denmark) and CD3 (Leica, Wetzlar, Germany). For morphological analysis of human cells from the secondary mice, cells were sorted based on GFP expression on a FACS Aria, deposited onto glass slides and stained using May Grünwald–Giemsa.
Flow cytometric analysis and cell sorting
Bones and spleens were crushed, cells resuspended and filtered to obtain a single-cell suspension that was analyzed by flow cytometry on a FACSCanto (BD Biosciences). Cells were stained with the following antibodies: CD45-APC, CD10-BV605, CD15-BV605 and glycophorin A-PE (from BD Biosciences) and CD33-BV421, CD19-PerCPCy5.5, CD34-APCCy7, CD68-PE, CD14-BV605, CD117-PECy7, IgM-PE, CD3-PECy7, FceRI-PE and CD25-BV421 (from BioLegend, San Diego, CA, USA). Control cells were stained with matching isotype controls. Sorting of cells was performed on a FACSAria (BD Biosciences).
For intracellular staining of phosphorylated STAT5 (signal transducer and activator of transcription 5), sorted pre-B cells were fixed in 1.6% paraformaldehyde for 10 min at room temperature. Cells were stored in 90% ethanol in −80 °C until analysis. Cells were washed two times in ice cold phosphate-buffered saline before resuspension in phosphate-buffered saline with 2% fetal calf serum. Cells were kept on ice and stained with antibodies against phosphorylated STAT5 (STAT5P-Alexa Flour 647 from BD) or matching isotype controls. Levels of phosphorylated STAT5 are presented as median fluorescence intensity, normalized to the median fluorescence intensity of isotype-stained cells.
Quantitative real-time PCR analysis
To analyze the expression of BCR-ABL1 in engrafted human cells, GFP+ and GFP− BM cells from BA mice were sorted into four populations: CD19+ B cells, CD3+ T cells, CD33+/CD14+/CD15+ myeloid cells and one population negative for all of these markers called ‘other'. RNA was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Real-time PCR analysis was performed using an ABI Prism 7500 analyzer (Applied Biosystems) and standard protocols. Extracted RNA was used as a negative control for the contamination of DNA. All samples were analyzed in three technical replicates. The primers and probes used for detection of BCR-ABL1 have been described by Gabert et al.16 The relative quantity of BCR-ABL1 was calculated based on the ddCt method17 and normalized to GAPDH.
RNA sequencing
Pro- and pre-B cells were sorted (between 40 000 and 1.7 × 106 cells) from BA or MIG control mice and RNA was extracted as described above. Library preparation and RNA sequencing of the samples were performed as described previously.18 Raw sequence reads were aligned to human reference genome hg19 using Tophat v.2.0.7 (ref. 19) and gene expression values were calculated as fragments per kilobase of transcript per million mapped reads using Cufflinks 2.1.1.20 For the analysis of the gene expression profile, additional samples were included: normal BM B-cell populations and pre-B childhood acute lymphoblastic leukemias (ALLs) positive for BCR-ABL1. Statistical analysis and visualization of the RNA sequencing data was performed using Qlucore Omics Explorer 3.0 (Qlucore, Lund, Sweden). Gene set enrichment analysis was performed with Qlucore Omics Explorer, using gene sets from the Molecular Signatures Database.21
Analysis of human BM using flow cytometry and Flow-Drop-FISH
BM from healthy donors or CML patients at the time of diagnosis were collected after informed consent according to a protocol approved by the regional ethics committee. The samples were separated on Lymphoprep (Axis-Shield PoC A/S) and MNCs were viably frozen. At the time of analysis, frozen human MNCs were thawed, stained with antibodies for cell sorting or analysis only (antibodies as described previously for analysis of human cells in mice), sorted onto glass slides, fixed and stained using dual-color, dual-fusion probes for BCR-ABL1 (Abbott Molecular, Des Plaines, IL, USA) as described previously.22 Cells were sorted into CD34+ progenitors, pro-B cells, pre-B cells and immature/mature B cells. Based on cell availability in the sample, between 116 and 3000 cells were acquired from each population and between 17 and 392 cells were scored by fluorescence in situ hybridization (FISH).
Statistics
GraphPad Prism Version 6.0a (GraphPad Software, La Jolla, CA, USA) was used to perform Student's t-test (unpaired, two-tailed). Mean values are accompanied by s.e.m. P-values ⩽0.05 were considered statistically significant.
Results
BCR-ABL1 expression leads to in vivo expansion of transduced cells and induces a lethal disorder in NSG mice
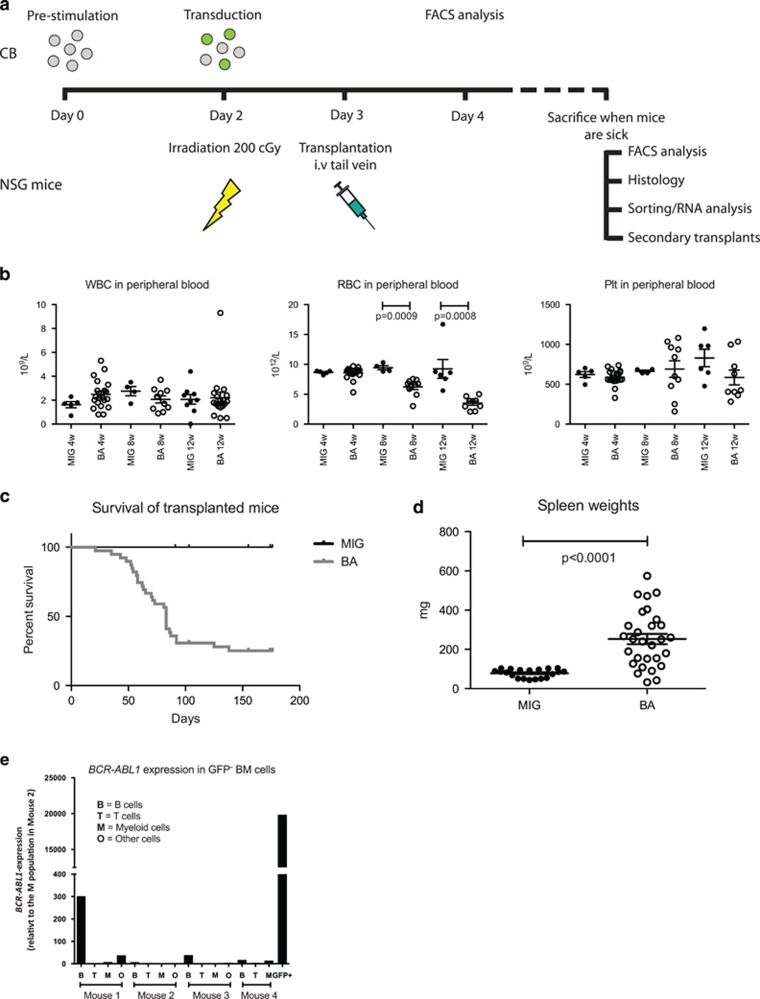
To establish a humanized mouse model of CML, NSG mice were transplanted with human CB CD34+ cells with ectopic expression of BCR-ABL1 (BA mice) (Figure 1a). Although the white blood cell and platelet counts did not change during the course of the experiment, a significant reduction of red blood cells was apparent already 8 weeks after transplantation (Figure 1b). Mice were killed when illness appeared, and as shown in Figure 1c, the majority of the mice developed signs of disease within 100 days after transplantation, something previously shown for this model.13 Analysis of the spleens showed a significant increase in spleen weight in BA mice (Figure 1d).
Figure 1.
Transplantation of BCR-ABL1-transduced CB cells to NSG mice induces a lethal disorder. (a) Flowchart showing generation of BCR-ABL1 transduction model and analysis of transplanted NSG mice. (b) Measurements of white blood cells (WBCs), red blood cells (RBCs) and platelets (Plts) in peripheral blood in transplanted mice at 4, 8 and 12 weeks after transplantation. (c) Survival of all transplanted mice. (d) Spleen weights of mice that succumbed to the disease. (e) Gene expression analysis of BCR-ABL1 in sorted GFP− BM cells from BA mice. Sorted BCR-ABL1+ GFP+ cells were used as a positive control.
At the time of killing, the presence of human cells in the BM and spleens of the transplanted mice was analyzed using flow cytometry. All mice showed robust human engraftment with a high proportion of GFP+ cells in the BM with no significant differences between the groups (Figure 2a). The frequency of human cells in the spleen was reduced in BA mice, but the frequency of GFP+ cells within the human cell population was not different from the control MIG mice (Figure 2a). To investigate if the BCR-ABL1-expressing cells had selectively expanded in vivo, we monitored the frequency of GFP+ cells among the human cells in vivo relative to the pretransplantation time point. A significant increase in BCR-ABL1-expressing cells was observed relative to the control (fold-change 1.77 vs 0.65), confirming that BCR-ABL1 alone is capable of expanding CB cells in an in vivo setting (data not shown).
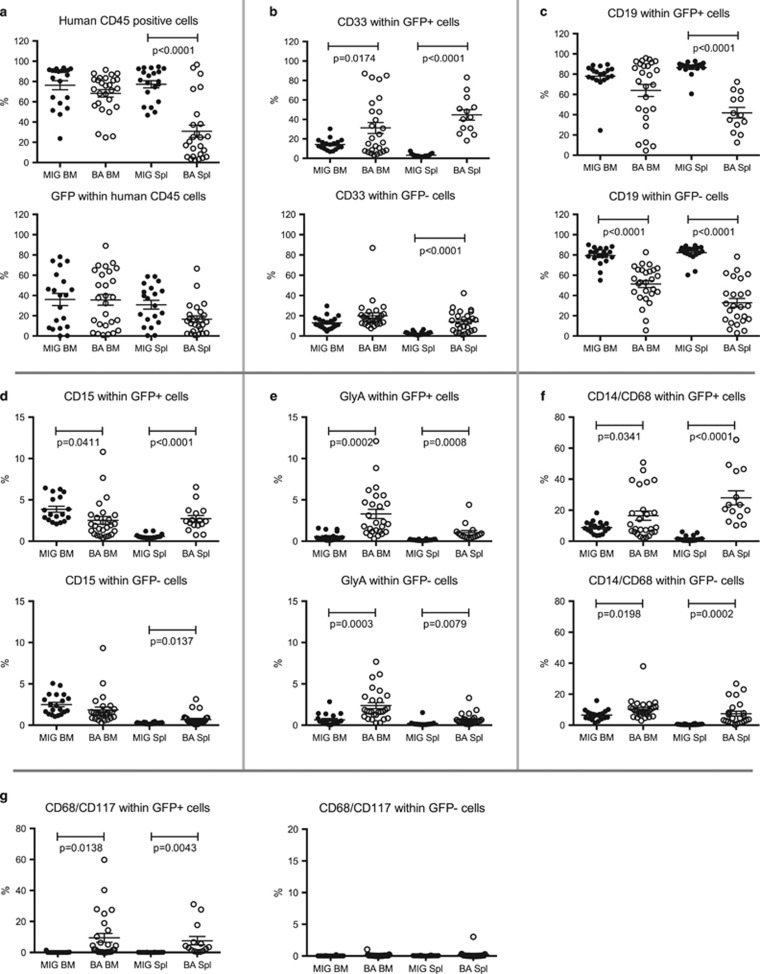
Figure 2.
Flow cytometric analysis reveals myeloid cell expansion in BA mice. BM and spleen cells from the transplanted mice were analyzed using flow cytometry for a range of human cell markers: (a) CD45 detecting human cells, (b) CD33 detecting myeloid cells, (c) CD19 detecting B cells, (d) CD15 detecting granulocytes, (e) glycophorin A (GlyA) detecting cells of the erythroid lineage, (f) CD14/CD68 detecting macrophages/histiocytes and (g) CD68/CD117 detecting mast cells.
BCR-ABL1 induces an increase of human CD33+ cells in both BM and spleen of transplanted mice
To investigate how the expression of BCR-ABL1 affected lineage distribution of the human cells in vivo, we performed flow cytometric analysis of cells from the BM and spleen. BCR-ABL1 expression reduced the frequency of CD34+ progenitors in the BM, and also among the GFP− cells, the frequency of CD34+ cells was significantly reduced in BA mice (Supplementary Figure 1). BCR-ABL1 induced a reduction of the frequency of CD19+ B cells in the spleen, and also among the GFP− cells, B-cell frequencies were lower both in BM and spleen in the BA mice (Figure 2c). A significant increase of human myeloid cells was observed among the BCR-ABL1+ cells (GFP+) within both the BM and the spleen (Figure 2b). However, in the spleens of BA mice, a similar skewing towards myeloid cells was observed for the GFP− cells (Figure 2b). This finding suggests that either GFP expression was silenced in BCR-ABL1-expressing cells or that microenvironmental factors in the BA mice also affected non-BCR-ABL1-expressing myeloid cells. To further investigate this, we sorted GFP+ and GFP− BM cells from BA mice and analyzed the level of BCR-ABL1 transcript. Although the GFP+ cells clearly expressed BCR-ABL1, the GFP− cells were negative/low for the fusion transcript (Figure 1e). The GFP− B-cell population in mouse 1 showed a weak expression of BCR-ABL1 (300 compared to 20 000 in the GFP+ sample), most likely caused by a small contamination of GFP+ cells at sorting. Thus, the increase of GFP− myeloid cells observed in the BA mice is likely caused by cell extrinsic mechanisms.
BCR-ABL1 induces expansion of mast cells in BA mice
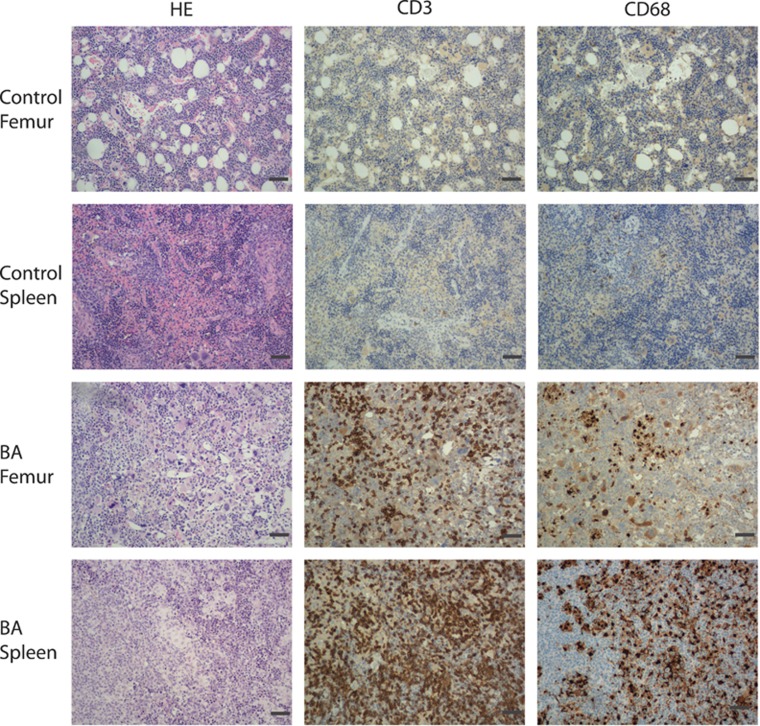
To characterize the myeloid sub-population that had expanded in the BA mice, the expression of several myeloid cell markers was analyzed. We observed an increase in the frequency of GFP+CD15+ cells in the spleen, consistent with an expansion of granulocytes during the CP of CML (Figure 2d). However, in BM the frequency of GFP+CD15+ cells was decreased (Figure 2d). We also observed a significant upregulation of GFP+ erythroid (glycophorin A-positive) cells both in the BM and spleen (Figure 2e), consistent with previous findings.11, 13 However, we also observed an expansion of both CD15+ and erythroid cells in the GFP population, suggesting that BCR-ABL1 has indirect effects on these cell populations. Moreover, we identified an increase in CD14 and CD68+/+ cells in the BM and spleen in BA mice, indicative of an expansion of monocytic/macrophage lineage cells (Figure 2f). This increase was particularly pronounced in the spleens and was even more evident when sections of the spleens were stained with an antibody directed against human CD68, visualizing large clusters of CD68+ macrophages/histiocytes (Figure 3). The observed increase of tissue macrophages/histiocytes, both GFP+ and GFP−, suggests that the presence of BCR-ABL1-expressing cells in BA mice induces an inflammatory response. Staining of human CD3 on the tissue sections showed massive infiltration of T cells, further supporting the inflammatory phenotype (Figure 3).
Figure 3.
Histology shows expansion of inflammatory cells in BA mice. Tissue sections from spleens and decalcified femurs from one representative MIG control and one BA mouse. Sections were stained with hematoxylin & eosin (HE), and antibodies targeting the T-cell marker CD3 and macrophage/histiocyte marker CD68. Scale bar, 140 μm.
A proportion of the CD68+ cells were CD14−, but coexpressed CD117, an immunophenotype defining mast cells. In contrast to the overall expansion of the other cell types, the mast cells were significantly expanded only within the GFP+ cells (Figure 2g). We conclude that several cell types of the myeloid lineage were expanded in the BA mice, but with the exception for mast cells, this expansion was not limited to BCR-ABL1-expressing cells.
Secondary transplantation shows engraftment of aberrant mast cells
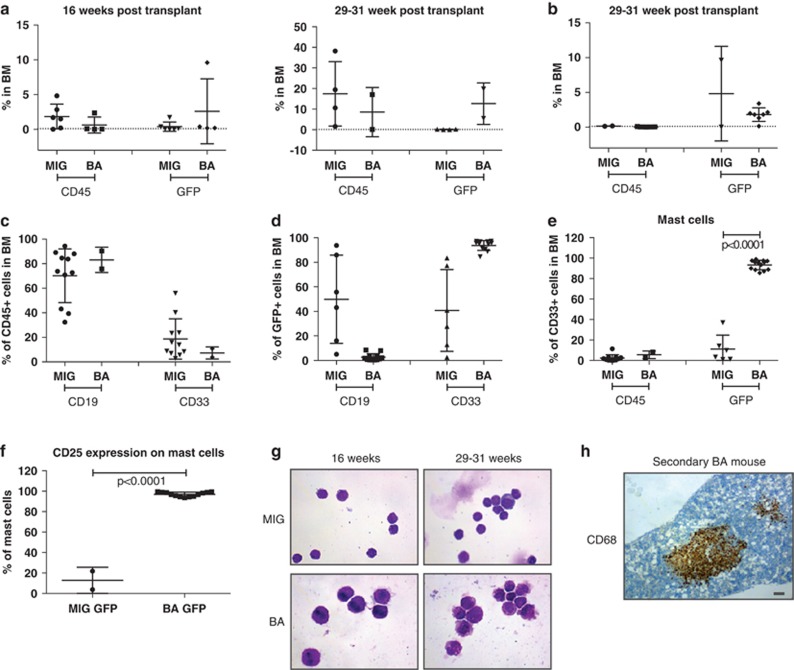
To evaluate whether cells from the primary recipients had the capacity to engraft and initiate myeloid disease upon serial transplantations, BM cells from primary mice were transplanted, either unsorted (Figure 4a) or sorted for GFP expression (Figure 4b), into sublethally irradiated NSG mice. Irrespectively of GFP sorting, these transplantations did not result in a lethal disease for the duration of the experiment. When the mice were killed, either at 16 or 29–31 weeks after transplant, the presence of human cells in the BM and spleen was analyzed (Figures 4a and b). In general, BM from primary MIG mice showed very poor GFP+ engraftment in secondary mice, whereas BM from primary BA mice generated secondary grafts that were dominated by GFP+ cells (Figures 4a and b). Engraftment levels above 0.1% were considered positive and samples from such mice were further analyzed for lineage distribution (Figure 4c).23 GFP+ cells from BA mice generated mainly CD33+ myeloid cells in secondary recipients, whereas mice engrafted with GFP+ MIG control cells exhibited similar patterns of lineage distribution as in the primary mice, that is, a dominance of B cells (Figure 4c). Further characterization of the CD33+ cells in secondary BA mice revealed that the vast majority of the cells were mast cells (here defined as CD68+CD117+ or FceRI+CD117+ cells; Figure 4d). Interestingly, the BCR-ABL1-expressing mast cells in the secondary mice expressed high levels of CD25 (IL2RA) (Figure 4e), a cell surface receptor known to be expressed on neoplastic mast cells.24 The expansion of aberrant mast cells, as defined by flow cytometry, could be confirmed by morphological assessment of sorted GFP+ cells from secondary mice. As shown in Figure 4f, the human cells in control mice were mainly of lymphocytic lineage, whereas cells from the secondary BA mice were densely granulated, consistent with mast cell morphology. In addition, femoral sections from secondary BA mice contained large foci of cells of macrophage origin (CD68+). These cells could not be detected by flow cytometry, most likely as they were lost by filtration of the BM before analysis. The presence of CD3+ cells in the BM and spleen and the clusters of macrophages/histiocytes in the spleen, indicative of an inflammatory phenotype that was evident in the primary BA mice, were not seen in the secondary BA mice (data not shown).
Figure 4.
Expansion of mast cells in secondary BA mice. Engraftment analysis of BM from secondary MIG and BA mice using flow cytometry measuring CD45+GFP− (CD45) and CD45+GFP+ (GFP) cell levels. Two groups of animals were analyzed: 16 and 29–31 weeks after transplantation. Mice were transplanted with either (a) unsorted or (b) GFP+ sorted cells and then killed at two different time points: 16 or 29–31 weeks after transplantation. Lineage distribution within (c) CD45+ cells or (d) GFP+ cells in secondary mice. (e) Mast cell frequency within CD45+ or GFP+ cells in secondary recipients. (f) CD25 expression on GFP+ mast cells. (g) Sorted GFP+ cells from MIG and BA mice killed at 16 or 29–31 weeks stained using MMG. (h) Sections of femur from BA secondary mice stained using a CD68 antibody. Scale bar, 100 μm.
In conclusion, secondary transplantations revealed long-term engraftment of aberrant CD25+ mast cells, whereas the pronounced inflammatory phenotype seen in the primary mice was not observed.
BCR-ABL1 expression results in a pre-B-cell differentiation block
Because CML can progress into lymphoid BC, we next explored if BCR-ABL1 expression in CB CD34+ cells would show a disturbance also of the lymphoid lineages. In BA mice, we found an increase of human T cells (CD3+) (Figures 3 and 5a). However, this increase was mainly seen among GFP− cells (Figure 5a), indicating that intrinsic expression of BCR-ABL1 partly inhibits T-cell development, but that BCR-ABL1 expression indirectly supports T-cell expansion.
Figure 5.
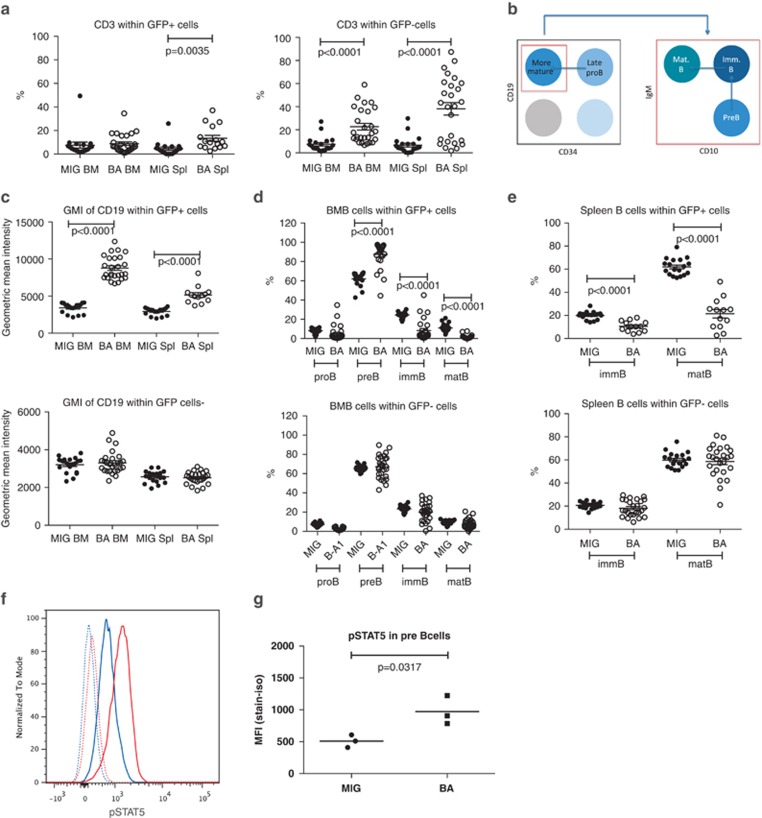
BA mice display a block in B-cell differentiation at the pre-B-cell stage. Flow cytometry analysis of BM and spleen cells from MIG and BA mice. (a) CD3 T cells among both GFP+ and GFP− cells, (b) cell surface markers used to define the different B-cell populations, (c) geometric mean intensity of CD19 expression on B cells within GFP+ and GFP− cells, (d) B-cell populations in BM and (e) B-cell populations in spleen. (f) Histogram showing pre-B cells from MIG mice stained with an isotype (dotted blue line) or pSTAT5 antibody (blue line) compared with pre-B cells from BA mice stained with an isotype (dotted red line) or pSTAT5 antibody (red line). (g) pSTAT5 analysis from three MIG and three BA mice shown as median fluorescent intensity (MFI). Isotype level was subtracted from the sample value.
Initial analysis of the B-cell lineage using the marker CD19 showed a decrease of CD19+ cells among BCR-ABL1-expressing cells, consistent with a previous study (Figure 2c).11 However, when measuring the geometric mean intensity of CD19 expression, we observed a higher CD19 expression on BCR-ABL1-expressing cells (Figure 5c). To further characterize the aberrant CD19 population, we used several B-cell markers to distinguish various differentiation stages from the late pro-B-cell stage to immature/mature B cells (Figure 5b).25 As shown in Figure 5d, this analysis revealed that BCR-ABL1 induced a partial block in B-cell differentiation at the pre-B-cell stage in NSG mice. This block was also evident in the spleen, resulting in fewer immature/mature B cells (Figure 5e). In CML, BCR-ABL1 directly activates the JAK2/STAT5 pathway, rendering STAT5 in a constantly activated state.26 Differentiation of B cells beyond the pre-B-cell stage requires dephosphorylation of STAT5,27 suggesting that the constant activation of STAT5 by BCR-ABL1 could potentially cause the block in B-cell differentiation seen in the BA mice. We therefore analyzed the phosphorylation state of STAT5 in pre-B cells from MIG and BA mice and could show that STAT5 was phosphorylated to a higher extent in the BA mice compared with the MIG mice (Figures 5f and g).
In conclusion, in the primary BA mice, BCR-ABL1 expression induced a block in B-cell differentiation at the pre-B-cell stage likely caused by increased STAT5 phosphorylation.
RNA sequencing of BCR-ABL1-expressing pre-B cells reveals an aberrant gene expression profile
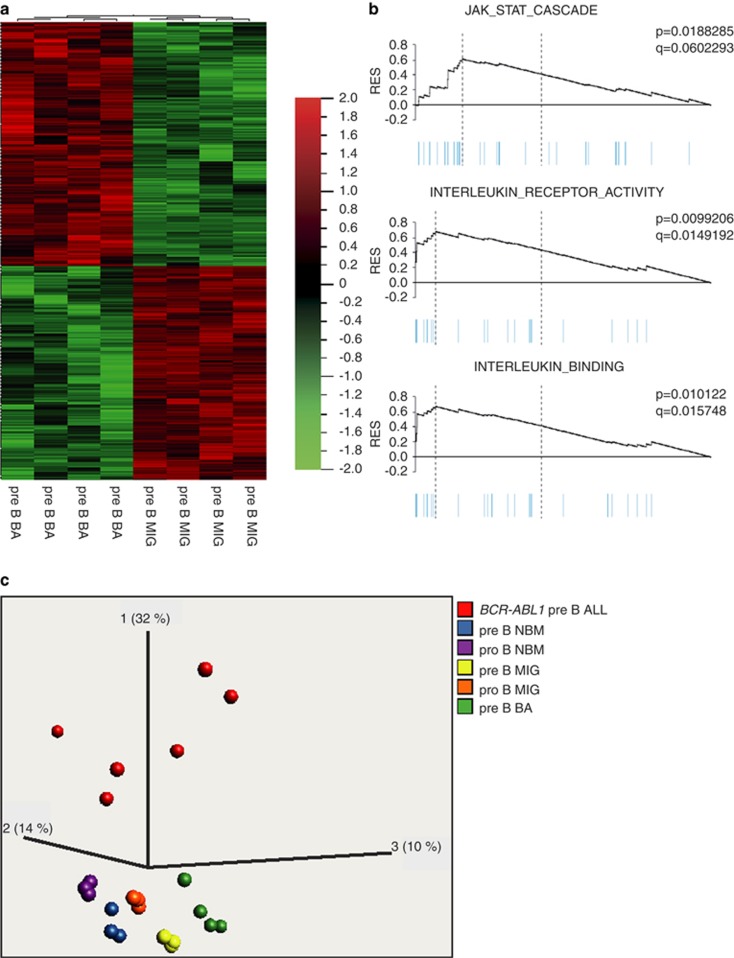
To investigate whether the accumulated pre-B cells displayed a similar gene expression profile as normal pre-B cells, we used RNA sequencing to determine the BCR-ABL1-induced global gene expression pattern in pre-B cells sorted from BA and MIG control mice. This analysis revealed that BCR-ABL1-expressing pre-B cells displayed a uniform gene expression profile distinct from that of MIG control pre-B cells (Figures 6a and b). In total, 704 genes with a significant (q=0.05) differential expression between MIG and BA pre-B cells could be identified (Figure 6a and Supplementary Table 1). Notably, CD25 (IL2RA) that was found to be aberrantly expressed on mast cells in secondary BA mice was among the top 10 upregulated genes in BA pre-B cells (Supplementary Table 1). CD25 has previously been shown to be expressed in a subset of ALL patients in which a majority of the cases harbored the BCR-ABL1 fusion gene.28 When comparing the gene expression profile of the pre-B cells from the BA mice to that of six BCR-ABL1+ primary childhood ALLs, we did not find a strong overlap between the two groups as determined by PCA analysis (Figure 6c).
Figure 6.
RNA sequencing shows a change in gene expression profile in BCR-ABL1+ pre-B cells. (a) Heat map of differentially expressed genes from RNA sequencing analysis of pre-B cells from BA and MIG control mice. (b) Gene set enrichment analysis of genes differentially expressed in (a) showing significant enrichment of different gene sets. (c) Using principal component analysis, the gene expression profile from BA and MIG pre-B cells were compared with those of B-cell populations from normal BM and childhood BCR-ABL1+ pre-B ALLs.
We also used the gene-expressing data to perform gene set enrichment analysis. By comparing BA pre-B cells to MIG and normal BM pre-B cells, a significant enrichment (q<0.25) was observed for the gene sets JAK-STAT-cascade, interleukin-receptor-activity and interleukin-binding (Figure 6b).
We conclude that BCR-ABL1 induces gene expression changes in pre-B cells that are enriched for gene sets including JAK-STAT signaling, but that gene expression profile is different from fully transformed BCR-ABL1+ ALL.
The block in B-cell differentiation is also present in patients with CP CML
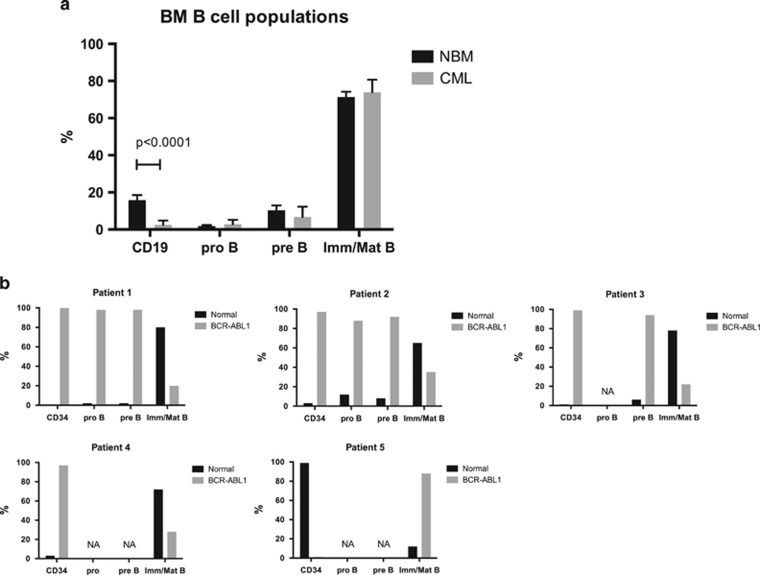
To investigate whether the B-cell differentiation block seen in the BA mice reflects a true feature of CP CML, BM MNCs from CML patients were analyzed using flow cytometry. Compared with normal BM, the frequency of total CD19+ cells among the MNCs was reduced but the frequencies of the different B-cell populations within the CD19+ cells were unchanged (Figure 7a). To determine the frequency of cells harboring the BCR-ABL1 rearrangement, total CD34+ progenitors, pro-B cells, pre-B cells and immature/mature B cells from CML patients were sorted onto slides and analyzed using FISH. Interestingly, all progenitor populations (total CD34+ cells, pro- and pre-B) were highly positive for BCR-ABL1, but the immature/mature B-cell population was dominated by normal, BCR-ABL1− cells (Figure 7b). This finding shows that the block in the transition from pre-B to mature B cells observed in the BA mice, causing an accumulation of pre-B cells, also exist in patients with CP CML.
Figure 7.
CML patients also display a block in B-cell differentiation. (a) Flow cytometric analysis of B-cell populations in BM MNCs from eight diagnostic samples from patients with CP CML (gray bars) and three normal healthy BM donors (black bars). (b) BM MNCs from five patients were sorted onto glass slides and stained with a FISH probe detecting the BCR-ABL1 fusion gene. Sorted populations are shown for each patient. For some of the patients, not all populations could be sorted owing to the lack of cells (NA). The frequency of BCR-ABL1+ cells (gray bars) and normal, BCR-ABL1− cells (black bars) within each cell population is presented in the graphs.
Discussion
In vivo models of CML are highly valuable to obtain critical insights into the disease pathogenesis of CML and to allow the assessment of novel therapies in a preclinical setting. In this study, we established and thoroughly characterized a humanized model of CML in NSG mice driven by retroviral BCR-ABL1 expression alone in CB CD34+ cells. Our findings confirm earlier studies,11, 12, 13 but importantly reveal several novel features not described previously in humanized CML mouse models.
Expansion of myeloid progenitors and mature granulocytes are archetypical features seen in CP CML patients. However, in our humanized mouse model, BCR-ABL1 did not expand the CD34+ hematopoietic progenitor population or the CD15+ granulocytes in the BM of BA mice and had only a modest positive effect on the number of granulocytes in the spleen. This is consistent with previously reported models where NOD/SCID, NOD/SCID-β2M or NSG mice were transplanted with human cells transduced with BCR-ABL1.11, 13 The reason for this lack of progenitor and granulocytic expansion is unknown, but could be a result of the xenograft setting in which human cells interact with the murine environment. In agreement with earlier reports,11, 12, 13 we observed a BCR-ABL1-induced increase of the human erythroid and monocyte/macrophage lineages in transplanted mice. However, BCR-ABL1 had both direct and indirect effects on the erythroid and monocyte/macrophage cell populations as both GFP+ and GFP− cells in BA mice were expanded. BCR-ABL1-expressing cells are known to produce a wide range of cytokines including interleukin-1 and granulocyte-colony-stimulating factor that could potentiallycontribute to these indirect effects.29
In contrast to previous reports,13 we obtained engraftment of BM from the primary BA mice in secondary recipients. We analyzed mice at 16 and 29–31 weeks after transplant and at both time points, the grafts were dominated by mast cells. Notably, in the primary BA mice the only myeloid cell type that was selectively expanded by BCR-ABL1 was mast cells. Because robust long-term multilineage engraftment of GFP+ control cells was observed in MIG secondary recipients, the skewed repopulation of BCR-ABL1-transduced cells in secondary recipients is unlikely to be caused by a failure to target stem/early progenitor cells at transduction, but rather is a consequence of BCR-ABL1-induced effects.
Interestingly, the mast cells in the secondary mice expressed CD25, a marker that has been shown to be expressed on neoplastic mast cells.24 The importance of aberrant mast cell development in CML has recently been shown in a study describing upregulation of CD25 and a role for interleukin-2 signaling using a BCR-ABL1 retroviral mouse model.28 CD25+ stem/progenitor cells from CML mice preferably generated mast cells in vitro and CD25 expression was also found on stem/progenitors (CD34+CD38− in patients with CP CML.28 Thus, the transplantable disease we observe with enrichment of aberrant CD25+ mast cells in secondary mice may represent a CML LSC giving rise to such cells in our secondary BA mice, although this remains to be proven. Unfortunately, the low number of CD34+ cells in our primary BA mice precluded isolation and transplantation of such cells to formally prove this hypothesis. Noteworthy, also other cell surface receptors, such as IL1RAP and ST2, have been shown to be upregulated on candidate CML stem cells, and intriguingly these receptors are also expressed by mast cells,22, 30 further suggesting a role of aberrant mast cell development in CML. However, we failed to identify a similar population of cells in viably frozen BM MNCs from three CP CML patients using flow cytometry (data not shown). In vivo mast cell expansion in humanized transduction models of CML has to our knowledge not been described previously. Interestingly, however, in a study by Dazzi et al.,31 CD34+ cells from CML patients were transplanted into NOD/SCID mice and engraftment analysis 12 weeks after transplant showed that all human cells were mast cells.31 Altogether, these observations indicate that mast cells are important in CML, but further studies will be needed to clarify the exact role of these cells in the disease pathology.
In addition to the BCR-ABL1-induced myeloid cell changes, we observed an increase in GFP− human T cells in BA mice. These findings indicate that BCR-ABL1 indirectly induces an expansion of untransduced human T cells, but that cell intrinsic expression of BCR-ABL1 is hampering T-cell development. This is consistent with studies in CML patients that have shown T cells to be polyclonal and lacking BCR-ABL1, suggesting that T-cell development is blocked by the expression of BCR-ABL1.32, 33 The increase of T cells observed in this study combined with increased numbers of macrophages/histiocytes suggests that the BCR-ABL1-expressing CB cells trigger an inflammatory reaction in the BA mice, possibly caused by paracrine effects of the transduced cells on the microenvironment, although this remains to be shown. In a previous study, we also observed similar clusters of macrophages/histiocytes when the BCR-FGFR1 fusion gene was expressed in CB cells transplanted into immunodeficient mice.15 This suggests that there might be common signaling pathways activated by both fusion proteins that trigger inflammation.
Previous reports have shown a decrease in frequency of B cells in the BM of CML patients.34 Although earlier studies using FISH have documented a reduced frequency of BCR-ABL1+ mature B cells in CML CP,32, 35 it has previously not been explored in detail how BCR1-ABL1 expression affects B-cell development in CML patients. Understanding how BCR-ABL1 affects B-cell development may provide critical insights into how CML progresses into a lymphoid BC.36 Here we report for the first time that the BCR-ABL1-induced block in B-cell development occurs at the pre-B-cell stage, both in the BA mice and in CML CP patients. Consistent with this, we were able to show that the level of STAT5 phosphorylation was higher in pre-B cells from BA mice compared with MIG mice. Because STAT5 is strongly activated by BCR-ABL126 and downregulation of STAT5 phosphorylation is critical for proper B-cell maturation beyond the pre-pro-B-cell stage,27, 37 we speculate that the increased activation of STAT5 caused by BCR-ABL1 prevents further differentiation of pre-B cells. RNA sequencing of BCR-ABL1-expressing pre-B cells showed that about 700 genes were differentially expressed compared with MIG pre-B cells, but failed to identify a strong transcriptional similarity to BCR-ABL1+ pre-B-ALL.
In conclusion, by expressing BCR-ABL1 in CB CD34+ cells transplanted into NSG mice, we observed skewing and expansion of human myeloid cells in combination with an inflammatory-like phenotype including increased numbers of macrophages/histiocytes accompanied by BCR-ABL1− T cells. Interestingly, mast cells were expanded in primary BA mice and completely dominated the graft in secondary mice. Moreover, we observed a BCR-ABL1-induced pre-B-cell block, a finding that was also validated in CML CP patients. This model should hence be valuable for further studies aimed at understanding the disease pathogenesis of CML.
Acknowledgments
We thank the staff at the Department of Obstetrics and Gynecology in Lund and Helsingborg, Sweden for collecting the umbilical cord blood, Professor Ravi Bhatia, Division of Stem Cell and Leukemia Research (City of Hope, Duarte, CA, USA) for providing the MIGR1 and MIGR1-p210 retroviral vectors, Josefin Persson at the Department of Clinical Pathology in Helsingborg, Sweden for the histology work and Ingbritt Åstrand-Grundström for morphological analysis of sorted cells from the secondary mice. This work was supported by the Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Inga-Britt and Arne Lundberg Foundation, the Gunnar Nilsson Cancer Foundation, the Medical Faculty of Lund University, the Swedish Research Council, the Swedish Society of Medicine and the Magnus Bergwall foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner CS, Koschmieder S, Iwasaki H, Iwasaki-Arai J, Radomska HS, Akashi K, et al. Inducible expression of BCR/ABL using human CD34 regulatory elements results in a megakaryocytic myeloproliferative syndrome. Blood. 2003;102:3363–3370. doi: 10.1182/blood-2003-03-0768. [DOI] [PubMed] [Google Scholar]
- Koschmieder S, Gottgens B, Zhang P, Iwasaki-Arai J, Akashi K, Kutok JL, et al. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 2005;105:324–334. doi: 10.1182/blood-2003-12-4369. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Lewis ID, McDiarmid LA, Samels LM, To LB, Hughes TP. Establishment of a reproducible model of chronic-phase chronic myeloid leukemia in NOD/SCID mice using blood-derived mononuclear or CD34+ cells. Blood. 1998;91:630–640. [PubMed] [Google Scholar]
- Eisterer W, Jiang X, Christ O, Glimm H, Lee KH, Pang E, et al. Different subsets of primary chronic myeloid leukemia stem cells engraft immunodeficient mice and produce a model of the human disease. Leukemia. 2005;19:435–441. doi: 10.1038/sj.leu.2403649. [DOI] [PubMed] [Google Scholar]
- Chalandon Y, Jiang X, Christ O, Loutet S, Thanopoulou E, Eaves A, et al. BCR-ABL-transduced human cord blood cells produce abnormal populations in immunodeficient mice. Leukemia. 2005;19:442–448. doi: 10.1038/sj.leu.2403650. [DOI] [PubMed] [Google Scholar]
- Rizo A, Horton SJ, Olthof S, Dontje B, Ausema A, van Os R, et al. BMI1 collaborates with BCR-ABL in leukemic transformation of human CD34+ cells. Blood. 2010;116:4621–4630. doi: 10.1182/blood-2010-02-270660. [DOI] [PubMed] [Google Scholar]
- Theocharides AP, Dobson SM, Laurenti E, Notta F, Voisin V, Cheng PY, et al. Dominant-negative Ikaros cooperates with BCR-ABL1 to induce human acute myeloid leukemia in xenografts Leukemia 2014(e-pub ahead of print 5 May 2014). [DOI] [PubMed]
- Krause DS, Fulzele K, Catic A, Sun CC, Dombkowski D, Hurley MP, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19:1513–1517. doi: 10.1038/nm.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerstam H, Jaras M, Andersson A, Johnels P, Hansen N, Lassen C, et al. Modeling the human 8p11-myeloproliferative syndrome in immunodeficient mice. Blood. 2010;116:2103–2111. doi: 10.1182/blood-2009-05-217182. [DOI] [PubMed] [Google Scholar]
- Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of 'real-time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lilljebjorn H, Agerstam H, Orsmark-Pietras C, Rissler M, Ehrencrona H, Nilsson L, et al. RNA-seq identifies clinically relevant fusion genes in leukemia including a novel MEF2D/CSF1R fusion responsive to imatinib. Leukemia. 2014;28:977–979. doi: 10.1038/leu.2013.324. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaras M, Johnels P, Hansen N, Agerstam H, Tsapogas P, Rissler M, et al. Isolation and killing of candidate chronic myeloid leukemia stem cells by antibody targeting of IL-1 receptor accessory protein. Proc Natl Acad Sci USA. 2010;107:16280–16285. doi: 10.1073/pnas.1004408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Dick JE. Engraftment of human hematopoietic stem cells is more efficient in female NOD/SCID/IL-2Rgc-null recipients. Blood. 2010;115:3704–3707. doi: 10.1182/blood-2009-10-249326. [DOI] [PubMed] [Google Scholar]
- Valent P, Cerny-Reiterer S, Herrmann H, Mirkina I, George TI, Sotlar K, et al. Phenotypic heterogeneity, novel diagnostic markers, and target expression profiles in normal and neoplastic human mast cells. Best Pract Res Clin Haematol. 2010;23:369–378. doi: 10.1016/j.beha.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Meffre E, LeDeist F, de Saint-Basile G, Deville A, Fougereau M, Fischer A, et al. A non-XLA primary deficiency causes the earliest known defect of B cell differentiation in humans: a comparison with an XLA case. Immunol Lett. 1997;57:93–99. doi: 10.1016/s0165-2478(97)00052-7. [DOI] [PubMed] [Google Scholar]
- Ilaria RL, Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi CI, Takubo K, Kobayashi H, Nakamura-Ishizu A, Honda H, Kataoka K, et al. The IL-2/CD25 axis maintains distinct subsets of chronic myeloid leukemia-initiating cells. Blood. 2014;123:2540–2549. doi: 10.1182/blood-2013-07-517847. [DOI] [PubMed] [Google Scholar]
- Zhang B, Ho YW, Huang Q, Maeda T, Lin A, Lee SU, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levescot A, Flamant S, Basbous S, Jacomet F, Feraud O, Anne Bourgeois E, et al. BCR-ABL-induced deregulation of the IL-33/ST2 pathway in CD34+ progenitors from chronic myeloid leukemia patients. Cancer Res. 2014;74:2669–2676. doi: 10.1158/0008-5472.CAN-13-2797. [DOI] [PubMed] [Google Scholar]
- Dazzi F, Hasserjian R, Gordon MY, Boecklin F, Cotter F, Corbo M, et al. Normal and chronic phase CML hematopoietic cells repopulate NOD/SCID bone marrow with different kinetics and cell lineage representation. Hematol J. 2000;1:307–315. doi: 10.1038/sj.thj.6200051. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Miura I, Saitoh K, Miura AB. Lineage involvement of stem cells bearing the philadelphia chromosome in chronic myeloid leukemia in the chronic phase as shown by a combination of fluorescence-activated cell sorting and fluorescence in situ hybridization. Blood. 1998;92:4758–4763. [PubMed] [Google Scholar]
- Kreutzman A, Juvonen V, Kairisto V, Ekblom M, Stenke L, Seggewiss R, et al. Mono/oligoclonal T and NK cells are common in chronic myeloid leukemia patients at diagnosis and expand during dasatinib therapy. Blood. 2010;116:772–782. doi: 10.1182/blood-2009-12-256800. [DOI] [PubMed] [Google Scholar]
- Rohon P, Porkka K, Mustjoki S. Immunoprofiling of patients with chronic myeloid leukemia at diagnosis and during tyrosine kinase inhibitor therapy. Eur J Haematol. 2010;85:387–398. doi: 10.1111/j.1600-0609.2010.01501.x. [DOI] [PubMed] [Google Scholar]
- Chandia M, Corrales A, Castellanos M, Matarraz S, Barrena S, Orfao A. Involvement of primary mesenchymal precursors and hematopoietic bone marrow cells from chronic myeloid leukemia patients by BCR-ABL1 fusion gene. Am J Hematol. 2014;89:288–294. doi: 10.1002/ajh.23626. [DOI] [PubMed] [Google Scholar]
- Hehlmann R. How I treat CML blast crisis. Blood. 2012;120:737–747. doi: 10.1182/blood-2012-03-380147. [DOI] [PubMed] [Google Scholar]
- Rolink A, Ghia P, Grawunder U, Haasner D, Karasuyama H, Kalberer C, et al. In vitro analyses of mechanisms of B-cell development. Semin Immunol. 1995;7:155–167. doi: 10.1016/1044-5323(95)90043-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.