Abstract
Changes in gene copy number are among the most frequent mutational events in all genomes and were among the mutations for which a physical basis was first known. Yet mechanisms of gene duplication remain uncertain because formation rates are difficult to measure and mechanisms may vary with position in a genome. Duplications are compared here to deletions, which seem formally similar but can arise at very different rates by distinct mechanisms. Methods of assessing duplication rates and dependencies are described with several proposed formation mechanisms. Emphasis is placed on duplications formed in extensively studied experimental situations. Duplications studied in microbes are compared with those observed in metazoan cells, specifically those in genomes of cancer cells. Duplications, and especially their derived amplifications, are suggested to form by multistep processes often under positive selection for increased copy number.
Gene duplications are one of the oldest and most frequent types of mutation. Genetic approaches are being used to dissect the mechanisms by which they form.
Gene duplications are among the oldest (Tice 1914; Morgan 1925; Bridges 1936) and perhaps the most frequent of mutation types (Anderson and Roth 1977; Reams and Neidle 2004; Lynch et al. 2008; Lipinski et al. 2011). Here we discuss duplications that form in real time as opposed to interspersed segmental duplications with ancient origins (Girirajan et al. 2011). We emphasize genetic approaches to understanding their formation and compare results in microbes to those in metazoan cells, especially those in cancer cells.
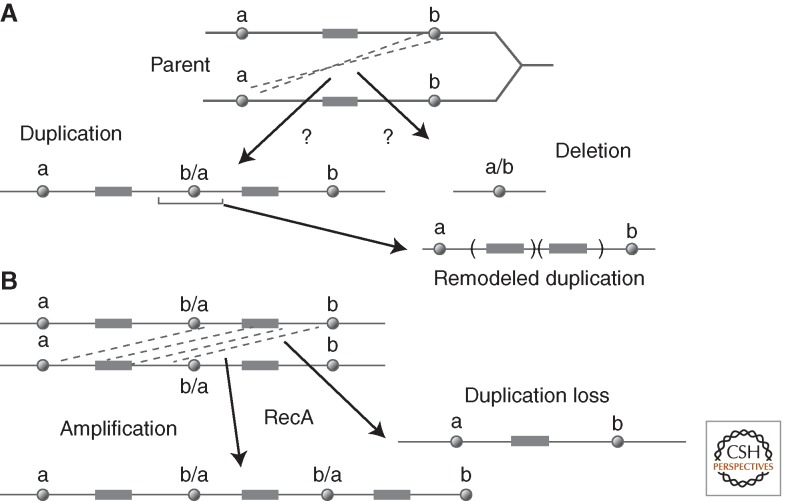
Figure 1A describes the formal process of forming a tandem duplication (or deletion) by a genetic exchange between separated sites in two sister chromosomes or homologs. When sites “a” and “b” are extensive direct-order sequence repeats (>100 bp), duplications can form by homologous recombination (Green 1963), described by Mehta and Haber (2014). In some situations, duplications appear to form by single-strand annealing (Reams et al. 2014), which is described by Morrical (2015). Most points in the bacterial chromosome are not flanked by extensive repeats and give rise to duplications whose junctions have short sequence repeats (4–12 base pairs) that are presumed to arise by some kind of single-strand annealing, template switching, or nonhomologous end joining. Several complex models suggest how junctions with short (or absent) repeated sequences can be generated by a multistep process (Hastings et al. 2009b; Kugelberg et al. 2010; Brewer et al. 2011; Elliott et al. 2013). In the absence of extensive repeats, duplications may form by aberrant recombination activities of topoisomerases (Shyamala et al. 1990).
Figure 1.
Duplication by exchanges between sister chromosomes. (A) a and b are sequence elements between which exchanges can be mediated by recombination, annealing, or transposition. Hybrid elements (b/a or a/b) are left at the rearrangement junctions. Duplications are subject to remodeling deletions that remove the junction. (B) Description of homologous recombination events that change copy number. These recombination events can increase or decrease copy number and make duplications prone to loss.
The junction sequence (b/a in Fig. 1A) is a key duplication feature that can shed light on the nature of the underlying formation event, but can also be misleading if an initial duplication has been remodeled, possibly under selection. As seen in Fig 1A, a finished duplication can be remodeled by deletions that remove the initial junction element and reduce the size of the repeated region. Such remodeling may be selectively favored if it reduces the fitness cost of the initial duplication while retaining the benefit provided by more copies of some included gene(s). A remodeling deletion creates a new join point that does not reflect the event that formed the initial duplication. Further amplification or loss of a duplication occurs by exchanges between the two extensive identical copies of the duplicated region (Fig. 1B). These events are frequent and depend heavily on homologous recombination.
Duplications can also form by mechanisms that do not involve an exchange between chromosomes, but rather depend on events occurring within a single chromosome. Several of these mechanisms rely on palindromic sequences and produce tandem inversion duplications (TIDs) with adjacent copies in inversion orientation. For all types of duplications, the nature of join points between copies can be informative, but can also reflect subsequent remodeling events rather than the initial exchange. These duplications, their formation, and remodeling will be discussed later in this article.
GENERAL PROBLEMS OF ASSESSING DUPLICATION RATES AND DEPENDENCIES
Detecting Duplications
In genetic approaches to duplication mechanisms, one measures the rate of duplication formation and observes how that rate is affected by mutations that eliminate various candidate functions. This requires observing changes in duplication frequency over time. The frequency of duplication-bearing cells in a population has been measured in several ways.
Selection for Cells with Increased Copy Number
This assay is probably the simplest but also the most problematic way to measure duplication. Because it involves selection, this assay may be the most biologically relevant. A population is placed under selective conditions that allow faster growth of cells with additional copies of some particular gene. This assay often suggests that duplication formation is heavily dependent on homologous recombination with 10- to 1000-fold fewer duplications in a RecA mutant strain (Petes and Hill 1988; Romero and Palacios 1997). This conclusion may be misleading if detection favors cells with more than two copies of the gene in question. The secondary amplification (and deamplification) events shown in Figure 1B are generally heavily dependent on RecA because they occur by exchanges between extensive perfect homologies. Thus, if selection favors cells with an amplification, the appearance of mutants may show heavy RecA dependence, even when formation of the initial duplication is independent of homologous recombination. Similarly, stringent selections that demand many gene copies (e.g., 20 copies) and do not allow slow growth and improvement of cells with a few copies (<10) may underreport the frequency of amplification.
Trapping Duplications
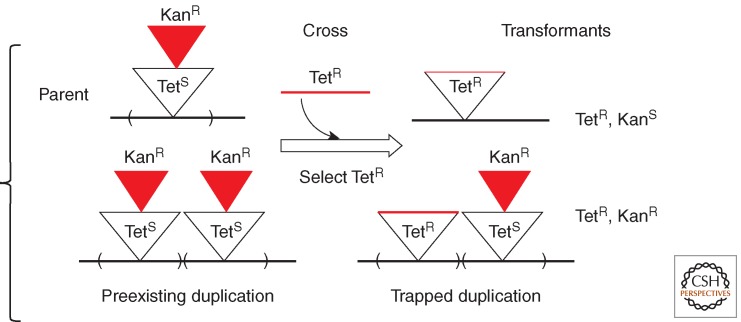
This assay detects duplication-bearing cells by their heterozygosity for two mutually exclusive, selectable genetic markers. It does not favor cells with higher amplification. For example, Figure 2 describes a strain with a tetracycline resistance (TetR) determinant whose expression is eliminated by the insertion of a kanamycin resistance (KanR) cassette. Into this TetS KanR strain, one can selectively introduce (by transduction or transformation) a fragment that restores TetR and removes the KanR insertion. Haploid cells that become TetR lose their preexisting KanR resistance. However, any recipient cell with a preexisting duplication of the TetS KanR region can acquire a TetR determinant in one copy and retain KanR in the other. Because the two selected markers occupy the same chromosomal site, they cannot both be retained without two copies of the region. Selection for resistance to both antibiotics thus “traps” a preexisting duplication and allows it to be selectively maintained. Trapping requires two copies of the region in question and does not favor recovery of cells with higher amplification. Duplication frequency is defined as the fraction of the TetR transformants that retain the original KanR phenotype. This method eliminates the contribution of selective amplification to detectability of duplications.
Figure 2.
Isolating duplications by trapping. This assay identifies and selectively maintains a preexisting duplication that formed under nonselective conditions. Frequency is defined as the fraction of recombinants that maintain two normally mutually exclusive markers. In this diagram, a KanR recipient receives TetR by a genetic cross. The duplication frequency in the recipient strain is the ratio of TetR KanR duplication transfomants to parental haploid TetR, KanS types. No amplification beyond duplication is selected. Using “recombineering” methods (Sawitzke et al. 2007), the cross that detects the duplication can be performed in recombination-deficient strains.
Trapping was first done by transduction crosses between two closely linked, complementary deletions within the his operon of Salmonella (Anderson et al. 1976). Most His+ recombinants from these crosses carry a duplication with one deletion allele in each copy. Because recombination between deletions is rare and preexisting duplications are common, a His+ phenotype is most often generated by acquiring the donor allele in one copy of a large preexisting duplication. The donor deletion allele replaces one copy of the recipient allele, generating a heterozygote whose His+ phenotype reflects complementation between deletions. In a later experiment, a wild-type his+ strain was crossed with a his::Tn10 (His− TetR) auxotroph and duplication frequency in the recipient was indicated by the fraction of transductants that retained both alleles His+, TetR; all such recombinations carried a duplication. Because these assays involve a transductional cross, they could detect duplications only in recombination-proficient cells and could not initially be used to test effects of recombination deficiencies on duplication formation. However, with the advent of recombineering (Court et al. 2002), the second marker could be introduced without RecA and the frequency of trapped duplications could be measured in recombination-deficient strains. The frequency of duplication-bearing cells (measured by trapping) was used to estimate the initial rates of duplication formation by determining the frequency attained by a population initiated by a single cell lacking a duplication and grown for a fixed small number of cell generations (Reams et al. 2010, 2014).
The trapping assay detects cells with two copies of the target site and places no restriction on the size of the amplified region (distance between a and b in Fig. 1), as long as that region includes the target. There is no bias toward higher amplification because a single copy of each alternate allele provides full ability to grow under selection. In determining the dependency of duplication rates on various recombination functions, the trapping assays have the advantage of being unaffected by growth rate or viability of the tested recombination mutant. That is, the slower growth and reduced viability of a recA mutant is not expected to reduce the measured duplication rate, which is based on the relative frequency of cells with and without a duplication. Both populations are subject to the same general growth and viability effects, even though the duplication rate may be strongly affected.
Identifying Duplications that Have a Particular Hybrid Junction Sequence
In these assays, the duplication&s hybrid join point (b/a in Fig. 1) provides a novel selectable or scorable phenotype. The two separated sequences (a and b) are allowed to recombine during nonselective pregrowth. Cells that acquire a new phenotype by virtue of their join-point sequence juxtaposition can be detected selectively or visually. One example below assesses the reciprocality of recombination during duplication formation by nonselectively scoring cells with an exchange between separated long lac repeats in the Salmonella chromosome that arise in a single clone (see the section Assessing Reciprocal Recombination in a Duplication/Deletion System). In another system (Anderson and Roth 1978), selection is used to detect the phenotype generated at the new duplication junction (see the section Duplication Formation Using Particular Short Recombining Sequences).
Determining Duplication Rates
In measuring any mutation rate, it is standard to determine the increase in mutant frequency during a period of nonselective growth. In the case of duplications, this method is complicated by the duplication’s associated fitness cost and frequent loss by recombination. That is, the rate of duplication formation may be underestimated because new duplication mutants are frequently lost by reversion. In addition, duplications and amplifications have inherent fitness costs that cause them to grow more slowly than the parent strain in which they arose, even under nonselective conditions (Reams et al. 2010). In measuring rates, one can correct for these effects, but only after independently measuring the loss rate and fitness cost. Applying these corrections is still complicated because duplications of a single site may vary widely in size and fitness cost (Reams et al. 2010).
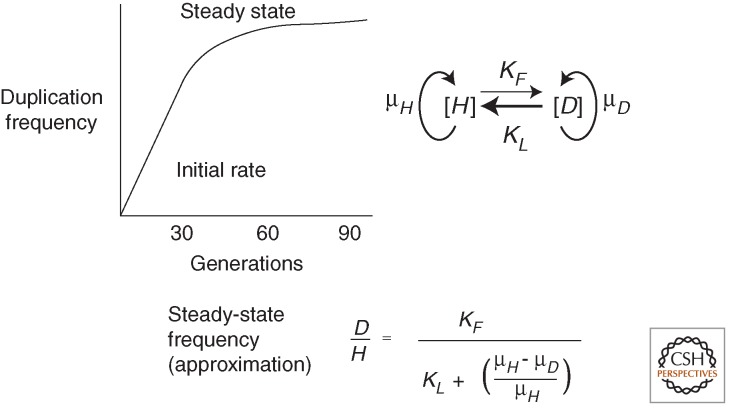
A related problem is the speed at which duplication frequency comes to a steady-state level (Reams et al. 2010). Point mutant frequencies increase linearly during nonselective growth and are subject to Luria–Delbrück fluctuation between populations. In contrast, the frequency of cells with duplications comes rapidly to a steady-state level dictated by the balance between the high formation rate on one hand, and the loss rate and fitness cost on the other. This steady state obscures Luria–Delbrück fluctuations, which are often used to measure mutation rates (Foster 2006). That is, duplications are immune to Luria–Delbrück fluctuations because their steady-state frequency is the same in all populations, regardless of whether the first duplication forms early or late in the culture.
The duplication formation rate can be assessed early in a culture (initial rate) before the steady state is reached. Alternatively, it can be inferred mathematically from the steady-state level using the equation in Figure 3 if fitness cost and loss rates are known. Most points in the Salmonella chromosome show a duplication formation rate of ∼10−5 per cell per generation, and a steady-state frequency of ∼10−3 (Anderson and Roth 1981). Typically, a bacterial culture initiated by a single cell lacking a duplication increases from 0% to within 70% of the steady-state level in ∼30 generations—which is the growth required for a single cell to generate a saturated overnight liquid culture (Reams et al. 2010). Using this method, a duplication hot spot (argH), flanked by 5-kb ribosomal RNA loci, rrnA and rrnE (100 kb apart), was found to duplicate at the surprisingly high rate of 10−3 per cell per generation (Reams et al. 2014). A cold spot (pyrD) flanked by no extensive repeats duplicated at the 1000-fold lower rate of 10−6 per cell per generation.
Figure 3.
Steady-state duplication frequencies. When a culture is started by a few cells with no duplication and is grown with no applied selection beyond viability, the duplication frequency increases to a steady state. This steady state is dictated in part by the relative rates of formation and loss (kF and kL). The loss rate is usually higher than the formation rate. The second factor is the fitness cost of the duplication, which is dictated by the difference between the higher growth rate of the parent strain (μH) and the lower growth rate of the haploid strain with the duplication (μD). The equation above approximates the steady-state frequency (Reams et al. 2010). In the equation, D and H denote the titer of duplication-bearing and haploid parent cells, respectively.
Comparing Rates of Duplication and Deletion Formation
As seen in Figure 1, duplication and deletion events appear formally similar and might be expected to occur at the same rate or be formed simultaneously by a reciprocal exchange. These rates are difficult to compare for several reasons. Deletions are limited to regions devoid of essential genes, whereas duplications show essentially no size limit. Regions between directly oriented 5-kb rrn loci duplicate at very high frequencies (10−3 per cell per division), whereas the corresponding deletions are all lethal. Rates of duplication and deletion can be compared only when recombining direct repeats are placed flanking nonessential genes (described below; M Carter, unpubl.).
An additional problem complicates assessing the contribution of single-strand annealing to deletion and duplication formation. Deletions can be generated by a double-strand break in one chromosome, when broken ends are resected and single-strand overhangs are allowed to anneal. In contrast, formation of a duplication by annealing requires two simultaneous breaks on opposite sides of the duplicated region in different sister chromosomes. These two simultaneous breaks are unlikely unless the recombining sequences are subject to frequent breakage or if damages are allowed to accumulate (Reams et al. 2014). The behavior of several particular systems is described below.
DUPLICATIONS BETWEEN LONG REPEATS
As outlined above, extended native direct sequence repeats can serve as the a and b elements in Figure 1 and initiate tandem duplications or deletions. Duplications between such repeats have often been detected by selection for cells with an increased copy number of an included gene. When measured in these assays, duplication shows a greater than 10-fold dependence on homologous recombination (RecA) (Petes and Hill 1988; Romero and Palacios 1997). However, as described below, some long repeats have been observed to produce duplications without participation of homologous recombination, suggesting formation by single-strand annealing (Reams et al. 2014). The RecA dependence seen in direct selection assays may reflect improved detection because of later recombination-dependent amplification events rather than the initial duplication exchange. To determine the role of recombination and avoid problems of higher amplification, several alternative assays were used to measure duplications formed between long repeats.
Assessing Reciprocal Recombination in a Duplication/Deletion System
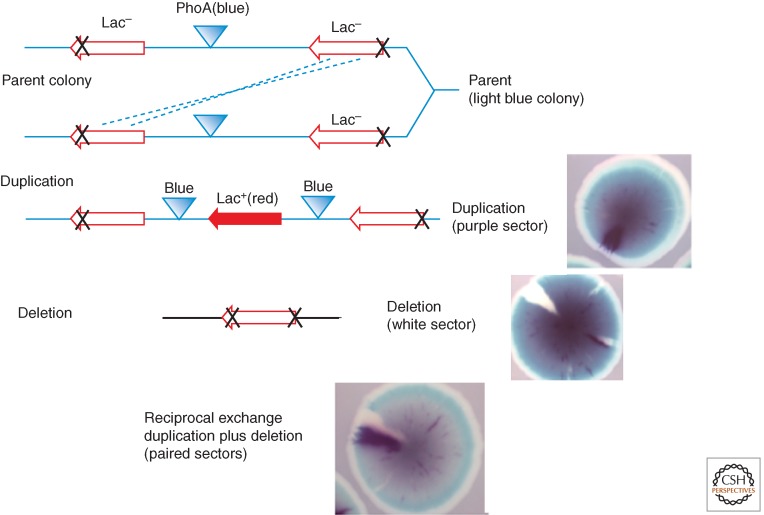
In this assay, direct 3-kb repeats of the Escherichia coli lac region are inserted 40 kb apart in the Salmonella chromosome (M Carter and AB Reams, unpubl.). This system assays the formation of both duplications and deletions between these repeats and estimates the frequency of reciprocal exchanges (see Fig. 4). The lac duplications are detected by the phenotype of an expressed recombinant lacZ+ gene at the join point. Because recombinants are scored visually and a single junction sequence is sufficient for nonselective visual detection of a duplication, this assay does not favor cells with higher amplification. In this assay, deletions are similarly detected visually by nonselected loss of an intervening marker.
Figure 4.
Visual detection of chromosomal duplications and deletions. The parent strain (top line) has separated inactive lac operons (3-kb repeats) and a central active phoA gene. The parent forms a light blue colony on rich medium containing chromogenic substrates Red-Gal and Blue-Pho. An unequal exchange can generate either a duplication (center line, purple sector) or deletion (bottom line, white sector). A reciprocal exchange forms twin sectors (bottom photograph).
In the assay strain (Fig. 4), the intact lacZ allele at the right is unexpressed because of a lack of a promoter and the lacZ allele on the left lacks the distal end of its coding sequence. Recombination between these defective lacZ alleles can generate a duplication with an expressed lacZ+ gene at the junction plus two copies of the central phoA+ gene. The reciprocal product is a deletion that lacks phoA and carries an untranscribed lacZ deletion allele at the deletion join point. The recombining sequences are 3 kb in length separated by 40 kb and have no added propensity to suffer nicks or breaks. All events are scored visually on nonselective medium.
Colonies of the tester strain grow on nonselective medium that includes indicators for both lac (Red-Gal) and phoA (Blue-Pho), where they form light blue colonies (phoA+ IacZ−). As these colonies form, an early duplication recombinant produces a purple sector (blue from two doses of phoA+ plus red from lacZ). A deletion recombinant forms a white sector (phoA− lac−). Reciprocal exchanges generate twin sectors—one purple and one white—above a light blue background (see Fig 4). In this assay, duplications and deletions form at similar rates, estimated at about ×10−4 per cell per generation, respectively. About 20% of duplication sectors are accompanied by a twin deletion sector, suggesting occasional reciprocality. In a recA mutant strain, deletion and duplication rates both dropped 10- and greater than 20-fold, respectively, suggesting that homologous recombination is responsible for most exchanges. Unexpectedly, single recB and recF mutations had little effect on either duplication or duplication rates. This behavior resembles earlier assays of duplication loss (Galitski and Roth 1997), but contrasts with the Rec-independent duplication of regions between rrn loci, which involve repeats with a similar size and separation (described below).
Duplications between Tandem Copies of the Ribosomal RNA (rrn) Genes
This assay resembles that described above, but assesses exchanges between native 5-kb ribosomal RNA loci (rrnB and rrnE) separated by ∼40 kb (Reams et al. 2014). Duplications of the rrnB–rrnE region were detected by the Kan Tet trapping assay described above (Fig. 2) and arose at a rate of ∼10−3 per cell per generation. Deletions of the intervening region are lethal and could not be recovered.
Surprisingly, the duplication rate of the rrnBE interval was essentially unaffected by recA, recB, recF, or recB-F double mutations, but dropped 30-fold in a ruvC-recG double mutant. This behavior was surprising in that recA, B, or F mutations impair homologous recombination pathways that operate in bacterial crosses (Kuzminov 2001; Spies and Kowalczykowski 2005). It was proposed that loss of homologous recombination was masked because these mutations activate an alternative pathway that can form duplications between rrn sequences by single-strand annealing. In the presence of recA, recB, or recF mutations, unrepaired gaps and breaks could accumulate. A sufficient accumulation would allow single-stranded ends from different rrn loci in different sister chromosomes to coexist and anneal, thus opening an alternative route to duplication formation. Achieving coincident single-strand ends would be facilitated by frequent damage to rrn sequences. The rrn loci are highly transcribed and their G/C skewed sequences include many quasipalindromes. These properties may combine to allow the nontranscribed strand to form secondary structures and be subject to consequent damage.
In recG, ruvC double mutants, duplication rates dropped, suggesting impairment of both homologous recombination and single-strand annealing. These mutations, particularly in combination, inhibit homologous recombination by preventing resolution of Holliday structures (Spies and Kowalczykowski 2005) but they do not prevent RecF and RecBC pathways from initiating repair of gaps and breaks. It was suggested that, in recG, ruvC mutants, the single-strand ends needed for annealing are still processed, but lead to accumulation of unresolved structures without an accumulation of free ends, thus ultimately blocking both pathways of duplication formation. Consistent with this idea, the duplication defect seen in recG, ruvC mutants is corrected by the addition of single recA, recB, or recF mutations. Each of these single mutations blocks an upstream step in homologous recombination and is expected to accumulate one or more sources of single-strand ends. The ability of these single mutations to correct the recG, ruvC duplication defect is thus explained if they leave ends available for the alternative annealing pathway (Reams et al. 2014).
Both the lac and rrn assays involve exchanges between substantial repeats (3 and 5 kb) separated by 100 and 40 kb, respectively, but the two assays showed very different dependencies on recombination. The RecA dependence of the lac assay suggests that less frequent or less persistent breaks initiate exchanges between different sisters that involve a single broken end in one chromosome and invading the other lac duplex (using RecA). Damage to both recombining sequences is rare. The Holliday structures generated by this invasion may lead to the occasional reciprocal exchanges seen in that assay. Reciprocality could not be tested for rrn flanked intervals because of the lethality of the deletions.
Reciprocity was tested for recombination between tandem 9-kb repeats of the rrn genes of yeast (Gangloff et al. 1996). About 100 such genes exist in tandem in the yeast chromosome (Linskens and Huberman 1988). As was seen for the lac system in the bacterial chromosome, reciprocality in yeast was seen in only ∼20% of exchanges between yeast rrn loci. This result was interpreted as evidence that the copy number change occurs by gene conversion-like events in which the two ends at a single break is repaired by invading an unbroken homolog at distant sites in the rrn array to increase the copy number in the broken chromosome. However, these events do not seem to require standard homologous recombination enzymes as expected for such exchanges (Houseley and Tollervey 2011). This may suggest that recombination defects and frequent damage to rrn sequences may activate alternative annealing pathways in yeast, as described above for bacteria (Reams et al. 2014). Such annealing events could often be nonreciprocal half-exchanges between sister chromosomes in which only one product is recovered (Yamamoto et al. 1992).
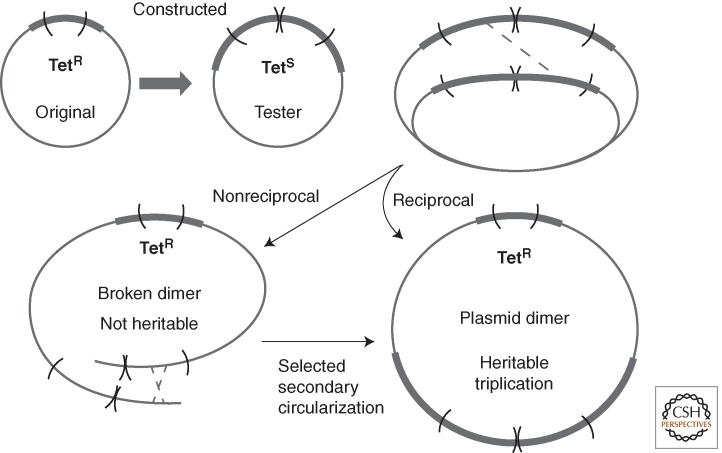
Evidence for Reciprocal Deletion/Duplication Events on a Small Plasmid
In two plasmid systems, a duplication is constructed within a gene (tet) whose restored function can be selected (Dianov et al. 1991; Mazin et al. 1991; Lovett et al. 1993; Kuzminov 1995; Bzymek and Lovett 2001). Selection is made for loss of the duplication and restoration of the Tet-resistance phenotype. This loss can occur without homologous recombination (RecA) when repeats of 21 or 42 bp are used, presumably by single-strand annealing and strand slippage (essentially by deletion). When larger repeats were used (165 or 401 bp), deletions decreased eightfold to tenfold in a recA strain. The deletion rate can be greatly enhanced by placing a 107-bp perfect palindrome between the repeats (Bzymek and Lovett 2001). Interestingly, placing either a perfect or imperfect palindrome between the recombining repeats caused a greater increase in deletions rates when RecA was absent. This system has been extremely revealing about the process of deletion formation and DNA repair events occurring near the replication fork, but the connection to duplication is less clear.
In this assay, the selected deletion is recovered in a circular plasmid dimer that carries the selected (tet+) gene in one plasmid unit and two or three copies of the defective TetR repeats in the second plasmid unit. This outcome has been interpreted as evidence for duplication by a single reciprocal exchange between nascent sister chromosomes. However, the same end point can be reached if the deletion arises by a nonreciprocal half-exchange (see Fig. 5). Such an exchange would produce a linear plasmid dimer with terminally redundant ends, which would not survive without closure. Genetic transmission (detection) of the functional tetA+ gene formed by deletion requires plasmid circularization—rejoining of the dimer ends. Reciprocality of the initial event in this assay is difficult to distinguish from required secondary recircularization involving one or even multiple plasmid copies. The reciprocality seen in the bacterial lac system above seems interpretable because the bacterial chromosomal dimer resolution mechanism permits recovery of a duplication or deletion recombinant in a single chromosome, regardless of whether it forms by a reciprocal or by a half-reciprocal exchange leaving a chromosome break.
Figure 5.
Deletion and duplication formation in a plasmid. The tetracycline resistance determinant (Tet) is disrupted by an internal duplication formed between nascent sister chromosomes. Selection for TetR demands loss of the duplication by a deletion event. Drug-resistant deletion mutants carry a plasmid dimer. Although this is consistent with a reciprocal recombination, a half-exchange would produce a linear dimer, whose TetR phenotype could only be maintained if the plasmid recircularized by a second exchange.
Duplications Formed by Transposable Elements
Transposable elements can contribute to duplications both by the act of transposition and by serving as portable regions of homology that support homologous recombination or annealing. The first example of a tandem duplication, Bar eye (Bridges 1936; Muller 1936), was much later shown to arise by an exchange between two preexisting transposable elements, one on either side of the duplicated region (Tsubota et al. 1989). The final duplication has a copy of the element at its join point but not at the outside end of either copy, suggesting that the parental strain (now lost) was heterozygous for two insertions flanking the region to be duplicated (Green 1985). Recombination between one element to the right of the region (in one parent) and a second element on the left side of the region (in the other parent) led to a duplication with a transposable element only at the junction (Tsubota et al. 1989). The phenotype of the Bar mutation is a result of the junction region structure, not to an increase in the gene copy number.
Similar duplications arise in bacteria by exchanges between repeated copies of insertion sequences (Peterson and Rownd 1985; Haack and Roth 1995). In the strains used for the Cairns selection (described below), duplications arise frequently (3 × 10−4 per cell per division) by homologous recombination between IS3 repeats that flank the lac operon on plasmid F′128 lac (Reams et al. 2012). In a recA mutant, the duplication rate is reduced only 100-fold and residual duplication ability depends heavily on IS3 transposase. Transposition is not involved because no novel DNA junction sequences are generated adjacent to IS3. The RecA independence suggests that these duplications arise by single-strand annealing, as was seen for rrn duplications described above. Transposase may contribute by nicking A at IS3 element ends and allowing nicks at different IS3 elements to provide the simultaneous single-strand ends needed for duplication by annealing.
Duplications can also form by acts of replicative transposition. An element close to one side of the gene being duplicated transposes replicatively into a target site on the opposite side of the duplicated region in a sister chromosome. This transposition generates, in one step, a duplication strain with one copy of the element at the junction and a parental copy on one end of the duplicated region. Multiple such duplications were detected as amplifications among unstable Lac+ revertants in the Cairns system described below (Kugelberg et al. 2006).
Very similar observations in Acinetobacter have been explained in a different way based on the transposition mechanism of the element involved. In the Acinetobacter system, growth on benzoate is allowed by co-amplification of the closely linked cat operons (Reams and Neidle 2003). Normally, a cat duplication occurs by RecA-independent nonhomologous end joining (NHEJ) or short homology-mediated annealing (Reams and Neidle 2003). These joint points have very short junction (SJ) repeats. However, when a copy of the transposable element IS1236 is placed close to one side of the cat operons, the majority of cat duplications carry a copy of IS1236 at their junction in addition to the original IS1236 insertion (Cuff et al. 2012). These duplications do not form by recombination following a simple transposition because no cells within the colony have a simple insertion at the downstream site predicted by the new duplication junction sequence. This behavior resembles duplication by replicative transposition described above. However, the IS1236 element is thought to transpose by excising a copy and adding an excised linear fragment to a distant target site—a mechanism that seems unlikely to generate a duplication join point (Duval-Valentin et al. 2004). It has been suggested that transposase introduces a nick at the distal end of the parent IS1236 element (farthest from cat) and this end initiates DNA replication on the sister chromosome at a site across from the cat operons. The proposed process resembles the template switching models described below (see MMBIR and TID).
TANDEM DUPLICATIONS BETWEEN SHORT OR NONEXISTENT REPEATS
Some duplications appear to arise by exchanges between very short preexisting sequence repeats (order of 10 base pairs) in the parent chromosome and have one copy of this sequence at their duplication join point (SJ duplications). Other duplications show no junction repeats. These duplications pose difficult mechanistic problems, but are likely to be important because most duplications are of this type. Most regions of the genome lack extensive nearby direct repeats and must duplicate in some other way. As expected, duplications in such regions have small or nonexistent repeats and form independent of homologous recombination. The models described below propose various ways to solve these problems and are not mutually exclusive.
SJ duplications of a particular gene (e.g., pyrD) arise at a rate of 10−6 per cell per division in the Salmonella chromosome. This rate is three orders of magnitude lower than that of regions between rrn sequence repeats, presumably because of a paucity of available flanking direct repeats. The pyrD duplication rate is, however, much higher than that of deletions, presumably because of a larger number of small site pairs that can be used for duplication (a and b in Fig. 1) without causing lethality. That is, even though any single pair of short sequences is used at a very low rate (∼10−9 per cell per division based on assays described below), the number of available sequence pairs is large. Duplication size and therefore frequency is less restricted by lethality than that of deletions. Several systems that detect duplication events between short sequences are described below with models proposed to explain them.
Selected Amplifications with Very Short Junction Sequences in Acinetobacter
Two closely linked operons (cat) are needed for growth on benzoate and require a single positive regulatory protein for their expression (Reams and Neidle 2003). Strains lacking this regulatory protein can grow on benzoate only when both cat operons are amplified. When 1010 mutant cells are plated on benzoate, ∼100 colonies accumulate over 4 weeks. Each colony is comprised of cells with an amplification (3–40 copies) of a region that includes both operons (Reams and Neidle 2004). These amplifications are generated by a multistep process. During growth before plating on selective medium, a RecA-independent exchange occurs between short repeats (0–6 base pairs) (Fig. 1A). On selective benzoate medium, these duplication-bearing cells grow slowly and improve their growth by cat amplification. This is the RecA-dependent extension of the process diagrammed in Figure 1B. The RecA-dependence of revertant colony yield reflects only the secondary selected amplification (AB Reams, unpubl.). The major puzzle of duplication formation between short repeats is how ends interact.
Amplifications Selected in E. coli and Salmonella during Long-Term Growth on Lactose
A system similar to the Acinetobacter system described above was designed by John Cairns to study the origin of mutations under selection (Cairns and Foster 1991). In this system, cells carrying a leaky lac frameshift mutation (2% residual function) on an F′ plasmid are pregrown on glycerol and then plated on minimal lactose medium, where they cannot grow. Over a weeks time, two types of revertant colonies accumulate. One colony type (10% of total) is similar to the amplification-carrying revertants described above for benzoate use in Acinetobacter. These colonies are composed of cells that carry a lac amplification with 10–100 tandem copies of the leaky mutant lac allele (Kugelberg et al. 2006). The initial duplication arises before plating and later amplifies under selection for faster growth on lactose. Growth is enhanced by the increased copy number of the mutant lac allele with no change in the lac sequence. When these Lac+ revertants are shifted to nonselective medium, the amplification and the Lac+ phenotype are no longer under selection and are progressively lost. Formation (and loss) of this unstable Lac+ phenotype depends on RecA, as expected for amplification (or loss) of a preexisting duplication. The duplications underlying these amplifications (SJ types) have very short junction sequences (4–12 bp) and highly variable repeat size. Among more than 30 independent lac amplifications sequenced, no two shared the same junction sequence (Kugelberg et al. 2006). These SJs seem unlikely to form by homologous recombination, which is required only for later amplification.
Nonindependent revertants (from a single-parent culture) do sometimes share a junction sequence, suggesting that they are sibs of a duplication-bearing cell that formed and divided before plating on selective medium (Kugelberg et al. 2006). During nonselective growth, these relatively small (∼20 kb) SJ duplications form at ∼6 × 10−6 per cell division, a 50-fold lower rate than seen for RecA-dependent lac duplications (130 kb) formed between 1-kb copies of IS3 on the same plasmid (Reams et al. 2012). Thus, in the Cairns lac system, as in the Acinetobacter benzoate system, unstable revertants arise from preexisting RecA-independent duplications that are subject to RecA-dependent amplification following exposure to selection.
Most of the unstable lac revertants seen in the Cairns system have an amplified region with tandem direct-order lac copies and are thought to arise by the process diagrammed in Figure 1. However, about one-third of the unstable revertants have a distinct type of amplification consisting of a series of repeats in alternating orientation, head-to-head and tail-to-tail (Kugelberg et al. 2006, 2010). These amplifications are expansions of an underlying TID that will be described later.
About 90% of the Lac+ revertant colonies in the Cairns system are not a result of amplification, but consist of cells with a stable lac+ revertant allele. These stable lac+ sequence changes were originally thought to arise during selective growth of the amplification strains described above (Kugelberg et al. 2006). However, a growing body of data suggests that these changes occur on selective medium in cells that are not growing (Hastings et al. 2004). Current evidence suggests that revertants arise from rare cells with multiple copies of the lac genes. Although these cells may be unable to divide on selective lactose medium, they can continuously replicate their F′ lac plasmid for several days as suggested previously (Galitski and Roth 1995).
Duplication Formation Using Particular Short Recombining Sequences
Even though exchanges between short repeats may underlie many spontaneous duplications, the rate and dependencies of forming specific SJs are difficult to study because the selected events can occur among a very wide variety of site pairs. This problem is minimized by assays that select for exchanges between specific short sequences that fuse two distant chromosomal regions and cause expression of a gene at the duplication junction. These assays severely limit the possible recombining sites to the few that form a join point, which provides the selectable phenotype. This method was first tested some time ago using a promoterless histidine operon with a silent hisD gene (Anderson and Roth 1978). Selection for hisD gene expression (growth on histidinol) revealed extremely rare mutants (frequency 10−9) with a duplication whose join point fused a downstream distant foreign promoter to the silent hisD gene. Most revertant duplications fused hisD gene to the end of the argAB operon 100 kb away (Anderson and Roth 1978; Shyamala et al. 1990). The responsible exchanges occurred between two nearly identical 28-bp repetitive extragenic palindromic (REP) elements present within both argAB and his operons. Other less-frequent duplications fused hisD to alternative active promoters by exchanges between less-similar REP elements. In this assay, a recA mutation reduced the yield of arg-hisD duplications only about sixfold and some arg-hisD fusions were recovered among the residual revertants that arose without RecA. The contribution of RecA to these exchanges may reflect enhanced recovery of mutants that amplified the join-point element (Conner 1993). It has been suggested that the RecA-independent duplications between REP elements may be generated by side activities of DNA gyrase or topoisomerase, which have been shown to bind and cleave REP elements (Shyamala et al. 1990; Gilson et al. 1991; Espeli and Boccard 1997).
MECHANISMS OF FORMING SJ DUPLICATIONS
Duplications with SJs form without homologous recombination. How can exchanges occur between such short sequences (sometimes zero base pairs)? Some models involve single-strand annealing between overhanging short complementary sequences from widely separated parts of the chromosome that can pair without need for strand invasion (RecA). Alternative models invoke template switching or illegitimate replicative extensions of 3′ ends as described below. The following mechanisms are not mutually exclusive, but some invoke discontinuous events, whereas others propose a multistep process. All try to solve the basic problem of recombination-independent joining of separated short sequences.
Illegitimate Recombination
Illegitimate recombination has been defined as exchanges between sequences with little or no similarity, whether or not they lead to duplication or some other rearrangement type. Hideo Ikeda and coworkers have measured such exchanges using a sensitive assay in which a λ prophage in the E. coli chromosome generates specialized transducing phages (Ikeda et al. 1995). Exchanges between one site within the prophage and a second site in the neighboring region of the E. coli chromosome excise a phage genome that lacks some phage genes and acquires some bacterial genes. These events have been placed into two classes. (1) Microhomology-independent events are thought to result from errors of topoisomerase and gyrase (Ashizawa et al. 1999). Gyrase-mediated exchanges might also contribute to the REP-mediated events described above because gyrase has been found to cause recombination in vitro and in vivo (Naito et al. 1984) and is known to bind to REP sequences (Yang and Ames 1988). (2) Microhomology-dependent events are attributed to single-strand annealing because they are stimulated by the RecE and RecT proteins of the Rac prophage (Shiraishi et al. 2006). At a double-strand end, RecE digests 5′ ends to reveal single-strand 3′ overhangs that are available for pairing catalyzed by the RecT protein. Given the sensitivity of these assays and their association with phage growth, it is not clear how heavily these pathways contribute to gene duplication in the bacterial chromosome. The events described by Ikeda and coworkers could also involve processes, such as template switching or TID modification as described below.
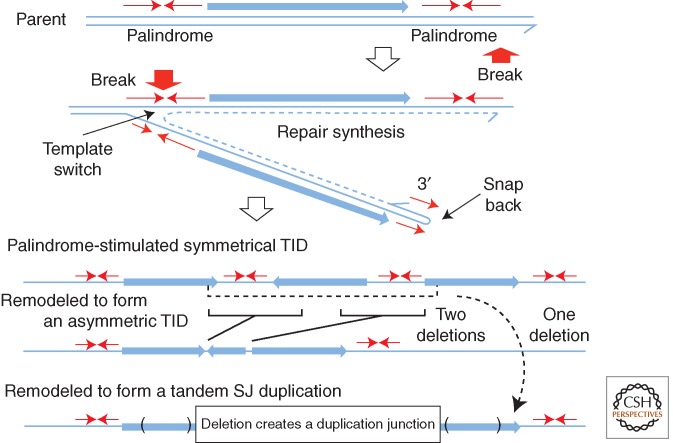
Formation and Processing of TIDs
This model produces duplications within a single chromosome without need for any genetic exchange between sister chromosomes (Kugelberg et al. 2010). The basic TID unit is actually a triplication of a region with copies in alternating orientation (head-to-head, tail-to-tail), whose formation is thought to be initiated at quasipalindromic sequences. A symmetrical TID is diagrammed in Figure 6. Two mechanisms have been suggested to explain TID formation and are outlined below. Once the basic TID is formed, it can amplify by recombinational exchanges between the direct-order repeats that flank the central inverse-order copy, much like the amplification drawn for standard tandem duplications in Fig 1B. Rearrangements of this type have been seen in two situations.
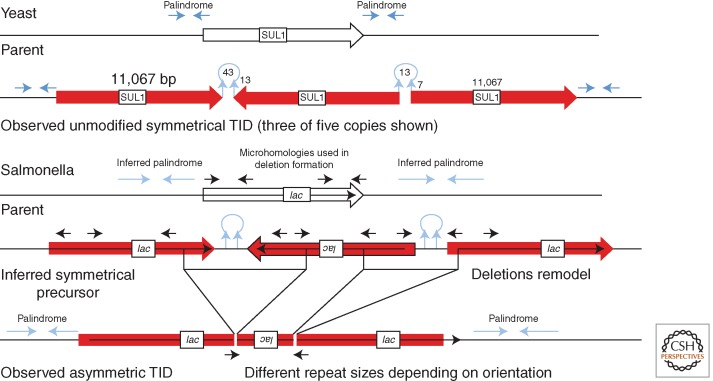
Figure 6.
Formation of a tandem inversion duplication (TID). This model proposes initiation of duplication by one palindromic sequence at which a 3′ end can snap back to prime repair synthesis. Template switching to the opposite strand by this replication track would be aided by a second palindrome or closely placed inverse repeat. Resolution or replication leaves three copies of the intervening region—two copies in direct order with a central third copy in inverse order. This same process can in principle operate at a single-strand nick far from a replication fork. The product is a symmetrical TID (sTID) whose two junctions have short parental palindromes that have been extended in the sTID and may be prone to remodeling by deletion (Kugelberg et al. 2010). It is proposed that observed asymmetric join points form when deletions remove the initial palindrome and leave an asymmetric join point generated at the site of the deletion. A single large deletion that removes both junctions and the central inverse-order copy can generate a simple-tandem repeat with a short-junction (SJ) sequence. Another model achieves the same end point by template switching across two diverging replication forks (Brewer et al. 2011). The same structures can be explained by the microhomology-mediated break-induced replication (MMBIR) model described below (Hastings et al. 2009a) in which template switches are not restricted to replication fork regions.
The simplest example is a TID amplification found in yeast after 300 generations of growth under selection for increased dosage of a sulfate transporter (Araya et al. 2010). The rearrangement has five tandem copies of the same chromosomal region in alternating orientations. The basic TID has two junction types, one between head-to-head copies and another between tail-to-tail copies. Each junction has a short quasipalindromic sequence that was present in the parent chromosome (see Fig. 6). In the symmetrical TID, these palindromes are extended through the entire inverse-order repeat. Two models to explain the origin of the TID are outlined below. In this yeast example, the initial symmetrical TID was presumably amplified further by subsequent recombination between direct-order repeats within the TID.
Amplifications with an asymmetric TID structure are seen in some unstable Lac+ revertants appearing under long-term selection in the Cairns system (described above). About 30% of the unstable Lac+ revertants in this system carry an amplification with repeated lac copies in alternating orientation (Kugelberg et al. 2006; Slack et al. 2006; Kugelberg et al. 2010). Unlike the symmetrical TID junctions seen in yeast, the asymmetrical bacterial TID junctions do not form extended symmetrical palindromes and have repeats in each orientation that are of different sizes (see Fig. 6). The asymmetric lac repeats can be explained as forming from an inferred initial symmetrical TID (as observed in yeast), but with junctions that are later remodeled by deletions that remove the origin palindromic junctions and render the join points asymmetric (see the bottom of Fig. 6). Formation of these deletions may be stimulated by the palindromic character of the junction (Sinden et al. 1991; Leach 1994). If the junctions are deleted individually, the final product is a TID with two asymmetric junctions (Kugelberg et al. 2010). The remodeling deletions arise between short direct repeats as is typical for deletion events. These short sequences were in inverse order in the parent but were brought into direct order by the TID.
The model we favor to explain formation of the initial TID (Fig. 6) suggests that a single-strand 3′ end (perhaps of the nascent leading strand) forms a snap-back structure that can initiate repair replication on the same strand. Replication continues away from the fork until a switch is made back to original leading strand template (see Fig. 6). This produces a branched structure whose replication or breakage (where indicated) leaves a symmetrical TID of the type described above and amplified in yeast. The extended palindromic junctions are subject to frequent deletions, especially in bacteria, where torsional supercoiling may favor hairpin extrusion (Sinden et al. 1991). If both junction types are rendered asymmetric by independent deletions, the final product is the asymmetric TID, which can subsequently be amplified under selection to produce the unstable TID amplifications that are 30% of the total unstable revertants (see the bottom of Fig. 6). If a single deletion removes both TID junctions with the central repeat, the product is a simple tandem (head-to-tail) SJ duplication, which can amplify and become an unstable revertant with a standard direct-order tandem repeat. According to the TID model, both reverse-orientation TID and tandem SJ amplifications form by this multistep process with short-lived intermediates.
The key to the TID model is the formation of snap-back structures and their use in priming repair synthesis. Considerable evidence supports such replication at snap backs in phage, bacteria, and yeast (Ripley 1982; Papanicolaou and Ripley 1991; Butler et al. 1996; Rattray et al. 2005; Dutra and Lovett 2006). Similar synthesis from snap-back palindromic sequences has been suggested for the breakage–fusion–bridge (BFB) model (described below).
A head-to-head amplification resembling the TID structure described above was found in yeast following prolonged growth under selection (Araya et al. 2010). This amplification arose between two short palindromes and was explained by a model in which a nascent leading single strand with a palindromic 3′ end anneals across the fork to initiate repair synthesis templated on the opposite sister chromosome (Brewer et al. 2011). This replication extends away from the fork of origin and toward another fork moving away at the opposite end of a replication bubble. There it switches back to the original leading strand template with the aid of a second palindrome. Switches at both diverging forks produces a head-to-head dimeric circle, which can be extracted and integrated into the chromosome to yield a symmetrical TID. Fork interactions at replication bubbles were also proposed to explain duplication by unequal translocation in human cancer cell lines (Howarth et al. 2011). The formation of TID duplications has also been explained using a template-switching model that does not restrict template switches to replication fork regions (Hastings et al. 2009b; Carvalho et al. 2011).
Duplication by Template Switching
In several models, joining of dissimilar sequences is attributed to replication template switching. Although these models can, in principle, explain the origin of TIDs with SJ sequences, they are mechanistically a bit tortuous and do little more than restate the features of the duplications they explain. One of the initial models (FoSTes) proposes that the 3′ end of a nascent Okazaki fragment anneals with the lagging strand template of a different replication fork on the opposite side of the gene being duplicated (Gu et al. 2008). A subsequent model, called microhomology-mediated break-induced replication (MMBIR), builds on break-induced replication (Anand et al. 2013). This model suggests that a double-strand end is resected to produce a 3′-ended single-strand overhang that can prime replication at a distant point in duplex DNA without benefit of a strand invasion protein. This replication start juxtaposes sequence from the priming strand with that of the newly synthesized template complement. If such forks collapse, the extended 3′ end can reinitiate elsewhere in the genome and thus produce complex rearrangements. This illegitimate initiation is said to become more likely during growth inhibition because of repression of the enzymes responsible for homologous recombination (Hastings et al. 2009b). The fork made in this way is unstable and subject to collapse during subsequent replication. Each collapse releases a new extended single 3′ end that can repeat the priming at a new distant point. The extending 3′ end thus accumulates sequence blocks from multiple genome regions in both orientations. The process of RecA-independent replication initiation has been shown in vitro (Li and Marians 2000; Kurth et al. 2013). These models may account for the complex rearrangements inferred to occur in some metazoan genomes (Liu et al. 2011, 2012), especially those thought to join sequences from different chromosomes. The models can also accommodate the TIDs found in bacteria and yeast.
Duplication and Amplification by BFB Cycles with and without Palindromes
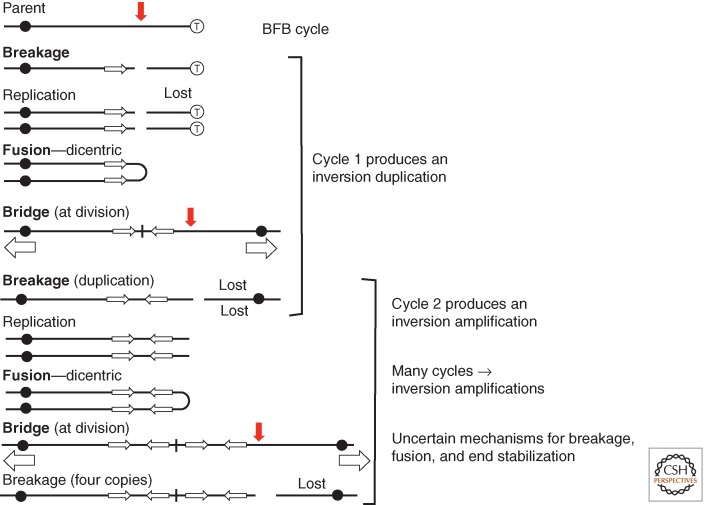
Some time ago, Barbara McClintock suggested the BFB cycle as a way of forming of inversion duplications and amplifications during mitosis (McClintock 1941). In her model, a chromosome breaks before replication and the broken ends of two copies fuse to generate a dicentric chromosome (see Fig. 7). At cell division, this dicentric breaks asymmetrically to form one chromosome with a terminal duplication and another with a corresponding deletion. The duplication-bearing chromosome lacks telomeres and replicates to form sisters that are subject to fusion and formation of another dicentric. Breakage then produces a chromosome with four copies of the repeat (two pairs of inverse-order repeats). Multiple mechanisms have been suggested to explain the breakage, fusion, and stabilization final product, but repetition of the BFB cycle continues to generate higher and higher copy number amplifications of inverse-order repeats.
Figure 7.
The breakage–fusion–bridge (BFB) cycle. Suggested many years ago by Barbara McClintock, this model explains the alternating orientation of copies seen in some amplification arrays. Issues are the source of the initial breaks, the forces that break a dicentric, the mechanisms of end fusions, and the stabilization of an array by blocking further end fusions. Several of these issues have been solved conceptually by the behavior of palindromic sequences.
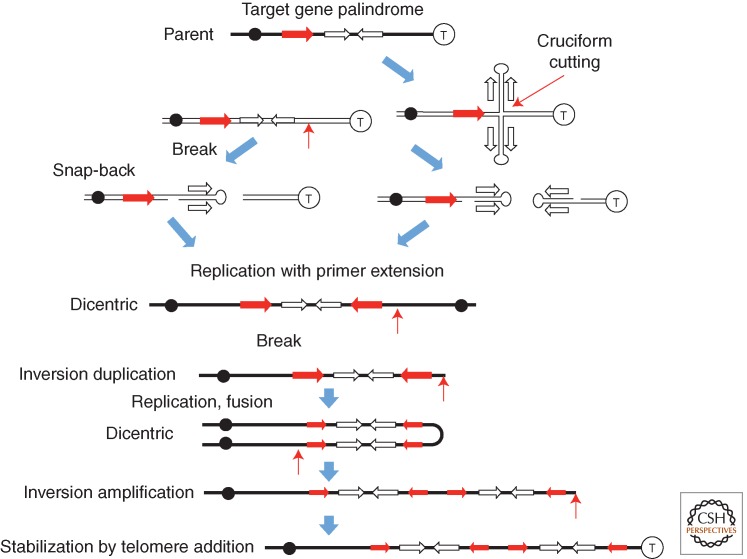
Based on the behavior of chromosomes during development of the Tetrahymena macronucleus, a model was proposed in which a palindromic sequence produces a break. Some possible mechanisms are in Figure 8. A terminal snap back at this end can prime replication leading to the formation of a dicentric chromosome that initiates the BFB cycle. Asymmetric breakage of the dicentric at cell division leaves an inversion duplication centered on the palindrome. The telomeric ends of the original chromosome are lost. The final product carries inverse-order repeats of various size regions, whose junctions are symmetrical and subject to remodeling to form asymmetric junctions. Gene amplification by BFB cycles has been brilliantly reviewed (Tanaka and Yao 2009). Palindrome-initiated amplification events of this type have been shown in yeast by Lobachev and coworkers (Lobachev et al. 1998; Narayanan and Lobachev 2007). It should be noted that snap-back primer extensions of the types suggested in Figure 8 may also occur at single-strand nicks that generate TIDs as described in Figure 7.
Figure 8.
Use of palindromic sequences for induction of breaks and fusions in the breakage–fusion–bridge (BFB) model. The frequent association of palindromic sequences with amplifications in mammalian amplification suggested various ways in which they might contribute to the events in the BFB model. A break generated near a palindrome (left side, top) can leave ends whose snap-back primes repair synthesis, and serves to generate a dicentric chromosome (left side). A cruciform structure can be cut to leave snap-back ends that can similarly prime replication to form a dicentric. Heavy black lines denote duplex DNA and lighter black lines denote single strands. Ends lacking telomeres are likely to be subject to fusion and continued rounds of the cycle.
DUPLICATION AND AMPLIFICATION BY MULTISTEP PROCESSES DURING GROWTH UNDER SELECTION
Most models for duplication formation, like those for point mutations, propose a single discontinuous event or a cascade of immediately sequential events with intermediate structures that cannot be inherited. However, several other duplication models described here involve multistep processes in which intermediate forms are heritable and therefore subject to remodeling and selection over multiple cell generations. This is notably true of the TID formation process in which the initial symmetrical duplication can be remodeled by deletion and amplified over multiple generations. This is also true of the BFB model in which multiple cell generations may be required to increase repeat copy number. In such processes, duplications can form over several generations. Selection can progressively favor steps in their initial formation and later modification, as they lead to their higher amplification.
The basic idea is that the initial duplication may form at a high rate and provide some modest selective benefit in excess of its cost. Cell growth is further improved if secondary rearrangements reduce the fitness cost, perhaps by removing selectively unimportant parts of the repeated unit. Lower cost allows higher selective amplification of a tandem duplication or a TID, thus improving growth. In the case of a symmetrical TID, junctions are extended quasi-palindromes that are subject to cutting. These junctions can be stabilized by deletions that render the junctions asymmetric or remove the entire central inverse repeat to form a simple tandem head-to-tail SJ duplication. Such nearly perfect palindromes are known to stimulate their own deletion (Sinden et al. 1991; Leach 1994). These deletions reduce duplication cost and increase the stability of the array. The intermediates in this process are all replicable, allowing duplications to be completed over several cell generations. The observed duplication and amplification junction sequences may reflect these secondary remodeling events rather than initial duplication formation.
The remodeling of palindromic junctions may be especially true in bacteria, where torsional supercoiling can drive hairpin formation and contribute to deletions of symmetrical TID junctions. This may explain the TID amplifications observed in the Cairns system. In eukaryotic chromosomes, toroidal supercoiling may minimize the rate of palindrome remodeling and allow quasipalindromic inversion junctions to persist. This could explain why yeast retains unmodified sTID junctions, whereas bacterial TID junctions are usually asymmetric. However, double hairpin structures are invoked in models for chromosome breakage at palindromes in yeast and mammalian cells (Akgun et al. 1997; Cunningham et al. 2003; Narayanan and Lobachev 2007). These results suggest that even in eukaryotes, palindromes may stimulate formation of deletions that remodel duplications and make them easier to amplify selectively. The selective remodeling of duplication junctions may make it difficult to infer formation mechanisms from the structure of ancient or even recent segmental duplications. The spacing and relative sizes of the repeated copies are likely to depend on the source of the breaks and the extent of selective modification.
AMPLIFICATIONS IN CANCER AND IN MICROBES
Amplifications are prominent genomic features of many types of malignant cells, particularly those of solid tumors (Albertson 2006). Similarly, amplifications often confer resistance to inhibiters used in cancer chemotherapy. Understanding how these amplifications are initiated and how they expand under selection is important to cancer prevention, therapy and in predicting the ultimate course of the disease.
The models described above emphasize genetic methods to isolate duplications and distinguish them from subsequent modification and amplification events. In contrast, cancer-associated amplifications are discovered in final form in genomes of malignant cells. Their structures are characterized and interpreted to suggest and support models for their formation. Many of the interpreted structures are likely to have developed during many generations under selection for improved growth. Data on amplifications in cancer cells come from different tumor types and from resistance selections of different stringencies imposed on cultured cell lines. These results may not all be interpretable in terms of a single consistent model. However, one hopes that some unifying principles may ultimately emerge. More importantly, one hopes that an understanding of amplification in microorganisms will help clarify the metazoan process. This seems possible because structures observed and characterized in bacteria and yeast are often similar to those in mammalian cells.
Much of the amplification literature on cancer and some on bacteria (Galhardo et al. 2007; Hastings 2007) treats formation of the amplification as a single discontinuous event rather than an evolutionary series of sequential occurrences—duplication, remodeling, and amplification. Ignoring selection leads to models that invoke sudden cataclysmic events and “genomic instability” to explain complex genomes with multiple rearrangements whose evolution may have required several events, occurring during many generations of selection for improved growth (Liu et al. 2011). This general viewpoint may also prompt models for sudden formation of amplified arrays rather than stepwise expansion of duplications under selection (Hyrien et al. 1988; Watanabe and Horiuchi 2005). Haber and Debatisse (2006) pointed out that secondary rearrangements complicate the interpretation of cancer cell amplifications. This problem increases if one considers that these structures can develop over many generations under selection. Selection would appear especially important for amplification in situations that allow frequent recombination between direct repeats in sister chromosomes or homologs. In these situations, amplifications are unstable and continuous copy loss is likely to restrict achievable copy number and be opposed by positive selection. Selection may favor changes that minimize loss rates and fitness costs so as to allow higher amplification of copy number.
In somatic cells, infrequent mitotic recombination may limit copy increases within a tandem array. However, this lower rate of copy number increase may be balanced by similar reduction in loss rate. In bacteria, the frequency of cells with an unselected duplication (and arguably the degree of amplification of any particular array) comes to a steady state. This occurs when the rate of copy gain (on one hand) balances the rate of copy loss and fitness cost (on the other hand) (Reams et al. 2010; S Maisnier-Patin and I Roush, unpubl.). Selection essentially reduces the fitness cost of these arrays and allows the steady-state level to increase. Thus, short amplifications may arise and persist in somatic cells before selection and then show rapid expansion and modification when selection is imposed, as might occur during tumor progression.
The effects of selection stringency and copy gain and loss rates may explain the evidence that amplifications are common in transformed cell lines but effectively absent from a normal somatic cell population (Tlsty 1990; Wright et al. 1990). We suspect that the selections used in these tests demand cells that already have many copies of the targeted gene. In transformed lines, a higher mitotic recombination rate or a higher level of preexisting duplications may allow cells to expand a short preexisting array and become fully resistant. Cells unable to expand their arrays many not survive the selection. That is, cells with more breaks, or less apoptosis in response to breaks, may show better amplification in response to stringent selection.
It has been claimed that amplifications in mammalian cells are only initiated after imposition of selection (Tlsty et al. 1989). This conclusion was based on the negative result of fluctuation tests designed to show preexisting genetic changes (Luria 1951). Unlike point mutations, duplications and short amplifications come to steady-state frequencies because of their high reversion rates and fitness cost as described above (Reams et al. 2010). The forces responsible for these steady states obscure frequency differences between cultures caused by differences in the timing of the initial duplication event. That is, any frequency elevation attributable to an early duplication event is returned to the steady state and fluctuation is not seen. This problem arose in the Cairns bacterial selection system (Cairns and Foster 1991), where the absence of fluctuation led to the initial conclusion that new mutants are initiated under selection. These tests could not detect preexisting copy number variants, which now seem likely to be responsible for initiating revertants (Sano et al. 2014).
The predominant model for gene duplication in mammalian cells is the BFB cycle described above in Figures 7 and 8. Support for this model reflects its ability to account for several troublesome features of mammalian gene amplifications.
Mammalian gene amplifications are often tandem arrays of copies in alternating orientation (TID). The BFB model explains the origin of such arrays. It can even explain expansion of the amplification without invoking selection. That is, an initiating break can start a cascade of mechanistically driven cycles that progressively add copies to the genome, even if amplification is deleterious. Now, however, similar tandem inversion expansions have been seen in both yeast and bacteria, where they are thought to arise in alternative ways. In microbes, formation of amplifications (of direct- or inverse-order repeats) seem to be driven by prolonged growth under selection for more gene copies rather than by repeated BFB cycles (Kugelberg et al. 2006; Slack et al. 2006; Araya et al. 2010).
Although not unique to the BFB model, double-strand breaks stimulate amplification, which are often associated with known fragile sites in DNA (Albertson 2006). The sequence features of fragile sites (A/T rich, G/C skewed) make them prone to cruciform formation (Mishmar et al. 1998; Zlotorynski et al. 2003). Thus, fragile sites are attractive as a source of breaks needed for BFB mechanism, but may also be a source of snap-back structures that contribute to amplification in other ways (Narayanan and Lobachev 2007).
Palindromic sequences are often associated with ends of amplified segments (Ford et al. 1985; Stark et al. 1989). The use of a palindrome in amplification was clearly shown in Tetrahymena and was interpreted in terms of the BFB cycle (Tanaka and Yao 2009). Palindromes fit with the BFB cycle because they can generate breaks and by hairpin cutting and snap-back replication can generate the dicentric chromosome called for by the BFB model. This may be the correct interpretation, but it is important to remember that other models for inversion duplication use palindromic sequences in independent ways.
The simplest BFB model generates inversion duplications with various repeat sizes separated by various size spacer regions because of variation in breaks sites and end joining. This structure reflects the positions of breaks at each turn of the cycle outlined in Figure 7 and diagrammed in Figure 8. Although this irregular structure is seen in some situations, arrays are often described as containing repeats of uniform size, separated by rather small regions that are extensions of one copy (Hyrien et al. 1988; Legouy et al. 1989; Okuno et al. 2004; Kitada and Yamasaki 2007). The actual joins between inverse-order sequences show very short junction repeats (order of 10 base pairs). This has been taken as support for duplication by BFB and the microhomologies attributed to NHEJ-dependent end fusions. One of these amplifications will be described below.
There is evidence in yeast that breakage and fusion aspects of BFB are explained not by NHEJ, but by processing of palindromes (Narayanan and Lobachev 2007). That is, a palindrome can assume a cruciform structure and be cut asymmetrically to generate two chromosome fragments, each with a cap formed by one arm of the cruciform (Fig. 8). This snap-back provides a primer that can stimulate replication of the chromosome. Thus, palindrome processing replaces the break and allows replication to generate a dicentric without fusion.
A simple yeast TID amplification recovered after prolonged selection for gene amplification is diagrammed in Figure 9 (top). One can account for its structure if the indicated palindromes (stems and loops) allowed a 3′ end to pair across a fork and prime a reverse-order copy as suggested (Araya et al. 2010; Brewer et al. 2011). Another explanation is snap-back repair synthesis as suggested for bacterial TID duplications (Kugelberg et al. 2010) and diagrammed in Figure 6. Regardless of the duplication mechanism, selection then favors further amplification of the basic three-copy TID unit using unequal sister-strand exchanges between tandem repeats that increase copy number from the three that are characteristic of a basic TID unit to the final five copies observed following selection. In this example, the basic unit appears to have amplified without prior modification. Each spacer between repeats includes only the closely spaced inverse-order sequences whose pairing is proposed to initiate the duplication.
Figure 9.
Amplifications characterized in yeast and bacteria. (Top) A yeast amplification was recovered following prolonged growth selecting for increases in the SUL1 product. The rearrangement appears to have arisen between two palindromic regions. A series of five identical copies of the amplified region are present in alternating orientation (only three copies are shown). It seems likely that formation involved a snap-back structure at each palindrome and that the initial structure has not been modified by subsequent rearrangement because the junction microhomologies are in inverse order. Unequal recombination caused the further amplification from three to five copies. (Bottom) A bacterial amplification, also recovered after prolonged selection, shows different-sized repeats in opposite orientation—here, the microhomologies are not palindromic. This is proposed to have arisen following replication from snap-back structures (as seen in yeast) to form asymmetrical inversion duplication with extended palindromes at each junction. The inferred initial inversion duplication (pictured) was then stabilized by asymmetric deletions that removed the initial junction palindrome and left the observed amplification.
In bacteria, the observed microhomologies at TID junctions do not include inverse repeats but rather a short microhomology found at widely separated points in the genome (see the bottom of Fig. 9). These junctions, like those of tandem duplications, have been attributed to secondary deletions that remove the original junction and shorten the duplicated region as diagrammed in Figure 7. It is inferred that the initial duplication in bacteria is likely to be an sTID like that seen in yeast. That is, the sTID occurs first and is then modified and probably stabilized by deletions that render it asymmetric. This process was diagrammed in Figure 7 and in more detail in Figure 9 (bottom). These deletions are frequent because of the stimulatory effects of the palindromic junctions, which are unstable in bacteria. The deletion process can leave different-sized repeats in opposite orientation, depending on the end points of the deletions (see bottom of Fig. 9).
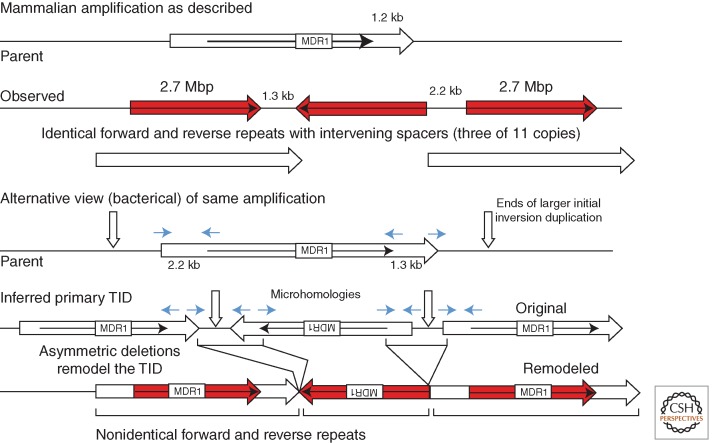
The BFB model predicts that mammalian amplifications consist of multiple pairs of large inverse repeats spaced at variable intervals and perhaps of variable size, depending on the positions of the several chromosome breaks. In contrast, somatic cell amplifications are often described as having multiple identical inverse-order repeats separated by a spacer of several kilobases in a regular tandem array (top of Fig. 10). The figure describes a particular amplification with 11 repeats characterized by Kitada and Yamasaki (2007). The junction between one side of the spacer and adjacent inverse-order sequence has microhomologies of a few base pairs (4 and 2 bp in this case), indicated by short arrows in the figure. The spacer regions and microhomologies can be attributed to a BFB cycle in which fusion was accomplished by NHEJ following the resection of one broken end.
Figure 10.
Two views of an amplification. The diagram describes three repeats of an 11-copy amplification selected in a human cell line for amplification of the MDR1 gene (Kitada and Yamasaki 2007). (Top) The inverse-order repeats are described as identical with spacers, but can also be viewed as distinct-sized repeats because the spacers are actually continuations of the amplified region. Formation of this structure can be explained in several ways, but is consistent with deletions modifying a larger initial inversion duplication.
However, the structure could also arise from a preexisting larger (possibly irregular) duplication formed by several BFB cycles that is later modified by deletion, as was suggested for bacterial TID amplifications (Kugelberg et al. 2010). This alternative view is diagrammed at the bottom of Figure 10, where the microhomologies are the point at which a deletion removed the junction region. This leaves inverse repeats that differ in length by the size of the spacer region. This sort of join-point deletion (“palindrome breaking”) has been suggested previously to explain such amplifications (Akgun et al. 1997; Cunningham et al. 2003; Kitada and Yamasaki 2007).
In the pictured mammalian amplification (Fig. 10), the increase in copy number from the basic three to 11 copies, like the yeast increase to five copies in Figure 9, seems likely to result from secondary unequal homologous recombination between direct-order sequences within the initial inversion duplication (see Fig. 1). The modifying deletions may allow higher amplification by removing unstable palindromes with significant fitness cost. In some cases, junctions may reflect deletions that formed between more substantial sequence repeats, such as Alu copies (Hyrien et al. 1987).
ACKNOWLEDGMENTS
We thank laboratory mates Eric Kofoid, Sophie Maisnier-Patin, Semarhy Quiñones, and Douglas Huseby, and colleagues Wolf Heyer, Neil Hunter, and Steve Kowalczykowski for advice and encouragement. This work is supported in part by National Institutes of Health (NIH) Grant GM27068.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M 1997. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 17: 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG 2006. Gene amplification in cancer. Trends Genet 22: 447–455. [DOI] [PubMed] [Google Scholar]
- Anand RP, Lovett ST, Haber JE 2013. Break-induced DNA replication. Cold Spring Harb Perspect Biol 5: a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RP, Roth JR 1977. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol 31: 473–505. [DOI] [PubMed] [Google Scholar]
- Anderson RP, Roth JR 1978. Tandem chromosomal duplications in Salmonella typhimurium: Fusion of histidine genes to novel promoters. J Mol Biol 119: 147–166. [DOI] [PubMed] [Google Scholar]
- Anderson P, Roth J 1981. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci 78: 3113–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RP, Miller CG, Roth JR 1976. Tandem duplications of the histidine operon observed following generalized transduction in Salmonella typhimurium. J Mol Biol 105: 201–218. [DOI] [PubMed] [Google Scholar]
- Araya CL, Payen C, Dunham MJ, Fields S 2010. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics 11: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashizawa Y, Yokochi T, Ogata Y, Shobuike Y, Kato J, Ikeda H 1999. Mechanism of DNA gyrase-mediated illegitimate recombination: Characterization of Escherichia coli gyrA mutations that confer hyper-recombination phenotype. J Mol Biol 289: 447–458. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Payen C, Raghuraman MK, Dunham MJ 2011. Origin-dependent inverted-repeat amplification: A replication-based model for generating palindromic amplicons. PLoS Genet 7: e1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB 1936. The bar “gene” a duplication. Science 83: 210–211. [DOI] [PubMed] [Google Scholar]
- Butler DK, Yasuda LE, Yao MC 1996. Induction of large DNA palindrome formation in yeast: Implications for gene amplification and genome stability in eukaryotes. Cell 87: 1115–1122. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Lovett ST 2001. Instability of repetititve DNA sequences: The role of replication in multiple mechanisms. Proc Natl Acad Sci 98: 8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Foster PL 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, et al. 2011. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet 43: 1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner CP 1993. Duplications between repetitive sequences in the chromosome of Salmonella, p. 76 University of Utah, Salt Lake City, UT. [Google Scholar]
- Court DL, Sawitzke JA, Thomason LC 2002. Genetic engineering using homologous recombination. Annu Rev Genet 36: 361–388. [DOI] [PubMed] [Google Scholar]
- Cuff LE, Elliott KT, Seaton SC, Ishaq MK, Laniohan NS, Karls AC, Neidle EL 2012. Analysis of IS1236-mediated gene amplification events in Acinetobacter baylyi ADP1. J Bacteriol 194: 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham LA, Cote AG, Cam-Ozdemir C, Lewis SM 2003. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol Cell Biol 23: 8740–8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov GL, Kuzminov AV, Mazin AV, Salganik RI 1991. Molecular mechanisms of deletion formation in Escherichia coli plasmids: I. Deletion formation mediated by long direct repeats. Mol Gen Genet 228: 153–159. [DOI] [PubMed] [Google Scholar]
- Dutra BE, Lovett ST 2006. Cis and trans-acting effects on a mutational hotspot involving a replication template switch. J Mol Biol 356: 300–311. [DOI] [PubMed] [Google Scholar]
- Duval-Valentin G, Marty-Cointin B, Chandler M 2004. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J 23: 3897–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott KT, Cuff LE, Neidle EL 2013. Copy number change: Evolving views on gene amplification. Future Microbiol 8: 887–899. [DOI] [PubMed] [Google Scholar]
- Espeli O, Boccard F 1997. In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: Genetic characterization of the target and identification of the cut site. Mol Microbiol 26: 767–777. [DOI] [PubMed] [Google Scholar]
- Ford M, Davies B, Griffiths M, Wilson J, Fried M 1985. Isolation of a gene enhancer within an amplified inverted duplication after “expression selection.” Proc Natl Acad Sci 82: 3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL 2006. Methods for determining spontaneous mutation rates. Methods Enzymol 409: 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM 2007. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol 42: 399–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T, Roth JR 1995. Evidence that F plasmid transfer replication underlies apparent adaptive mutation. Science 268: 421–423. [DOI] [PubMed] [Google Scholar]
- Galitski T, Roth JR 1997. Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics 146: 751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Zou H, Rothstein R 1996. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J 15: 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M 1991. The BIME family of bacterial highly repetitive sequences. Res Microbiol 142: 217–222. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, Eichler EE 2011. Human copy number variation and complex genetic disease. Annu Rev Genet 45: 203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MM 1963. Unequal crossing over and the genetical organization of the white locus of Drosophila melanogaster. Z Vererbungsl 94: 200–214. [Google Scholar]
- Green MM 1985. The role of mobile DNA elements in unequal and intrachromosomal crossing-over in Drosophila melanogaster. In Aneuploidy (ed. Dellarco V, Voytek P, Hollaender A), pp. 353–362 Plenum, New York. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR 2008. Mechanisms for human genomic rearrangements. Pathogenetics 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack KR, Roth JR 1995. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics 141: 1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Debatisse M 2006. Gene amplification: Yeast takes a turn. Cell 125: 1237–1240. [DOI] [PubMed] [Google Scholar]
- Hastings PJ 2007. Adaptive amplification. Crit Rev Biochem Mol Biol 42: 271–283. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Slack A, Petrosino JF, Rosenberg SM 2004. Adaptive amplification and point mutation are independent mechanisms: Evidence for various stress-inducible mutation mechanisms. PLoS Biol 2: e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009a. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009b. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2011. Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res 39: 8778–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth KD, Pole JC, Beavis JC, Batty EM, Newman S, Bignell GR, Edwards PA 2011. Large duplications at reciprocal translocation breakpoints that might be the counterpart of large deletions and could arise from stalled replication bubbles. Genome Res 21: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Debatisse M, Buttin G, de Saint Vincent BR 1987. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A+T-rich DNA. EMBO J 6: 2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Debatisse M, Buttin G, de Saint Vincent BR 1988. The multicopy appearance of a large inverted duplication and the sequence at the inversion joint suggest a new model for gene amplification. EMBO J 7: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Shimizu H, Ukita T, Kumagai M 1995. A novel assay for illegitimate recombination in Escherichia coli: Stimulation of λbio transducing phage formation by ultra-violet light and its independence from RecA function. Adv Biophys 31: 197–208. [DOI] [PubMed] [Google Scholar]
- Kitada K, Yamasaki T 2007. The MDR1/ABCB1 regional amplification in large inverted repeats with asymmetric sequences and microhomologies at the junction sites. Cancer Genet Cytogenet 178: 120–127. [DOI] [PubMed] [Google Scholar]
- Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc Natl Acad Sci 103: 17319–17324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E, Kofoid E, Andersson DI, Lu Y, Mellor J, Roth FP, Roth JR 2010. The tandem inversion duplication in Salmonella enterica: Selection drives unstable precursors to final mutation types. Genetics 185: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, Georgescu RE, O’Donnell ME 2013. A solution to release twisted DNA during chromosome replication by coupled DNA polymerases. Nature 496: 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A 1995. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol 16: 373–384. [DOI] [PubMed] [Google Scholar]
- Kuzminov A 2001. DNA replication meets genetic exchange: Chromosomal damage and its repair by homologous recombination. Proc Natl Acad Sci 98: 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach DR 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16: 893–900. [DOI] [PubMed] [Google Scholar]
- Legouy E, Fossar N, Lhomond G, Brison O 1989. Structure of four amplified DNA novel joints. Somat Cell Mol Genet 15: 309–320. [DOI] [PubMed] [Google Scholar]
- Li X, Marians KJ 2000. Two distinct triggers for cycling of the lagging strand polymerase at the replication fork. J Biol Chem 275: 34757–34765. [DOI] [PubMed] [Google Scholar]
- Linskens MH, Huberman JA 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol 8: 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski KJ, Farslow JC, Fitzpatrick KA, Lynch M, Katju V, Bergthorsson U 2011. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr Biol 21: 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR 2012. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev 22: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148: 1507–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST, Drapkin PT, Sutera VA Jr, Gluckman-Peskind TJ 1993. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics 135: 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE 1951. The frequency distribution of spontaneous bacteriophage mutants as evidence for the exponential rate of phage reproduction. Cold Spring Harb Symp Quant Biol 16: 463–470. [DOI] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, et al. 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Kuzminov AV, Dianov GL, Salganik RI 1991. Mechanisms of deletion formation in Escherichia coli plasmids: II. Deletions mediated by short direct repeats. Mol Gen Genet 228: 209–214. [DOI] [PubMed] [Google Scholar]
- McClintock B 1941. The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 6: a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, et al. 1998. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci 95: 8141–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan LV 1925. Polyploidy in Drosophila melanogaster with two attached X chromosomes. Genetics 10: 148–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrical SW 2015. DNA pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ 1936. Bar duplication. Science 83: 528–530. [DOI] [PubMed] [Google Scholar]
- Naito A, Naito S, Ikeda H 1984. Homology is not required for recombination mediated by DNA gyrase of Escherichia coli. Mol Gen Genet 193: 238–243. [DOI] [PubMed] [Google Scholar]
- Narayanan V, Lobachev KS 2007. Intrachromosomal gene amplification triggered by hairpin-capped breaks requires homologous recombination and is independent of nonhomologous end-joining. Cell Cycle 6: 1814–1818. [DOI] [PubMed] [Google Scholar]
- Okuno Y, Hahn PJ, Gilbert DM 2004. Structure of a palindromic amplicon junction implicates microhomology-mediated end joining as a mechanism of sister chromatid fusion during gene amplification. Nucleic Acids Res 32: 749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou C, Ripley LS 1991. An in vitro approach to identifying specificity determinants of mutagenesis mediated by DNA misalignments. J Mol Biol 221: 805–821. [DOI] [PubMed] [Google Scholar]
- Peterson BC, Rownd RH 1985. Recombination sites in plasmid drug resistance gene amplification. J Bacteriol 164: 1359–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD, Hill CW 1988. Recombination between repeated genes in microorganisms. Annu Rev Genet 22: 147–168. [DOI] [PubMed] [Google Scholar]
- Rattray AJ, Shafer BK, Neelam B, Strathern JN 2005. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev 19: 1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reams AB, Neidle EL 2003. Genome plasticity in Acinetobacter: New degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol Microbiol 47: 1291–1304. [DOI] [PubMed] [Google Scholar]
- Reams AB, Neidle EL 2004. Gene amplification involves site-specific short homology-independent illegitimate recombination in Acinetobacter sp. strain ADP1. J Mol Biol 338: 643–656. [DOI] [PubMed] [Google Scholar]
- Reams AB, Kofoid E, Savageau M, Roth JR 2010. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184: 1077–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reams AB, Kofoid E, Kugelberg E, Roth JR 2012. Multiple pathways of duplication formation with and without recombination (RecA) in Salmonella enterica. Genetics 192: 397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reams AB, Kofoid E, Duleba N, Roth JR 2014. Recombination and annealing pathways compete for substrates in making rrn duplications in Salmonella enterica. Genetics 196: 119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley LS 1982. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc Natl Acad Sci 79: 4128–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Palacios R 1997. Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet 31: 91–111. [DOI] [PubMed] [Google Scholar]
- Sano E, Maisnier-Patin S, Aboubechara J, Quiñones-Soto S, Roth JR 2014. Plasmid copy number underlies adaptive mutability in bacteria. Genetics 198: 919–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL 2007. Recombineering: In vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol 421: 171–199. [DOI] [PubMed] [Google Scholar]
- Shiraishi K, Imai Y, Yoshizaki S, Tadaki T, Ogata Y, Ikeda H 2006. The role of UvrD in RecET-mediated illegitimate recombination in Escherichia coli. Genes Genet Syst 81: 291–297. [DOI] [PubMed] [Google Scholar]
- Shyamala V, Schneider E, Ames GF 1990. Tandem chromosomal duplications: Role of REP sequences in the recombination event at the join-point. EMBO J 9: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Zheng GX, Brankamp RG, Allen KN 1991. On the deletion of inverted repeated DNA in Escherichia coli: Effects of length, thermal stability, and cruciform formation in vivo. Genetics 129: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet 2: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Kowalczykowski SC 2005. Homologous recombination by the RecBCD and RecF pathways. In The bacterial chromosome (ed. Higgins N), pp. 389–404 ASM, Washington, D.C. [Google Scholar]
- Stark GR, Debatisse M, Giulotto E, Wahl GM 1989. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 57: 901–908. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yao MC 2009. Palindromic gene amplification—An evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer 9: 216–224. [DOI] [PubMed] [Google Scholar]
- Tice SC 1914. A new sex-linked character in Drosophila. Biol Bull 26: 221–230. [Google Scholar]
- Tlsty TD 1990. Normal diploid human and rodent cells lack a detectable frequency of gene amplification. Proc Natl Acad Sci 87: 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlsty TD, Margolin BH, Lum K 1989. Differences in the rates of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luria-Delbruck fluctuation analysis. Proc Natl Acad Sci 86: 9441–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota SI, Rosenberg D, Szostak H, Rubin D, Schedl P 1989. The cloning of the Bar region and the B breakpoint in Drosophila melanogaster: Evidence for a transposon-induced rearrangement. Genetics 122: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Horiuchi T 2005. A novel gene amplification system in yeast based on double rolling-circle replication. EMBO J 24: 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JA, Smith HS, Watt FM, Hancock MC, Hudson DL, Stark GR 1990. DNA amplification is rare in normal human cells. Proc Natl Acad Sci 87: 1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Kusano K, Takahashi NK, Yoshikura H, Kobayashi I 1992. Gene conversion in the Escherichia coli RecF pathway: A successive half crossing-over model. Mol Gen Genet 234: 1–13. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ames GF 1988. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci 85: 8850–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B 2003. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol 23: 7143–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]