Abstract
AIM: To investigate the association between single nucleotide polymorphism (SNP) in promoter of the DNA methyltrans-ferase 3B (DNMT3B) gene and risk for development and lymphatic metastasis of gastric cardiac adenocarcinoma (GCA).
METHODS: The hospital based case-control study included 212 GCA patients and 294 control subjects without overt cancer. The DNMT3B SNP was genotyped by PCR and restriction fragment length polymorphism (RFLP) analysis.
RESULTS: The C/C genotype was not detected in both GCA patients and controls. In control subjects, the frequency of T/T and C/T genotypes was 94.9% and 5.1% respectively, and that of T and C alleles was 97.4% and 2.6%, respectively. The genotype and allelotype distribution in the GCA patients was not significantly different from that in controls (P = 0.34 and 0.33, respectively). When stratified by smoking status and family history of upper gastrointestinal cancer, significant difference in the genotype distribution was not observed between GCA patients and controls. The distribution of DNMT3B genotypes in GCA patients with or without lymphatic metastasis did not show significant difference (P = 0.42).
CONCLUSION: The distribution of DNMT3B SNP in North China is distinct from that in Caucasians. Although this SNP has been associated with susceptibility to lung, head, neck and breast cancer, it may not be used as a stratification marker to predict susceptibility and lymphatic metastasis of GCA, at least in the population of North China.
Keywords: DNA methyltransferase, Single nucleotide polymorphism, Gastric cardiac adenocarcinoma, Suscep-tibility, Lymphatic metastasis
INTRODUCTION
Aberrant DNA methylation is one of the most epigenetic changes in human cancers. Generally, the overall level of DNA methylation is lower in cancer cells than in normal cells, while a number of tumor suppressor genes are silenced by DNA methylation on CpG islands around their promoter regions in cancer cells[1-5]. DNA methylation is mediated by a family of DNA methyltransferases (DNMTs), three active forms (DNMT1, DNMT3A and DNMT3B) of DNMT have been identified in mammalian cells[6-8]. DNMT1 is required for maintaining methylation whereas DNMT3A and DNMT3B are responsible for de novo methylation[7,9]. DNMT3B plays a crucial role in tumorigenesis. Significant overexpression of DNMT3B has been observed in tumors while DNMT1 and DNMT3A are only modestly overexpressed with lower frequency[10]. The DNMT3B gene located on chromosome 20q 11.2, contains a C→T single nucleotide polymorphism (SNP) in the 149-base pair upstream of the transcription start site of the promoter region (C46359T, GenBank accession no. AL035071), which may result in a 30% increase in promoter activity[11] . Few studies suggested that the DNMT3B SNP may modify susceptibility to tumors, although conflicting results are reported in different tumor types. Carriers of the DNMT3B T allele, particularly of the heterozygous genotype, have a significant increase in risk of developing lung, head, and neck cancer[11,12]. However, genotypes with the C allele are associated with increased risk of breast cancer[13].
Gastric cardiac adenocarcinoma (GCA), one of the common tumor types in some areas of North China, has drawn increasing attention for its incidence elevation in recent years. Several polymorphic genes have been correlated with modification of susceptibility to GCA[14-17]. To our knowledge, the association between the DNMT3B SNP and the risk of development of upper gastrointestinal carcinoma has not been reported. In addition, the prevalence of DNMT3B SNP in Asian population has not been documented so far. Since the SNP may influence the function of DNMT3B, which plays an important role in carcinogenesis of upper gastrointestinal carcinoma, we hypothesized that DNMT3B SNP may modify susceptibility and disease progression of GCA. Therefore, a hospital-based case-control study was performed in a population of North China with the aim of testing the hypothesis.
MATERIALS AND METHODS
Subjects
The study recruited 212 patients with gastric cardiac cancer and 294 control subjects without overt cancer. The cancer patients were outpatients for endoscopic examination or inpatients hospitalized for surgery in the Fourth Affiliated Hospital of Hebei Medical University between 2001 and 2003. Histological tumor typing was carried out on the basis of biopsies or resected specimens in the Department of Pathology of the same hospital. Gastric cardiac carcinomas were all adenocarcinomas with their epicenters at the gastroesophageal junction, i.e., from 1 cm above to 2 cm below the junction between the end of the tubular esophagus and the beginning of the saccular stomach[18]. The healthy subjects having no history of cancerous or genetic diseases, visited the same hospital for regular physical examination and volunteered to join the epidemiology survey in the same period. All the cancer patients and control subjects were unrelated Han nationality and from Shijiazhuang city or its surrounding regions. Information on gender, age, smoking habits and family history was obtained from cancer patients and healthy controls by two professional interviewers. The definition of smokers and family history of upper gastrointestinal cancer (UGIC) was described previously[17,19]. Informed written consent was obtained from all the recruited subjects. The study was approved by the Ethics Committee of Hebei Cancer Institute.
DNA extraction
Five milliliters of venous blood from each subject was drawn in vacuum tubes containing EDTA and stored at 4 °C. Genomic DNA was extracted within one week after sampling by using proteinase K digestion followed by a salting out procedure[20].
DNMT 3B genotyping
The transition of C→T of DNMT3B SNP creates an Avr II restriction site, which could be exploited for genotyping by PCR and subsequent restriction fragment length polymorphism (RFLP) analysis. PCR was performed in a 25-µL volume containing 100 ng of DNA template, 2.5 µL of 10× buffer, 1 U of Taq-DNA-polymerase (BioDev-Tech., Beijing, China), 200 μmol/L of dNTPs and 200 nmol/L of sense primer (5’-TGCTGTGACAGGCAGAGCAG-3’) and antisense primer (5’- GGTAGCCGGGAACT-CCACGG-3’)[11]. For amplification, an initial denaturation step at 95 °C for 5 min was followed by 35 cycles at 95 °C for 30 s, at 66 °C for 30 s, at 72 °C for 1 min, and a final step at 72 °C for 5 min to allow for the complete extension of all PCR fragments. Subsequently, the PCR products were digested overnight with 10 units of Avr II (TakaRa Biotechnology Co., Ltd, Dalian, Liaoning, China) at 37 °C and separated on a 2% agarose gel. RFLP bands were visualized by ethidium bromide staining under UV light. Depending upon the existence of the Avr II recognizing site, the DNMT3B T/T genotype was expected to show two DNA bands at the positions of 207 and 173 bp, the C/C genotype was expected to show a single band (380 bp), while the heterozygote was expected to have three bands (380, 207 and 173 bp). For a negative control, distilled water was used instead of template DNA in the reaction system in each panel of PCR reactions. For 20% of the samples, the PCR-RFLP reaction was repeated once for quality control.
DNA sequencing analysis
DNA sequencing analysis was used to confirm the result of DNMT3B genotyping by PCR-RFLP analysis in 12 randomly selected representative samples. The PCR fragments were recovered from agarose gel followed by purification with DNA clean-up Kit (Wizard SV Gel and PCR clean-up System, Promega). DNA sequences of the PCR products were determined using the PCR sense primer with applied biosystems model 377 sequencer (PE Applied Biosystems, Warrington, UK).
Statistical analyses
Statistical analyses was performed using the SPSS11.5 software package (SPSS company, Chicago, IL, USA). Hardy-Weingberg equilibrium assumption was performed to compare the observed and expected genotype frequencies using χ2 test. Comparison of the DNMT3B genotype and allelotype distribution in the study groups was performed by means of two-sided contingency tables using χ2 test or Fischer’s exact test. The odds ratio (OR) and 95% confidence interval (CI) were calculated using an unconditional logistic regression model and adjusted by age and gender accordingly. P<0.05 was considered statistically significant.
RESULTS
As shown in Table 1, the mean age of GCA patients was comparable to that of healthy controls (P = 0.09). The gender distribution in GCA patients was also similar to that in healthy controls (P = 0.10). Information on smoking status was not obtained from 56 control subjects and 16 GCA patients, and family history of UGIC from 129 control subjects and 26 GCA patients was unclear or failed to be recorded. The proportion of smokers in GCA patients (58.2%) was significantly higher than that in healthy controls (43.3%) (χ2 = 9.53, P = 0.002). In addition, the frequency of individuals with positive family history of UGIC in GCA patients (30.6%) was significantly higher than that in healthy controls (5.5%) (χ2 = 36.34, P <0.0001). Thus, smoking and family history of UGIC significantly increased the risk of developing of GCA in this population (the age and gender adjusted OR = 1.80 and 9.60, 95%CI = 1.17-2.77 and 4.34-21.22, respectively).
Table 1.
Demographic characteristics of GCA patients and healthy individuals.
| Groups | Control n (%) | GCA n (%) | P |
| Sex | |||
| Male | 201 (68.4) | 159 (75.0) | 0.10 |
| Female | 93 (31.6) | 53 (25.0) | |
| Mean age in years (SD) | 59.1 (10.2) | 60.4 (8.4) | 0.09 |
| Smoking status | |||
| Ex- or current smoker | 103 (43.3) | 114 (58.2) | 0.0025 |
| Non-smoker | 135 (56.7) | 82 (41.8) | |
| Family history of UGIC | |||
| Positive | 9 (5.5) | 57 (30.6) | <0.0001 |
| Negative | 156 (94.5) | 129 (69.4) |
All the patients and controls were successfully genotyped for the DNMT3B polymorphism (Figure 1). The results of re-genotyped samples consistently matched the original ones. The genotyping by PCR-RFLP analysis was completely confirmed by DNA sequencing analysis (Figure 2). The genotype distribution in GCA patients and healthy controls was consistent with that expected by Hardy-Weinberg equilibrium. In addition, the DNMT3B genotype distribution did not correlate with gender and age both in patients and in controls (data not shown). The DNMT3B C/C genotype was not detected either in GCA patients or in control subjects. In healthy controls, the frequency of T/T and C/T genotypes was 94.9% and 5.1%, respectively, and that of T and C alleles was 97.4% and 2.6%, respectively.
Figure 1.
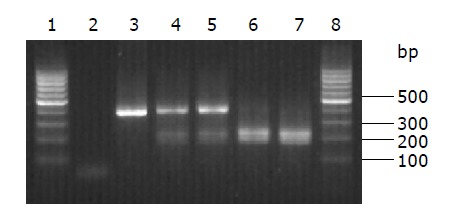
DNMT3B genotyping patterns by PCR-RFLP analysis. Lanes 1 and 8: 100 bp molecular marker; lane 2: negative control for PCR reaction; lane 3: PCR product; lanes 3 and 4: C/T genotype; lanes 6 and 7: T/T genotype.
Figure 2.
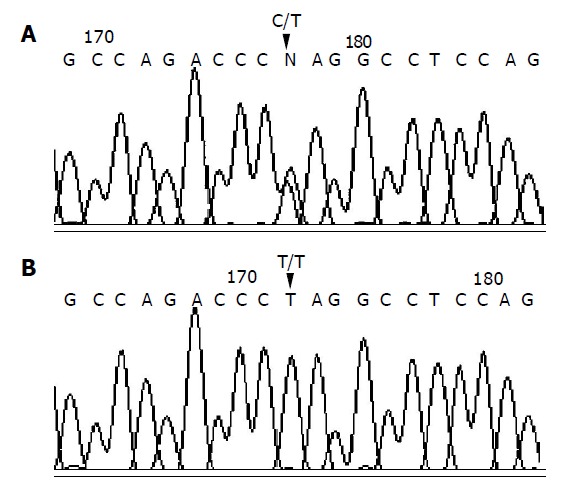
C/T genotype (A) and T/T genotype (B) confirmed by sequencing.
The frequency of DNMT3B T/T and C/T genotypes in GCA patients was 96.7% and 3.3% while that of T and C alleles was 98.3% and 1.7%, respectively. The genotype and allelotype distribution in the overall GCA patients was not significantly different from that in healthy controls (χ2 = 0.96 and 0.94, P = 0.33 and 0.33, respectively). When stratified by smoking status and family history of UGIC, the genotype distribution in GCA patients and healthy controls still did not show any significant difference (Table 2). Compared to T/T, in the study population, the C/T genotype did not significantly modify the risk of developing of GCA (the age and gender adjusted OR = 0.66, 95%CI = 0.26-1.67). The similar trend was observed in stratification analysis according to the smoking status and family history of UGIC (Table 2).
Table 2.
Influence of DNMT3B SNP on the risk of development and lymphatic metastases of GCA.
| Group | DNMT3B genotype T/T | n (%) C/T | aOR (95%CI) | P |
| Overall | ||||
| Control | 279 (94.9) | 15 (5.1) | ||
| GCA | 205 (96.7) | 7 (3.3) | 0.66 (0.26-1.67) | 0.38 |
| Nonsmoker | ||||
| Control | 130 (96.3) | 5 (3.7) | ||
| GCA | 78 (95.1) | 4 (4.9) | 1.43 (0.37-5.56) | 0.61 |
| Smoker | ||||
| Control | 98 (95.1) | 5 (4.9) | ||
| GCA | 111 (97.4) | 3 (2.6) | 0.61 (0.14-2.69) | 0.51 |
| Family history of UGIC in | ||||
| GCA patients | 82 (96.5) | 3 (3.5) | 0.40(0.05-3.14) | 0.38 |
| Positive | 97 (96.0) | 4 (4.0) | 0.94 (0.35-2.50) | 0.91 |
| Negative | ||||
| GCA with LM data | ||||
| LM- (n = 62) | 58 (93.5) | 4 (6.5) | ||
| LM+(n = 69) | 67 (97.1) | 2 (2.9) | 0.45 (0.08-2.63) | 0.38 |
LM: lymphatic metastasis.
To investigate the significance of DNMT3B genotyping in the progression of GCA, influence of DNMT3B polym-orphism on the occurrence of lymphatic metastasis was analyzed in 131 GCA patients whose clinical information was available. The distribution of DNMT3B genotype in GCA patients with or without lymphatic metastasis was not significantly different (2.9% versus 6.5%, Fisher’s exact test, P = 0.42). The C/T genotype did not significantly influence the risk of lymphatic metastasis compared to the T/T genotype (the age and gender adjusted OR = 0.45, 95% CI = 0.08-2.63). Thus, the effect of DNMT3B SNP on lymphatic metastasis of GCA was not observed in this study.
DISCUSSION
The present study shows a distinct difference in distribution of DNMT3B SNP between Chinese and Caucasians[11,13]. The C/C genotype in Chinese population is absent or rare, as demonstrated by the frequency of zero in both the GCA patients and healthy controls. Additionally, the frequency of the C/T genotype is very low in Chinese population (5.1% in the control group) compared to that in Caucasians. The frequency of T/T, C/T and C/C genotypes has been reported to be 23.2%, 41.8%, 35% in Americans[11] and 23.3%, 45%, and 31.8% in British population[13] respectively. The significance of great diversity in DNMT3B SNP distribution in different ethnic populations remains unknown. It was reported that the C to T transition induces a 30% increase in promoter activity of the DNMT3B gene[11,13]. If the observation could be verified in in vivo studies, the distinct prevalence of the T/T genotype in Chinese might imply the high level of DNMT3B activity in this population, which might result in a different methylation status between Caucasians and Chinese. In this study, we designed experiments to assess the association between DNMT3B genotypes and DNMT3B transcription and expression in resected tumor tissues and normal epithelia. Unfortunately, we could not conduct the experiments due to the absence of C/C genotype and the insufficient number of samples with C/T genotype. Nevertheless, to understand the significance of the great diversity in DNMT3B SNP distribution in different populations, the correlation between genotype and phenotype in different ethnic groups needs to be comparatively investigated.
The present study demonstrates that the association between DNMT3B SNP and susceptibility to GCA may not exist, at least in the study population. In addition, the DNMT3B SNP appears not to be a stratification marker for the risk of lymphatic metastasis of GCA. Although the sample size in the present study may be too small to detect the minor effects, the very similar distribution of DNMT3B genotypes in cancer patients and healthy controls suggests that DNMT3B SNP may not independently modify the risk of development and lymphatic metastasis of GCA. The present result, and those from lung and breast cancer which are the only published data on association between DNMT3B SNP and cancer development, can show the different possible roles of DNMT3B in different cell types. In lung cancer, the combined variant genotypes (CT+TT) of DNMT3B SNP have been associated with a nearly two-fold increased risk of cancer development compared to the C/C genotype[11]. In breast cancer, however, the combined genotypes of C/T and C/C have shown significantly higher risk of developing lung cancer compared to the T/T genotype[13]. Since the different splice variants of DNMT3B which may alter catalytic activity are expressed in a tissue specific manner[9], and repression of DNMT3B activity does not result in the re-expression of all hypermethylated tumor suppressor genes in some cell system[21,22], it is therefore important to explore the complex interplay of DNMTs in different tumor types. In addition, the absent association between DNMT3B SNP and GCA may be explained by the infrequency of C allele in the study population. To clarify the role of DNMT3B SNP in the development and progression of GCA, investigations in other populations need to be performed. Moreover, discovery of other polymorphisms in DNMT genes may facilitate further exploration of associations between genetic alteration and function of DNMTs.
In conclusion, the distribution of DNMT3B C46359T polymorphism in North China is distinct from that in Caucasians, and DNMT3B SNP may not be used as a stratification marker to predict susceptibility and lymphatic metastasis of GCA.
ACKNOWLEDGMENTS
The authors thank Mr. Li-Wei Zhang and Xiao-Qing Guo, Mr. Ming He and Ming-Li Wu at the Fourth Affiliated Hospital of Hebei Medical University, China, for their assistance in recruiting study subjects.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30371591 and the Natural Science Foundation of Hebei Province, No. C20040062
References
- 1.Laird PW, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3 Spec No:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 3.Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 4.Szyf M, Pakneshan P, Rabbani SA. DNA demethylation and cancer: therapeutic implications. Cancer Lett. 2004;211:133–143. doi: 10.1016/j.canlet.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 6.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–2113. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 8.Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 10.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002;62:4992–4995. [PubMed] [Google Scholar]
- 12.Wang L, Rodriguez M, Kim ES, Xu Y, Bekele N, El-Naggar AK, Hong WK, Mao L, Oh YW. A novel C/T polymorphism in the core promoter of human de novo cytosine DNA methyltransferase 3B6 is associated with prognosis in head and neck cancer. Int J Oncol. 2004;25:993–999. [PubMed] [Google Scholar]
- 13.Montgomery KG, Liu MC, Eccles DM, Campbell IG. The DNMT3B C--& gt; T promoter polymorphism and risk of breast cancer in a British population: a case-control study. Breast Cancer Res. 2004;6:R390–R394. doi: 10.1186/bcr807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao X, Yu C, Tan W, Xiong P, Liang G, Lu W, Lin D. A functional polymorphism in the matrix metalloproteinase-2 gene promoter (-1306C/T) is associated with risk of development but not metastasis of gastric cardia adenocarcinoma. Cancer Res. 2003;63:3987–3990. [PubMed] [Google Scholar]
- 15.Miao X, Xing D, Tan W, Qi J, Lu W, Lin D. Susceptibility to gastric cardia adenocarcinoma and genetic polymorphisms in methylenetetrahydrofolate reductase in an at-risk Chinese population. Cancer Epidemiol Biomarkers Prev. 2002;11:1454–1458. [PubMed] [Google Scholar]
- 16.Stolzenberg-Solomon RZ, Qiao YL, Abnet CC, Ratnasinghe DL, Dawsey SM, Dong ZW, Taylor PR, Mark SD. Esophageal and gastric cardia cancer risk and folate- and vitamin B(12)-related polymorphisms in Linxian, China. Cancer Epidemiol Biomarkers Prev. 2003;12:1222–1226. [PubMed] [Google Scholar]
- 17.Zhang J, Li Y, Wang R, Wen D, Sarbia M, Kuang G, Wu M, Wei L, He M, Zhang L, et al. Association of cyclin D1 (G870A) polymorphism with susceptibility to esophageal and gastric cardiac carcinoma in a northern Chinese population. Int J Cancer. 2003;105:281–284. doi: 10.1002/ijc.11067. [DOI] [PubMed] [Google Scholar]
- 18.Siewert JR, Stein HJ. Classification of adenocarcinoma of the oesophagogastric junction. Br J Surg. 1998;85:1457–1459. doi: 10.1046/j.1365-2168.1998.00940.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Schulz WA, Li Y, Wang R, Zotz R, Wen D, Siegel D, Ross D, Gabbert HE, Sarbia M. Association of NAD(P)H: quinone oxidoreductase 1 (NQO1) C609T polymorphism with esophageal squamous cell carcinoma in a German Caucasian and a northern Chinese population. Carcinogenesis. 2003;24:905–909. doi: 10.1093/carcin/bgg019. [DOI] [PubMed] [Google Scholar]
- 20.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soejima K, Fang W, Rollins BJ. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene. 2003;22:4723–4733. doi: 10.1038/sj.onc.1206510. [DOI] [PubMed] [Google Scholar]
- 22.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]


