Abstract
AIM: To explore the new target genes transactivated by hepatitis C virus (HCV) core protein and to elucidate the pathogenesis of HCV infection.
METHODS: Reverse transcribed cDNA was subjected to microarray assay. The coding gene transactivated by HCV core protein was cloned and analyzed with bioinformatics methods.
RESULTS: The expressive vector of pcDNA3.1(-)-core was constructed and confirmed by restriction enzyme digestion and DNA sequencing and approved correct. mRNA was purified from HepG2 and HepG2 cells transfected with pcDNA3.1(-)-core, respectively. The cDNA derived was subjected to microarray assay. A new gene named HCTP4 was cloned with molecular biological method in combination with bioinformatics method.
CONCLUSION: HCV core is a potential transactivator. Microarray is an efficient and convenient method for analysis of differentially expressed genes.
Keywords: Hepatitis C virus, Core protein, Microarray assay
INTRODUCTION
Hepatitis C virus (HCV) causes chronic liver disease, including chronic active hepatitis, liver cirrhosis and hepatocellular carcinoma[1-4]. About 170 million persons are infected with HCV worldwide and about 3.2% people are positive for anti-HCV in China. The pathogenesis of HCV infection is not clear[5,6].
The HCV core gene contains the most conserved sequence in the coding region of most HCV genotypes, which implies an important biological function. Since suitable viral culture systems are usually not available[7-9], analysis of HCV genome organization and viral-product function is important to understand the viral life cycle and the pathogenesis of HCV infection. In order to understand the pathogenesis of HCV infection, we investigated the transactivating effect of HCV core protein by microarray assay. Among 1152 genes, 95 genes transregulated by HCV core protein are involved in signal transduction, cell proliferation, differentiation, apoptosis, immunosuppression. One new gene, HCTP4 was studied by microarrary assay.
MATERIALS AND METHODS
Construction and identification of expression vectors of HCV core
Plasmid pBRTM/HCV-1 (provided by Rice CM, USC Rockfeller University) containing full-length HCV cDNA (9401 nt) was used to design polymerase chain reaction (PCR) primers for core (342-914 nt ) of HCV. PCR product was cloned into pGEM-T. After its accuracy was verified, sequences of the genes of HCV core were ligated into plasmid pcDNA3.1(-)-core containing full-length of HCV core gene. pcDNA3.1(-) obtained from Invitrogen Co. was digested by EcoRI and BamHI (Takara). PCR primers were as follows: sense primer, 5’-GAA TTC AAT GAG CAC GAA TCC TAA-3’; antisense primer, 5’-GGA TCC AGG CTG AAG CGG GCA CA-3’ (Shanghai BioAsia Biotechnology Co., Ltd, China).
Expression of pcDNA3.1(-)-core in HepG2 cells
HepG2 cells were transiently transfected with pcDNA3.1(-)-core using lipofectamine. At the same time, empty vectors transfected into cells served as control. HepG2 cells were plated at a density of 1×106 in RPMI 1640 containing 100 U/mL of penicillin, 100 µg/mL of streptomycin, and 100 mL/L heat-inactivated fetal bovine serum (FBS). Twenty-four hours after the cells growth reached 40-50% confluence, the cells were transfected with plasmids by using lipofectamine according to the manufacturer’s protocol (Gibco Co., USA).
mRNA and cDNA isolation
Total cellular RNA was isolated using TRIzol (Invitrogen Co., USA) according to the manufacturer’s instructions. Then mRNA was reverse transcribed to generate Cy3 and Cy5 fluorescent-labeled cDNA probes.
Hybridization conditions
Hybridization of the fluorescent probe to the microchip was performed in 1×UniHyb solution at 37 °C for 30 min. DNA Probe was denatured before hybridization at 95 °C for 1 min and chilled on ice. A 2- to 3-µL spot from each probe was applied to the microarray and covered with a plastic cover slip (5 mm×5 mm) to prevent drying of the probe during incubation in the hybridization cassette (TeleChem International, Inc., USA). After hybridization, the slides were washed once with 2×SSC+0.2% SDS for 10 min at room temperature, once with 0.1×SSC+0.2% SDS for 10 min, and once with 0.1×SSC for 10 min and dried at room temperature.
Scanning and quantitation of microarrays
Fluorescent images of the microarrays were generated by scanning the slides using a ScanArray 3000 (General Scanning). The fluorescent signals from each spot were measured and compared using ImaGene 3.0 software. Analysis of collected data was performed on the basis of total fluorescence intensity measured from a fixed circular area of each oligonucleotide spot. Fluorescent signals with a statistically significant difference (P<0.01) from the background level were considered to be positive and the results were expressed as a ratio.
Cloning and identification of new gene HCTP4
Among 95 different genes, we found a new gene and named it HCTP4. The HCTP4 gene was amplified by PCR using HpG2 cell DNA. PCR primers were as follows: sense primer, 5’-CCA TGG ATG TCA CAA GTT AAA AGC TC-3’; antisense primer, 5’-GGA TCC TTA GCA GTG GAA TCG AGT GG-3’ (Shanghai BioAsia Biotechnology Co., Ltd).
Study of HCTP4 by microarray assay
Briefly, the recombined expression plasmid pcDNA3.1(-)-HCTP4 was constructed, and HepG2 cells were transfected. Total mRNA was isolated from the HepG2 cells transfected with pcDNA3.1(-) and pcDNA3.1(-)-HCTP4, respectively. Microarray was conducted for screening of up- and down-regulated genes of HepG2 cells. Fluorescent signals with a statistically significant difference (P<0.01) from the background level were considered to be positive and the results were expressed as a ratio.
RESULTS
Identification of expression vector
Restriction enzyme analysis of pcDNA3.1(-)-core plasmid with EcoRI/BamHI yielded two bands: 4 900 bp pcDNA3.1(-) and 573 bp HCV core. Analysis of PCR reaction products by agarose gel electrophoresis got a clear band of the expected size (573 bp). Sequence of the PCR product was correct (Figure 1).
Figure 1.
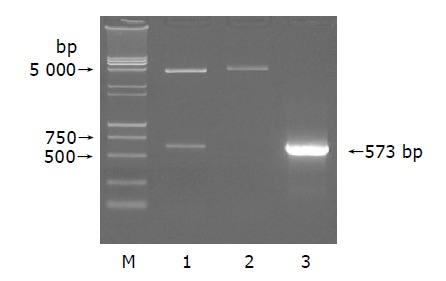
Eletrophoresis of pcDNA3.1(-)-core plasmid(A), cDNA(B) and HCTP4 (C) in 1% agarose gel. A: Lane 1: EcoRI/BamHI; lane 2: HindIII; lane 3: plasmid; M: DNA Marker (15000+2000 bp).
Identification of HCV core transient expression
After being reverse-transcribed by three different Oligo dT, identification of cDNA by PCR yielded a common 573 bp band (Figure 2).
Figure 2.
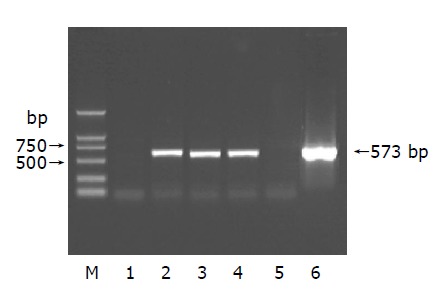
Eletrophoresis of cDNA in 1% agarose gel. Lane 1: negative control; lanes 2-4: total RNA; lane 5: blank control; lane 6: positive control; M: DNA Marker (2000 bp).
Result of HCV core by microarray analysis
Approximately 45 up-regulated and 50 down-regulated genes were identified by HCV core in HepG2 cells. Some up- and down-regulated genes are shown in Tables 1 and 2.
Table 1.
Up-regulated genes by HCV core.
| Accession numbers | Protein | Cy5/Cy3 |
| NM_005657 | Tumor protein p53 binding protein 1, TP53BP1 | 2.004 |
| D50683 | TGF-betaIIR alpha | 2.072 |
| NM_006595 | Apoptosis inhibitor 5, API5 | 2.199 |
| NM_000612 | Insulin-like growth factor 2, IGF2 | 2.232 |
| NM_002530 | Neurotrophic tyrosine kinase, receptor | 2.233 |
| NM_000760 | Colony stimulating factor 3 receptor, CSF3R | 2.253 |
| NM_014350 | TNF-induced protein, GG2-1 | 2.358 |
| NM_006290 | Tumor necrosis factor, alpha-induced protein 3, TNFAIP3 | 2.359 |
| NM_003151 | Signal transducer and activator of transcription 4, STAT4 | 2.390 |
| U07139 | Voltage-gated calcium channel beta subunit | 2.423 |
| NM_000014 | Alpha-2-macroglobulin, A2M | 2.526 |
| NM_012112 | Chromosome 20 open reading frame 1, C20orf1 | 2.689 |
| NM_002736 | Protein kinase, cAMP- dependent, regulatory | 2.710 |
| NM_014575 | Schwannomin interacting protein 1, SCHIP1 | 2.737 |
| U47077 | DNA-dependent protein kinase catalytic subunit, DNA-PKcs | 2.787 |
| AF352051 | Proliferation potential-related protein | 2.827 |
| NM_003998 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105, NF-kB1 | 3.005 |
| NM_004728 | DEAD/H box polypeptide 21 | 3.149 |
| NM_005100 | A kinase anchor protein 12, AKAP12 | 3.897 |
| NM_005171 | Activating transcription factor 1, ATF1 | 4.142 |
| BC008959 | Histocompatibility 13 | 4.760 |
| NM_014863 | B cell RAG associated protein, BRAG | 5.769 |
| NM_002291 | Laminin, beta 1, LAMB1 | 10.334 |
Table 2.
Down-regulated genes by HCV core.
| Accession numbers | Protein | Cy5/Cy3 |
| NM_000535 | PMS2 postmeiotic segregation increased 2 | 0.122 |
| NM_012096 | Adaptor protein containing pH domain, PTB domain and leucine zipper motif, APPL | 0.386 |
| NM_003844 | Tumor necrosis factor receptor superfamily, member 10a | 0.388 |
| NM_001226 | Caspase 6, apoptosis-related cysteine protease | 0.400 |
| NM_001229 | Caspase 9, apoptosis-related cysteine protease | 0.429 |
| NM_002287 | Leukocyte-associated Ig-like receptor 1 | 0.440 |
| AF090693 | Apoptosis-related RNA binding protein | 0.462 |
| NM_021020 | Leucine zipper, putative tumor suppressor 1 LZTS1 | 0.464 |
| NM_000062 | Serine or cysteine proteinase inhibitor | 0.471 |
| NM_000575 | Interleukin 1, alpha | 0.472 |
| AF016266 | TRAIL receptor 2 | 0.477 |
| NM_003796 | RNA polymerase II subunit 5 RPB5-mediating protein, RMP | 0.485 |
| NM_000629 | Interferon alpha, beta and omega receptor 1,IFNAR1 | 0.494 |
Identification of RT-PCR products from HCTP4
Among 95 different genes, we found a new gene and named it HCTP4. The nucleotide sequence data of HCTP4 reported in this paper appear in the GenBank nucleotide sequence database under the following accession numbers AY734680. The production of HCTP4 PCR is 2244 bp. (Figure 3).
Figure 3.
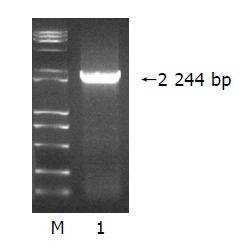
Eletrophoresis of HCTP4 in 1% agarose gel. Lane 1: HCTP4; M: DNA marker (15000 bp).
Result of HCTP4 by microarray analysis
DNA microarray showed that 56 genes were up-regulated by HCTP4 in HepG2 cells (Table 3) and 52 genes were down-regulated by HCTP4 in HepG2 cells (Table 4).
Table 3.
Up-regulated genes by HCTP4 protein.
| Accession numbers | Protein | Cy5/Cy3 |
| NM_002599 | Phosphodiesterase 2A, cGMP-stimulated (PDE2A) | 2.011 |
| NM_053274 | FKBP-associated protein (FAP48), transcript variant 1 | 2.014 |
| NM_012319 | LIV-1 protein, estrogen regulated (LIV-1) | 2.017 |
| NM_007047 | Butyrophilin,subfamily 3, member A2 (BTN3A2) | 2.030 |
| NM_021950 | Membrane-spanning 4-domains, subfamily A, member 1 (MS4A1) | 2.033 |
| NM_000817 | Glutamate decarboxylase 1 (GAD1) | 2.048 |
| NM_033625 | Ribosomal protein L34 (RPL34) | 2.055 |
| NM_006526 | Zinc finger protein 217 (ZNF217) | 2.058 |
| NM_112219 | Esterase D | 2.068 |
| NM_000386 | Bleomycin hydrolase (BLMH) | 2.079 |
| NM_006330 | Lysophospholipase 1 (LYPLA1) | 2.088 |
| D21262 | KIAA0035 gene | 2.089 |
| NM_003796 | RPB5-mediating protein (RMP) | 2.091 |
| NM_004904 | cAMP response element-binding protein (CRE-BPa) | 2.096 |
| AF012086 | Ran binding protein 2 (RanBP2alpha) | 2.118 |
| NM_005836 | Translational inhibitor protein p14.5 (UK114) | 2.126 |
| NM_003998 | Heat shock 70 ku protein 8 (HSPA8) | 2.138 |
| NM_000816 | Gamma-aminobutyric acid (GABA)A receptor, gamma 2 (GABRG2) | 2.174 |
| NM_003129 | Squalene epoxidase (SQLE) | 2.174 |
| NM_022171 | T-cell leukemia translocation altered gene (TCTA) | 2.211 |
| NM_006644 | Heat shock 105 ku (HSP105B) | 2.256 |
| AF070674 | Inhibitor of apoptosis protein-1 (MIHC) | 2.281 |
| NM_001892 | Casein kinase 1, alpha 1 (CSNK1A1) | 2.285 |
| NM_001539 | Heat shock protein, DNAJ-like 2 (HSJ2) | 2.303 |
| NM_013943 | Chloride intracellular channel 4 (CLIC4) | 2.325 |
| NM_006407 | Vitamin A responsive; cytoskeleton related (JVVA) | 2.370 |
| NM_014637 | T-cell receptor rearranged beta chain gene V-region (V-D-J)V-beta-AT | 2.395 |
| M11952 | Nuclear receptor subfamily 3, group C, member 1 (NR3C1) | 2.396 |
| NM_007268 | Ig superfamily protein (Z391G) | 2.445 |
| NM_021129 | Pyrophosphatase (inorganic) (PP), nuclear gene encoding mitochondrial protein | 2.498 |
| NM_002731 | Protein kinase, cAMP-dependent, catalytic, beta (PRKACB) | 2.525 |
| D85606 | Gene for cholecystokinin type-A receptor | 2.635 |
| NM_024824 | Hypothetical protein FLJ11806 (FLJ11806) | 2.730 |
| AF072928 | Myotubularin related protein 6 | 2.802 |
Table 4.
Down-regulated genes by HCV core.
| Accession numbers | Protein | Cy5/Cy3 |
| NM_001961 | Eukaryotic translation elongation factor 2 (EEF2) | 0.040 |
| NM_002313 | Actin binding LIM protein 1 (LIM), transcript variant ABLIM-1 | 0.214 |
| NM_014680 | KIAA0100 gene product (KIAA0100) | 0.234 |
| NM_003682 | MAP-kinase activating death domain (MADD) | 0.259 |
| NM_001226 | Prosaposin | 0.266 |
| NM_004728 | DEAD/H box polypeptide 21 (DDX21) | 0.313 |
| NM_013975 | Ligase III, DNA, ATP-dependent (LIG3) | 0.317 |
| NM_014889 | Metalloprotease 1 (MP1) | 0.347 |
| NM_005770 | Small EDRK-rich factor 2 (SERF2) | 0.348 |
| NM_005167 | Ras homolog gene family, member C (ARHC) | 0.368 |
| NM_000208 | Insulin receptor (INSR) | 0.369 |
| NM_003330 | Thioredoxin reductase 1 (TXNRD1) | 0.373 |
| NM_005243 | Ewing sarcoma breakpoint region 1 (EWSR1) | 0.396 |
| NM_016250 | N-myc downstream-regulated gene 2 (NDRG2) | 0.404 |
| NM_001250 | Tumor necrosis factor receptor superfamily, member 5 (TNFRSF5) | 0.406 |
| NM_003313 | Tissue specific transplantation antigen P35B (TSTA3) | 0.416 |
| NM_004127 | G protein pathway suppressor 1 (GPS1) | 0.416 |
| NM_002708 | Protein phosphatase 1, catalytic subunit, alpha isoform (PPP1CA) | 0.431 |
| NM_001777 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer | 0.444 |
| NM_002199 | Interferon regulatory factor 2 (IRF2) | 0.454 |
| NM_002087 | Granulin (GRN) | 0.469 |
| NM_000660 | Transforming growth factor, beta 1 (TGFB1) | 0.475 |
| NM_054012 | Argininosuccinate synthetase (ASS) | 0.478 |
| NM_002084 | Glutathione peroxidase 3 (GPX3) | 0.493 |
DISCUSSION
Diverse functional activities of the HCV putative core protein are noted in a number of investigations[10-13]. We cotransfected HepG2 cells with pcDNA3.1(-)-core and pSV-lacZ and demonstrated that the HCV core was successfully expressed in transfected HepG2 cells. Expression of β-gal was 5.4-fold higher in cotransfected pcDNA3.1(-)-core and pSV-lacZ than in cotransfected empty pcDNA3.1(-) and pSV-lacZ. HCV core had a significant transactivating effect on early promoter of SV40, and increased the expression of downstream gene lacZ. This result indicates that the HCV core protein expressed in HepG2 cells retains its biological activity in terms of transcriptional activation, which is inconsistent with previous reports[14].
To understand the trans-action mechanism of the core protein, a microarray assay was used to identify the relative transactivating target genes of HCV core protein. Approximately 45 up-regulated and 50 down-regulated genes were identified by HCV core protein in HepG2 cells. The up-regulated genes include tumor protein p53 binding protein 1, apoptosis inhibitor 5, TGF-βIIR alpha, insulin-like growth factor 2, tumor necrosis factor α-induced protein 3, signal transducer and activator of transcription 4, α-2-macroglobulin and proliferation potential-related protein. The down-regulated genes include member 10 of a tumor necrosis factor receptor superfamily, apoptosis-related cysteine protease, leukocyte-associated Ig-like receptor 1, apoptosis-related RNA binding protein, leucine zipper, interleukin 1, interferon α, α and ω receptor 1. The results show that HCV core protein has multiple regulatory functions in host-cell transcription, apoptosis, cell transformation and lipid metabolism and may play a role in suppressing host immune response[15-19].
The transregulation of HCV core protein is displayed extensively, one of the mechanisms of transregulation is that HCV core protein interacts with the promoters of genome in infected cells and affects the expression of gene. Another mechanism is that HCV core protein interacts with transcription factor in nuclei of infected cells and indirectly affects the expression of gene. HCV core protein interacts with various proteins which may be an important reason for hepatocellular damage and development of hepatocellular carcinoma. HCTP4 was identified and deposited in GenBank; the access number is AY734680. In order to investigate the function of HCTP4, cDNA microarray technology was employed. Approximately 56 up-regulated and 52 down-regulated genes were identified in HepG2 cells.
In the up-regulated genes by HCTP4, CLIC4 is differentially regulated in fibroblasts and its expression contributes to a collective stationary myofibroblast phenotype[20]. ZNF217 is a candidate oncogene on chromosome 20q13.2. ZNF217-transduced cultures give rise to immortalized cells. Overexpression of ZNF217 may be responsible for the development of hepatomas[21]. Inhibitor of apoptosis protein-1(MIHC) has effects on apoptosis[22]. JVVA is vitamin A-responsive and might be associated with cytoskeleton, which may play a role in the regulation of cell differentiation[23]. cAMP is an important signaling molecule for a variety of cellular functions and exerts its effects by activating the cAMP-dependent protein kinase (AMPK), which transduces the signal through phosphorylation of different target proteins. The inactive holoenzyme of AMPK is a tetramer composed of two regulatory and two catalytic subunits. cAMP causes dissociation of the inactive holoenzyme into a dimer of regulatory subunits bound to four cAMP and two free monomeric catalytic subunits[24,25].
In the down-regulated genes by HCTP4, TGFB is a multifunctional peptide that controls proliferation, differentiation, and other functions in many cell types. TGFB acts synergistically with TGFA (MIM 190170) in inducing transformation and as a negative autocrine growth factor. Dysregulation of TGFB activation and signaling may result in apoptosis[26]. LIM domains found in over 60 proteins, play key roles in the regulation of developmental pathways and function as protein-binding interfaces, mediating specific protein-protein interactions, thus becoming a candidate tumor suppressor gene[27]. HCTP4 may have effects on development of hepatomas. Saposins (sphingolipid activator proteins) A-D are 80-amino acid lysosomal glycoproteins encoded by a single gene, termed prosaposin. The proteolytic processing of prosaposin to individual saposins occurs predominantly in acidified compartments including lysosome. The physiological importance of this locus has been demonstrated by the genetic deficiencies of individual saposins or prosaposin that lead to various glycosphingolipid storage diseases[28,29]. Insulin is a pleiotropic hormone with multiple integrated metabolic and mitogenic signaling pathways upon binding to the cell surface insulin receptor[30]. HCTP4 interacts with prosaposin and insulin receptor and influences their biological functions. These results are associated with the nonregulation of sugar and lipid metabolism by HCV core[4,31]. Eukaryotes, in contrast to prokaryotes, contain more than one DNA ligase, and these enzymes have distinct roles in DNA metabolism. Five DNA ligase activities have been purified from mammalian cell extracts. Ligase III is more closely related to DNA ligase encoded by pox viruses rather than replicative DNA ligases such as mammalian DNA ligase 1, and may be involved in DNA repair and recombination[32]. Thioredoxin and thioredoxin reductase 1 (TXNRD1) are redox proteins that have been implicated in cellular events such as cell proliferation, transformation, and apoptosis[33,34]. DEAD box proteins characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD) are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, cellular growth and division. This gene encodes a DEAD box protein, which is an antigen recognized by autoimmune antibodies, unwinds double-stranded RNA, folds single-stranded RNA, and may play an important role in ribosomal RNA biogenesis, RNA editing, RNA transport, and general transcription[35]. MADD is intimately involved in anti-apoptotic and cell-survival processes[36]. N-myc downstream-regulated gene 2(NDRG2) is a member of the N-myc downregulated gene family, which belongs to the alpha/beta hydrolase superfamily. The protein encoded by this gene is a cytoplasmic protein that may play a role in neurite outgrowth. This gene may be involved in glioblastoma carcinogenesis[37]. TNFRSF5 is a member of the TNF-receptor superfamily. This receptor has been found to be essential in mediating a broad variety of immune and inflammatory responses including T cell-dependent immunoglobulin class switching, memory B cell development, and germ center formation[38,39].
In conclusion, HCV core protein and HCTP4 are related to chronic liver disease, liver cirrhosis and hepatocellular carcinoma.
Footnotes
Supported by the National Natural Science Foundation of China, No. 39970674
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Di Bisceglie AM, Lyra AC, Schwartz M, Reddy RK, Martin P, Gores G, Lok AS, Hussain KB, Gish R, Van Thiel DH, et al. Hepatitis C-related hepatocellular carcinoma in the United States: influence of ethnic status. Am J Gastroenterol. 2003;98:2060–2063. doi: 10.1111/j.1572-0241.2003.t01-1-07641.x. [DOI] [PubMed] [Google Scholar]
- 3.Siavoshian S, Abraham JD, Kieny MP, Schuster C. HCV core, NS3, NS5A and NS5B proteins modulate cell proliferation independently from p53 expression in hepatocarcinoma cell lines. Arch Virol. 2004;149:323–336. doi: 10.1007/s00705-003-0205-7. [DOI] [PubMed] [Google Scholar]
- 4.Bataller R, Paik YH, Lindquist JN, Lemasters JJ, Brenner DA. Hepatitis C virus core and nonstructural proteins induce fibrogenic effects in hepatic stellate cells. Gastroenterology. 2004;126:529–540. doi: 10.1053/j.gastro.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 5.McHutchison JG. Understanding hepatitis C. Am J Manag Care. 2004;10:S21–S29. [PubMed] [Google Scholar]
- 6.Poynard T, Yuen MF, Ratziu V, Lai CL. Viral hepatitis C. Lancet. 2003;362:2095–2100. doi: 10.1016/s0140-6736(03)15109-4. [DOI] [PubMed] [Google Scholar]
- 7.Trujillo-Murillo Kdel C, Garza-Rodríguez Mdel L, Martínez-Rodríguez HG, Barrera-Saldaña HA, Bosques-Padilla F, Ramos-Jiménez J, Rivas-Estilla AM. Experimental models for hepatitis C virus (HCV): new opportunities for combating hepatitis C. Ann Hepatol. 2004;3:54–62. [PubMed] [Google Scholar]
- 8.Ito T, Mukaigawa J, Zuo J, Hirabayashi Y, Mitamura K, Yasui K. Cultivation of hepatitis C virus in primary hepatocyte culture from patients with chronic hepatitis C results in release of high titre infectious virus. J Gen Virol. 1996;77(Pt 5):1043–1054. doi: 10.1099/0022-1317-77-5-1043. [DOI] [PubMed] [Google Scholar]
- 9.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 10.Sacco R, Tsutsumi T, Suzuki R, Otsuka M, Aizaki H, Sakamoto S, Matsuda M, Seki N, Matsuura Y, Miyamura T, et al. Antiapoptotic regulation by hepatitis C virus core protein through up-regulation of inhibitor of caspase-activated DNase. Virology. 2003;317:24–35. doi: 10.1016/j.virol.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Kao CF, Chen SY, Lee YH. Activation of RNA polymerase I transcription by hepatitis C virus core protein. J Biomed Sci. 2004;11:72–94. doi: 10.1007/BF02256551. [DOI] [PubMed] [Google Scholar]
- 12.Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi SI, Ichikawa M, Kajita T, Moradpour D, Wands JR, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray RB, Steele R, Basu A, Meyer K, Majumder M, Ghosh AK, Ray R. Distinct functional role of Hepatitis C virus core protein on NF-kappaB regulation is linked to genomic variation. Virus Res. 2002;87:21–29. doi: 10.1016/s0168-1702(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 14.Chang J, Yang SH, Cho YG, Hwang SB, Hahn YS, Sung YC. Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J Virol. 1998;72:3060–3065. doi: 10.1128/jvi.72.4.3060-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology. 2004;126:586–597. doi: 10.1053/j.gastro.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Ohkawa K, Ishida H, Nakanishi F, Hosui A, Ueda K, Takehara T, Hori M, Hayashi N. Hepatitis C virus core functions as a suppressor of cyclin-dependent kinase-activating kinase and impairs cell cycle progression. J Biol Chem. 2004;279:11719–11726. doi: 10.1074/jbc.M308560200. [DOI] [PubMed] [Google Scholar]
- 17.Crosse K, Umeadi OG, Anania FA, Laurin J, Papadimitriou J, Drachenberg C, Howell CD. Racial differences in liver inflammation and fibrosis related to chronic hepatitis C. Clin Gastroenterol Hepatol. 2004;2:463–468. doi: 10.1016/s1542-3565(04)00162-4. [DOI] [PubMed] [Google Scholar]
- 18.Alonzi T, Agrati C, Costabile B, Cicchini C, Amicone L, Cavallari C, Rocca CD, Folgori A, Fipaldini C, Poccia F, et al. Steatosis and intrahepatic lymphocyte recruitment in hepatitis C virus transgenic mice. J Gen Virol. 2004;85:1509–1520. doi: 10.1099/vir.0.19724-0. [DOI] [PubMed] [Google Scholar]
- 19.Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, Matsuura Y, Miyamura T, Bréchot C, Barba G. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 20.Rønnov-Jessen L, Villadsen R, Edwards JC, Petersen OW. Differential expression of a chloride intracellular channel gene, CLIC4, in transforming growth factor-beta1-mediated conversion of fibroblasts to myofibroblasts. Am J Pathol. 2002;161:471–480. doi: 10.1016/s0002-9440(10)64203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nonet GH, Stampfer MR, Chin K, Gray JW, Collins CC, Yaswen P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2001;61:1250–1254. [PubMed] [Google Scholar]
- 22.Horrevoets AJ, Fontijn RD, van Zonneveld AJ, de Vries CJ, ten Cate JW, Pannekoek H. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood. 1999;93:3418–3431. [PubMed] [Google Scholar]
- 23.Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, Doi T, Shimotohno K, Harada T, Nishida E, et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 24.Cartier C, Hemonnot B, Gay B, Bardy M, Sanchiz C, Devaux C, Briant L. Active cAMP-dependent protein kinase incorporated within highly purified HIV-1 particles is required for viral infectivity and interacts with viral capsid protein. J Biol Chem. 2003;278:35211–35219. doi: 10.1074/jbc.M301257200. [DOI] [PubMed] [Google Scholar]
- 25.Zidovetzki R, Wang JL, Chen P, Jeyaseelan R, Hofman F. Human immunodeficiency virus Tat protein induces interleukin 6 mRNA expression in human brain endothelial cells via protein kinase C- and cAMP-dependent protein kinase pathways. AIDS Res Hum Retroviruses. 1998;14:825–833. doi: 10.1089/aid.1998.14.825. [DOI] [PubMed] [Google Scholar]
- 26.Fukuchi M, Nakajima M, Fukai Y, Miyazaki T, Masuda N, Sohda M, Manda R, Tsukada K, Kato H, Kuwano H. Increased expression of c-Ski as a co-repressor in transforming growth factor-beta signaling correlates with progression of esophageal squamous cell carcinoma. Int J Cancer. 2004;108:818–824. doi: 10.1002/ijc.11651. [DOI] [PubMed] [Google Scholar]
- 27.Kim AC, Peters LL, Knoll JH, Van Huffel C, Ciciotte SL, Kleyn PW, Chishti AH. Limatin (LIMAB1), an actin-binding LIM protein, maps to mouse chromosome 19 and human chromosome 10q25, a region frequently deleted in human cancers. Genomics. 1997;46:291–293. doi: 10.1006/geno.1997.5029. [DOI] [PubMed] [Google Scholar]
- 28.Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Privé GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci USA. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Qi X, Grabowski GA. Saposin C is required for normal resistance of acid beta-glucosidase to proteolytic degradation. J Biol Chem. 2003;278:31918–31923. doi: 10.1074/jbc.M302752200. [DOI] [PubMed] [Google Scholar]
- 30.He HJ, Kole S, Kwon YK, Crow MT, Bernier M. Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:27096–27104. doi: 10.1074/jbc.M301003200. [DOI] [PubMed] [Google Scholar]
- 31.Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Tsukamoto K, Kimura S, Moriya K, Koike K. Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology. 2004;126:840–848. doi: 10.1053/j.gastro.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 32.Bordone L, Campbell C. DNA ligase III is degraded by calpain during cell death induced by DNA-damaging agents. J Biol Chem. 2002;277:26673–26680. doi: 10.1074/jbc.M112037200. [DOI] [PubMed] [Google Scholar]
- 33.Lechner S, Müller-Ladner U, Neumann E, Spöttl T, Schlottmann K, Rüschoff J, Schölmerich J, Kullmann F. Thioredoxin reductase 1 expression in colon cancer: discrepancy between in vitro and in vivo findings. Lab Invest. 2003;83:1321–1331. doi: 10.1097/01.lab.0000085189.47968.f8. [DOI] [PubMed] [Google Scholar]
- 34.Anestål K, Arnér ES. Rapid induction of cell death by selenium-compromised thioredoxin reductase 1 but not by the fully active enzyme containing selenocysteine. J Biol Chem. 2003;278:15966–15972. doi: 10.1074/jbc.M210733200. [DOI] [PubMed] [Google Scholar]
- 35.Henning D, So RB, Jin R, Lau LF, Valdez BC. Silencing of RNA helicase II/Gualpha inhibits mammalian ribosomal RNA production. J Biol Chem. 2003;278:52307–52314. doi: 10.1074/jbc.M310846200. [DOI] [PubMed] [Google Scholar]
- 36.Lim KM, Chow VT. Induction of marked apoptosis in mammalian cancer cell lines by antisense DNA treatment to abolish expression of DENN (differentially expressed in normal and neoplastic cells) Mol Carcinog. 2002;35:110–126. doi: 10.1002/mc.10082. [DOI] [PubMed] [Google Scholar]
- 37.Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu X, Au WS, Wang J, Li F, Ji S, et al. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342–347. doi: 10.1002/ijc.11228. [DOI] [PubMed] [Google Scholar]
- 38.Contin C, Pitard V, Itai T, Nagata S, Moreau JF, Déchanet-Merville J. Membrane-anchored CD40 is processed by the tumor necrosis factor-alpha-converting enzyme. Implications for CD40 signaling. J Biol Chem. 2003;278:32801–32809. doi: 10.1074/jbc.M209993200. [DOI] [PubMed] [Google Scholar]
- 39.Eeva J, Postila V, Mättö M, Nuutinen U, Ropponen A, Eray M, Pelkonen J. Kinetics and signaling requirements of CD40-mediated protection from B cell receptor-induced apoptosis. Eur J Immunol. 2003;33:2783–2791. doi: 10.1002/eji.200324227. [DOI] [PubMed] [Google Scholar]


