Abstract
Monobodies are antibody alternatives derived from fibronectin that are thermodynamically stable, small in size, and can be produced in bacterial systems. Monobodies have been engineered to bind a wide variety of target proteins with high affinity and specificity. Using alanine-scanning mutagenesis simulations, we identified two scaffold residues that are critical to the binding interaction between the monobody YS1 and its ligand, maltose-binding protein (MBP). Steered molecular dynamics (SMD) simulations predicted that the E47A and R33A mutations in the YS1 scaffold substantially destabilize the YS1-MBP interface by reducing the bond rupture force and the lifetime of single hydrogen bonds. SMD simulations further indicated that the R33A mutation weakens the hydrogen binding between all scaffold residues and MBP and not just between R33 and MBP. We validated the simulation data and characterized the effects of mutations on YS1-MBP binding by using single-molecule force spectroscopy and surface plasmon resonance. We propose that interfacial stability resulting from R33 of YS1 stacking with R344 of MBP synergistically stabilizes both its own bond and the interacting scaffold residues of YS1. Our integrated approach improves our understanding of the monobody scaffold interactions with a target, thus providing guidance for the improved engineering of monobodies.
High affinity proteins are utilized in a wide spectrum of applications ranging from chemical and biological threat detection1 to protein-based therapeutics2. Although monoclonal antibodies have traditionally been favored as therapeutic biomolecules, they are large in size, require eukaryotic expression for production3, and generally offer poor thermal stability4. As a result, synthetic antibody mimetic proteins based on molecular scaffolds have gained popularity. Utilizing a conserved protein scaffold as a platform and combinatorial engineering techniques, selections for high affinity binding or conformational stability can be performed5,6. Synthetic domains have also been engineered to produce biosensors7,8 and achieve binding to a wide array of molecules9,10. Engineered protein scaffolds have been explored for use as both therapeutics11 and diagnostics12. An increased understanding of how scaffold structure affects interactions with ligands will facilitate the engineering of improved scaffold proteins.
The protein of interest in this study is derived from the tenth fibronectin III domain (FNfn10) scaffold13,14. Similar to the immunoglobulin (Ig) complementarity determining region, the ~94 amino acid peptide contains a β-sheet backbone and three relevant loop domains (BC, DE, and FG)13,14. The three loops have been diversified using phage or yeast display combinatorial libraries to produce proteins known as monobodies, with low nanomolar9,15 to picomolar Kd values16 and the capability to bind to targets such as small ubiquitin-related modifiers (SUMO)17, maltose-binding protein (MBP)9,15, lysozyme16, and fyn kinase18. Comparable binding affinity to antibodies coupled with the absence of disulfide bonds, ease of production in bacterial systems13,19, and high thermal stability20 are reasons why monobodies have become attractive alternatives to antibodies as therapeutic biomolecules.
A better understanding of monobody interactions with their ligands (paratope/epitope binding) will allow for improved monobody design. Here, we have studied the interaction of monobody YS1 with its ligand, maltose-binding protein (MBP), with a focus on scaffold interactions. YS1 was developed by Koide et al.9 utilizing a Y/S binary combinatorial library platform to diversify amino acids on the BC, DE, and FG loops of the FNfn10 scaffold. The monobody was originally named MBP-74 but was subsequently renamed YS115. The X-ray crystal structure of YS1 bound to MBP shows the convex paratope of YS1 binding to the sugar binding pocket of MBP9,15. Based on the crystal structure, the interacting paratope of YS1 includes both loop and scaffold proteins. Alanine-scanning mutagenesis indicates that the BC loop of the monobody does not significantly contribute to binding, but that alanine mutations at seven of the nine residues of the FG loop result in greater than 10-fold decrease in affinity15. Although it has been speculated that the contacts with the scaffold residues are a crystallization artifact9, the effect of mutations on these scaffolds has not been reported. Previous studies on monobodies have mainly focused on altering the BC, DE, and FG loops to achieve high binding affinities and increased protein stability9,15,21. While scaffold modifications have been considered in modifying monobody structural stability22,23 and in combination with loop modifications17, little work has focused solely on how interacting scaffold residues affect binding kinetics. Utilizing computational modeling and biophysical analyses we have explored how scaffold modifications affect YS1-MBP binding kinetics.
Structure-based design of therapeutic molecules is becoming increasingly important with the growth of structural databases and increased computing speeds24,25. Whereas the x-ray crystal structure reveals interactions in the context of a static crystal, molecular dynamics simulations can show instantaneous molecular movement and are useful for determining the preferred motion of proteins26. Through steered molecular dynamics (SMD) an external force is applied to a binding pair and the dissociation interactions are measured with respect to time27. As a result, the structural mechanics of the unbinding process can be explored. Single molecule force spectroscopy is a biophysical method to experimentally examine such processes, and has been utilized to measure binding kinetics of cell to cell interactions and protein-protein interactions between single molecules28,29,30,31,32,33,34.
Typically only a small fraction of buried residues contributes to the majority of the binding affinity in binding interactions. These residues are referred to as hot spots35,36. Hot spots have been studied in a wide range of human antibodies and other natural proteins, and modification of these residues has a substantial destabilizing effect on the protein interface35,36. We sought to identify the hot spots of the YS1-MBP interface on the previously unstudied scaffold interactions using computational alanine-scanning mutagenesis. By coupling molecular simulations and biophysical tools we have characterized the critical interacting β-sheet residues of YS1 and assessed their influence on binding kinetics. With the increasing interest in the development of scaffold based mimetic proteins, an understanding of scaffold hot spots will lead to the creation of better-targeted therapeutics.
Results
Computational simulations predict R33 and E47 as critical scaffold framework residues
To identify the critical YS1 residues involved in YS1-MBP binding, we first executed a computational alanine-scanning mutagenesis analysis utilizing molecular dynamics and Robetta and DrugScorePPI simulations to analyze the binding interface. Specifically, each of the YS1 interacting residues was individually mutated to alanine. The change in binding free energy (ΔΔGbind) and the protein stability of the mutated complex partner in isolation (ΔGpartner) were calculated37,38. ΔGpartner represents the predicted stabilizing or destabilizing effect of the alanine mutation on free YS1 and is a component of ΔΔGbind with large values signifying decreased YS1 stability and the potential of decreased binding. Additionally, the degree of buriedness was calculated for interfacial residues38 as a high degree of residue burial is necessary albeit not sufficient for hot spot identification36. The three calculations were taken together to identify pivotal binding residues.
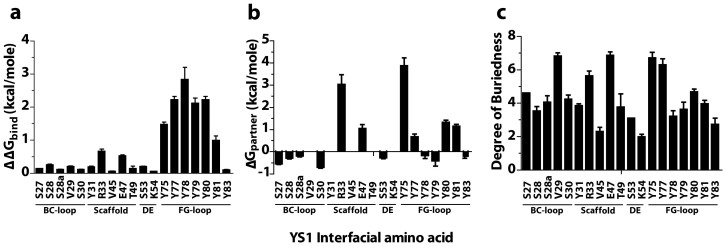
Simulations predict that YS1-MBP binding hot spots are localized to the FG-loop and the scaffold (Fig. 1). Amino acid mutations in both the BC-loop and DE-loop were predicted to have an insignificant effect on protein complex stability as alanine mutations in both loops caused minimal negative perturbations of ΔΔGbind (less than about 0.25 kcal/mol) and ΔGpartner (< 0 kcal/mol). Six amino acids in the FG-loop (Y75, Y77, Y78 Y79, Y80 and Y81) and two in the scaffold framework (R33 and E47) were predicted to be important binding residues. The FG-loop amino acids were identified based on their high ΔΔGbind (> 1.5 kcal/mol) or their high ΔGpartner value (> 1 kcal/mol). In a previous experimental alanine-scanning mutagenesis study of the FG loop, the Y75A, Y77, Y78, Y79A, Y80A, Y81A and Y82A mutations caused the largest reduction in affinity (≥ 1.5 kcal/mol)15. The two scaffold residues were identified based on their high ΔGpartner value (> 1 kcal/mol), their high degree of buriedness (greater than 5.5) and because they had the highest ΔΔGbind of any non-FG- loop residues (Fig. 1). As common in interfacial hot spots, the residues are grouped in close proximity to one another and make contact with the target protein MBP (Fig. 2a, b)35,36.
Figure 1. Computational simulations predict the critical residues of YS1 at the YS1-MBP binding interface.
(a) DrugscorePPI webserver prediction of changes in binding free energy (ΔΔGbind) upon alanine mutation. (b) Robetta prediction of the effect of alanine mutation on mutated protein complex partner in isolation (ΔGpartner). Residues V29, Y31, V45, T49 and K54 were omitted as Robetta predictions failed to identify them as being present in 50% of the predicted binding complexes. (c) DrugscorePPI webserver prediction of the degree of buriedness of the interfacial residues at the YS1-MBP interface. Results are the mean ± s.e.m. of 6-8 pairs of randomly selected monobody-MBP complexes.
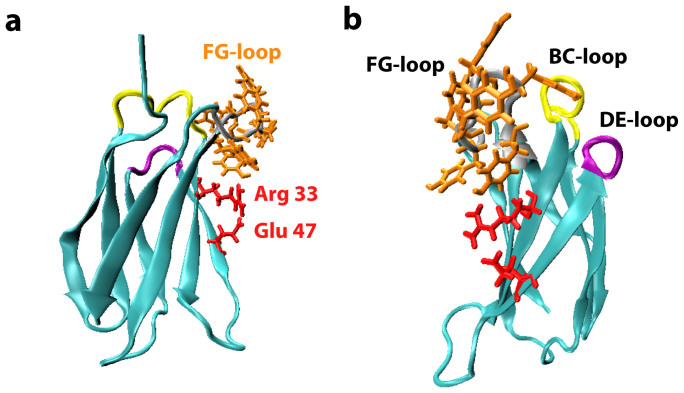
Figure 2. Two perpendicular views of the crystal structure of monobody YS1 (cyan).
DrugscorePPI simulation predicted that binding energy between YS1 and its target MBP is provided by two groups of interfacial residues: Tyr75, Tyr77, Try78, Try79, Tyr80 and Y81 (orange) which are located in the FG-loop; and Arg33 and Glu47 (red) which belong to the framework of the scaffold. The PDB ID is 2OBG. The figure was made with the software VMD (DeLano Scientific, San Francisco, CA).
Since monobodies feature a structure similar to the antibody variable domain, most YS1 studies focus on the modification of three exposed loops corresponding to the three complementarity determining regions in antibodies9,15. Here, we explored the impact of modifications to the R33 and E47 residues, which reside in the C and D β-sheets of the scaffold, respectively. Based on the large value of ΔGpartner (3.04 kcal/mol) and a larger ΔΔGbind, we anticipated that R33A mutation would have a greater destabilizing effect on the YS1-MBP interaction than E47A mutations (ΔGpartner 1.04 kcal/mol). We analyzed the effect of these mutations using SMD simulation as well as two biophysical tools: surface plasmon resonance (SPR) and single molecule force spectroscopy.
Use of Steered Molecular Dynamics predicts a crucial role of R33 in scaffold stabilization
SMD simulations were performed using six randomly selected pairs of monobody-MBP complexes at equilibrium. Pulling was performed at a constant speed of 10 Å/ns with a spring stiffness of 70 pN/Å on residue V72 of the monobody and was designed to pull the monobody away from the center of mass of MBP. Water molecules were simulated by the generalized born implicit solvent (GBIS) model implemented in NAMD. Each simulation lasted 4 ns with data points saved every 0.5 ps for analysis. SMD enabled the quantification of the contribution of each residue to the YS1-MBP binding interaction27.
In order to reduce computational time we utilized the generalized born implicit solvent in NAMD for water modeling. The GBIS electrostatics calculation first determines and then utilizes the Born radius of each atom to quantify the atom's exposure to solvent and its dielectric screening from other atoms. GBIS simulations of the repulsive and the attractive components of the nonpolar solvation free energy of small molecules are in agreement with explicit models39. NAMD's GBIS capability has been utilized for simulating several large protein structures40,41 and validated for the simulation of a ribosome undergoing two major conformational changes42. In this instance, GBIS and TIP3P explicit solvent structures closely agree, with a root-mean-square deviation between models of 1.5 Å42.
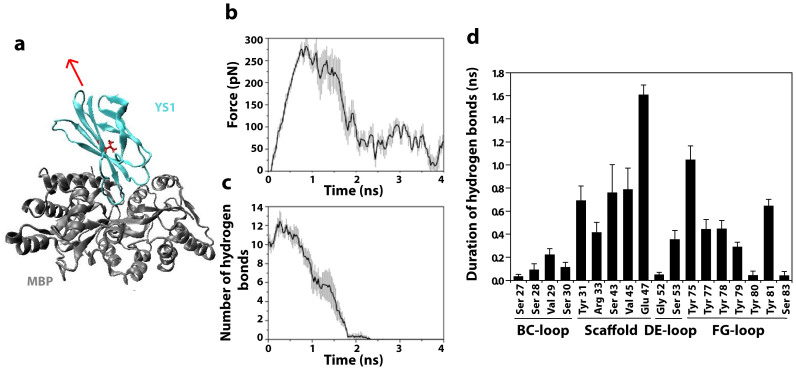
The mechanics of bond rupture can be separated into three distinct regions (Fig. 3b). First, as the pulling force increases linearly, the number of hydrogen bonds remains relatively constant. As the applied force reaches the rupture force the hydrogen bonds begin to break one by one. When all bonds have broken and YS1 and MBP are completely dissociated, the force reaches background levels. The lifetimes of hydrogen bonds involving specific residues of the YS1 scaffold are comparable to those of the FG loop (Fig. 3d). Thus, predictions from SMD simulations indicating the importance of scaffold residues in the YS1-MBP interaction are in line with the findings from computational alanine-scanning mutagenesis.
Figure 3. Steered molecular dynamics (SMD) simulates the unbinding of monobody YS1 from its target MBP.
(a) A schematic illustration showing the monobody YS1 (cyan) being pulled away from MBP (grey) under a constant pulling velocity on Val72, which is indicated in red. The retracting direction was selected to pull the center of mass of the monobody away from that of MBP as indicated by the red arrow. (b) Force-time course of SMD simulated unbinding of monobody YS1 from its target MBP. (c) The time evolution of the average number of hydrogen bonds and (d) hydrogen bond duration measured for each interacting residue between monobody YS1 and MBP during unbinding simulations.
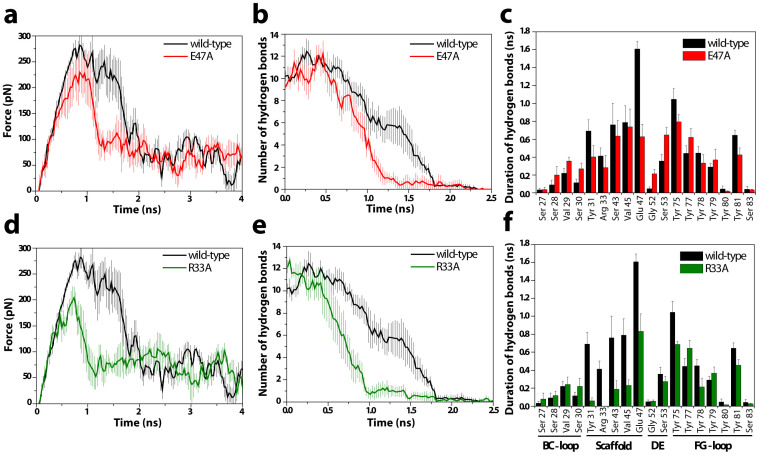
Destabilizing mutations, which decrease bond duration time between the interacting residues in the presence of force, reduce the overall interaction time between YS1 and its ligand, thereby indicating weaker binding. Based on the SMD simulation, the E47A mutation reduces the interaction lifetime between the YS1 and MBP by 30% and causes the hydrogen bonds to break earlier and at a faster rate (Fig. 4a, b). Coupled with the moderate decrease in binding force, this result suggests that the E47A mutation decreases the overall strength of the interaction. The R33A mutation has a more profound effect than E47A, reducing the overall bond lifetime by 50% and the binding force by ~30% (Fig. 4d, e). Both E47A and R33A mutations have little effect on the duration of hydrogen bonds involving residues of the monobody loops. Instead, they affect the duration of hydrogen bonds between the monobody scaffold residues and MBP (Fig 4.c, f). The E47A mutation decreases hydrogen bond duration in the E47 residue alone, and has little effect on the other scaffold residues, likely resulting in the modest decrease in the overall YS1-MBP bond duration observed with the E47A mutation. In contrast, the R33A mutation not only reduces the duration of the hydrogen bond at R33 but also at the other interacting residues of the scaffold (Fig. 4f). Consequently, this mutation substantially destabilizes the bond and results in a more significant loss of intermolecular recognition. This destabilization and subsequent synergistic bond weakening likely contributes to the larger ΔΔGbind and ΔGpartner predicted in the computational alanine-scanning mutagenesis analysis and further supports a major role for R33 in the YS1-MBP interaction.
Figure 4. Steered molecular dynamics simulations of unbinding of (a-c) E47A and (d-f) R33A mutants from the target MBP.
Equilibrated mutant YS1-MBP pairs were pulled apart at a constant speed and spring stiffness. During the dissociation event, (a and d) pulling force, (b and e) the number of hydrogen bonds between interacting residues, and (c and f) the duration of hydrogen bonds for each interacting residue were determined for both mutants. Results are the mean ± s.e.m. of six pairs of randomly selected monobody-MBP complexes.
Single-molecule force spectroscopy supports the role of E47 and R33 in YS1-MBP interaction
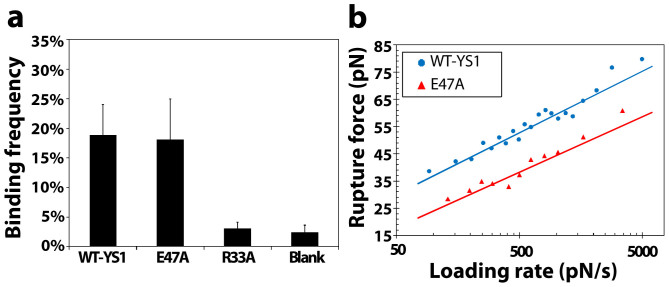
Single-molecule force spectroscopy was utilized to characterize the binding kinetics of the R33A and E47A mutant variants of YS1 with MBP. MBP was properly oriented in a lipid bilayer via an attached transmembrane domain from the human CD44 transmembrane protein as in our previous study of the interaction of MBP with a DARPin28. YS1, YS1(E47A), and YS1(R33A) coated cantilevers were brought in contact with MBP for a constant dwell time and retracted at a predetermined retraction speed. The tensile strength of the bond (bond rupture force) and loading rates of single binding events were recorded over a range of retraction velocities28,29. The concentrations of YS1 on the cantilever and MBP in the lipid bilayer were selected to yield a binding event percentage of ~20% (Fig. 5a) as this frequency will ensure that the majority of binding events (>89%) are caused from the breakage of single bonds (based on the Poisson distribution)29,30.
Figure 5. Characterization of the kinetic and micromechanical properties of monobody YS1 binding to MBP using single-molecule force spectroscopy.
(a) Frequency of binding events between MBP and YS1, YS1(E47A) or YS1(R33A). The last column (blank) represents a control experiment using a blank bilayer. Data represent the mean ± s.e.m. of at least three independent experiments. (b) Rupture force as a function of loading rate for the binding of YS1 (blue circles) and YS1(E47A) (red triangles) to MBP. The lines are a nonlinear least-squares fitting of the Bell dissociation model for determining the unstressed off-rate (k0off) and reactive compliance (xβ) of a bond.
The tensile strength measured for YS1 binding to MBP was in the range of 35–80 pN over a wide range of loading rates (Fig. 5b). The E47A mutation reduces the rupture force by approximately 10–15 pN over a similar range of loading rates, indicating that the E47 residue plays an auxiliary role in this interaction. Using the Bell model43, the values for the unstressed off-rate 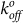 (second−1) and reactive compliance xβ (nm) for YS1(E47A) were determined by plotting mean rupture force against the logarithm of the loading rate and fitting the data with a least-squares regression line (Fig. 5b). The E47A mutant increased
(second−1) and reactive compliance xβ (nm) for YS1(E47A) were determined by plotting mean rupture force against the logarithm of the loading rate and fitting the data with a least-squares regression line (Fig. 5b). The E47A mutant increased 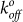 of the binding interaction 3-fold compared to the YS1 (Table 1). Larger values of
of the binding interaction 3-fold compared to the YS1 (Table 1). Larger values of 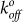 are indicative of shorter unstressed bond lifetime and weaker binding. Interestingly the reactive compliance, which reflects the susceptibility of the bond to rupture under stress, was similar for both the wild-type and mutant monobodies. In accordance with the Bell Model43, an applied force (f) will alter the bond off rate (koff) by the following equation: koff/
are indicative of shorter unstressed bond lifetime and weaker binding. Interestingly the reactive compliance, which reflects the susceptibility of the bond to rupture under stress, was similar for both the wild-type and mutant monobodies. In accordance with the Bell Model43, an applied force (f) will alter the bond off rate (koff) by the following equation: koff/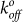 = exp(fxβ/kβT). Therefore, the similar xβ values observed here and in other studies28 indicate a similar sensitivity to rupture in the presence of force. Collectively, the E47A mutation decreases the strength of the YS1-MBP bond as indicated by its lower tensile stress and shorter unstressed bond lifetime.
= exp(fxβ/kβT). Therefore, the similar xβ values observed here and in other studies28 indicate a similar sensitivity to rupture in the presence of force. Collectively, the E47A mutation decreases the strength of the YS1-MBP bond as indicated by its lower tensile stress and shorter unstressed bond lifetime.
Table 1. Comparison of experimentally determined kinetic constants for mutations at critical scaffold residues as determined by scanning alanine mutagenesis simulations.
| DrugscorePPI | Robetta | Single-molecule force spectroscopy | Suface plamon resonance | ||||
|---|---|---|---|---|---|---|---|
| sample | ΔΔGbind (kcal/mol) | ΔΔGpartner (kcal/mol) |
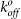 (× 10−3s−1) (× 10−3s−1) |
xβ(nm) | kon(× 105M−1s−1) |
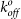 (× 10−3s−1) (× 10−3s−1) |
Kd (nM) |
| Wild-type | 0.24 ± .06 | 0.42 ± .03 | 4.4 ± 0.7 | 40.6 ± 3.2 | 100 ± 18 | ||
| E47A | .53 ± .03 | 1.01 ± .18 | 0.76 ± .39 | 0.46 ± .05 | 3.6 ± 0.3 | 38.7 ± 15 | 101 ± 41 |
| R33A | .67 ± .06 | 3.04 ± .44 | N.B. | — | N.B. | — | — |
N.B. represents no binding. This No binding is defined as having a binding frequency under 5% for single molecule force spectroscopy or displaying an <5 RU response up to 1 μM concentration in SPR. Data represent the mean ± s.e.m. of at least three independent experiments.
In concert with both scanning alanine mutagenesis and SMD simulations, force-spectroscopy also revealed that the R33A mutation had a pronounced destabilizing effect on the monobody-MBP interaction, as evidenced by the dramatic decrease in binding frequency down to basal levels (Fig. 5a). Of note, the auxiliary role of the E47A mutation is further substantiated by our findings showing that this mutation did not significantly affect binding frequency of monobody-MBP interaction (Fig. 5a).
Surface Plasmon Resonance confirms that the pivotal role of the scaffold residue R33 in YS1-MBP interaction
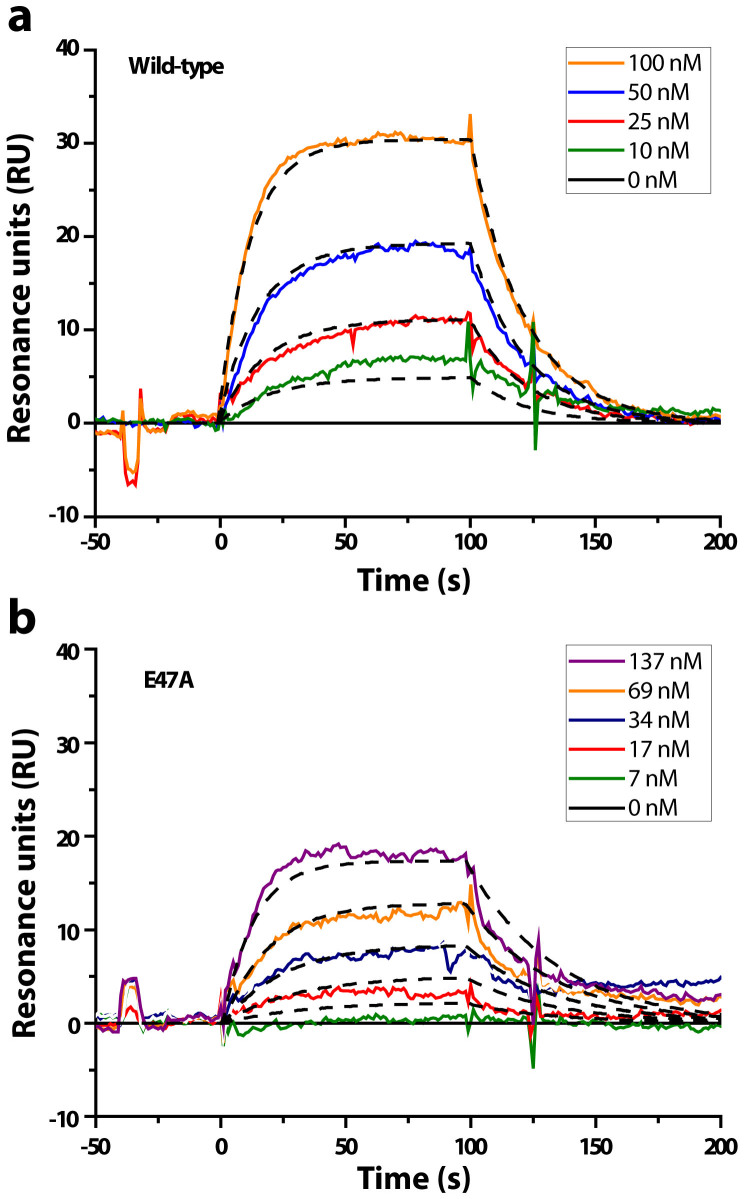
SPR experiments were performed to determine the binding kinetic constants (Kd, kon, koff). YS1, YS1(R33A), and YS1(E47A) were flowed over a chip sensor coated with biotinylated MBP (Fig. 6). In accordance with the single molecule force spectroscopy results, the R33A mutation reduced the binding signal to non-significant levels, further demonstrating the critical role of R33 in the YS1-MBP interaction. Binding of MBP to both YS1 and YS1(E47A) fit a 1:1 binding model and had equivalent Kd values of about 100 nM (Table 1). The E47A mutation resulted in no significant change in koff or kon. The similar kinetic constants found here are consistent with the similar binding frequency detected between MBP and YS1 or YS1(E47A) using single-molecule force spectroscopy. The discrepancy in the dissociation rate constants obtained by SPR versus force spectroscopy between YS1-MBP and YS1(E47A)-MBP binding is likely due to differences in binding geometries (3D vs. 2D)28. Overall, SPR data correlate well with both SMD simulations and single-molecule force spectroscopy data and indicate the R33A mutation substantially destabilizes YS1-MBP binding while the E47A mutation plays a less important role.
Figure 6. Binding interactions of monobodies and MBP analyzed using SPR.
Representative sensograms were generated by perfusing (a) wild-type YS1 or (b) YS1(E47A) over biotinylated MBP immobilized SA chip at concentrations ranging from 0-137 nM for 100 seconds, followed by washing with buffer for 10 minutes. To generate the kon, koff and Kd a 1:1 Langmuir binding model was fit for YS1- and YS1(E47A) -MBP interaction. χ2 vales were 0.88 and 1.92 for YS1 and YS1(E47A), respectively. Data were analyzed using BIAevaluation 3.0 (BIAcore).
Discussion
A better understanding of how monobodies associate with their ligands will facilitate the development of robust binding pockets for higher affinity recognition and allow for the generation of improved monobodies. We used computational alanine-scanning mutagenesis, SMD, single-molecule force spectroscopy and SPR to identify and characterize the effect of monobody scaffold residues on the binding interaction between monobody YS1 and MBP. Molecular simulations identified E47 and R33 as scaffold residues that contribute to monobody-MBP recognition. Mutation at E47 resulted in a weaker bond and reduced interaction duration, thereby suggesting an auxiliary role for E47 in monobody-MBP recognition. On the other hand, the R33 residue plays a key role in this interaction as R33A mutation abrogated binding in both experimental assays, an effect likely caused by the synergistic destabilization of the interacting scaffold residues (Fig. 4f).
Our Kd value for YS1-MBP binding obtained by SPR falls between the reported values of 73 nM and 135 nM of prior work, which also used SPR9,15. Those studies and ours indicate 1:1 binding kinetics. Previous mutagenesis work combined shotgun scanning mutagenesis with alanine-scanning mutagenesis for an analysis of YS1-MBP binding, but focused only on the BC and FG loops15. That study concluded that the BC loop of YS1 is robust to mutation whereas the FG is sensitive15. A similar trend was also observed in our in situ scanning alanine analysis (Fig. 1), where DrugScorePPI predicts the interfacial FG loop residues that were most important for the interaction9.
Although previous studies have primarily focused on varying loop residues to achieve higher binding affinities, recent work has demonstrated the importance of monobody scaffold residues in binding17,44. Interactions between monobodies and small ubiquitin-related modifier (SUMO) proteins were found to rely heavily on both FG loop and scaffold binding, and scaffold modifications were required to generate the desired isoform specificity of the monobody17. A focus on scaffold residue diversity has generated monobodies capable of binding to the Abl SH2 domain with low nanomolar affinities due primarily to scaffold binding44. Koide et al. noted that the scaffold interactions found in the YS1-MBP crystal structure were possibly due to the lattice packing because the contacts were polar and charged and the region showed little to no chemical shift perturbation in an NMR study15. However, R33 has been a point of interest in several monobody studies. R33 was found at multiple binding interfaces, and its mutation has led to either increased or deceased binding strength when coupled with additional scaffold and loop amino acid modifications17,44. In our study, molecular dynamics simulations, single-molecule force spectroscopy and SPR studies indicate that R33 is critical for the YS1-MBP interaction.
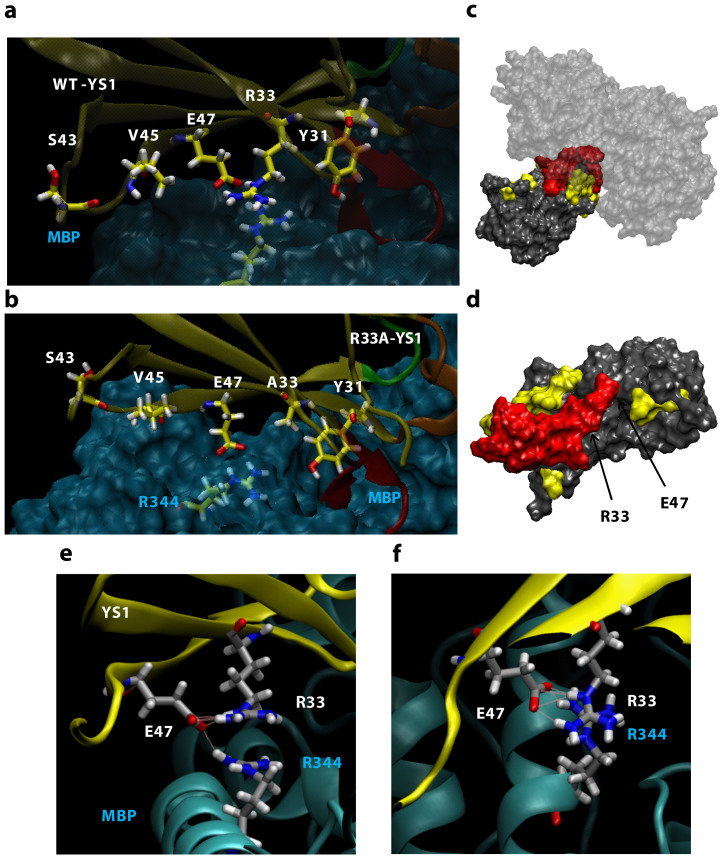
Hot spots in natural proteins tend to have nonrandom compositions with the second most abundant amino acid being arginine36,45. Its presence at hot spots is thought to occur due to its ability to form several favorable interactions including electron delocalization through the guanidinium motif and the ability to form up to five hydrogen bonds36. As predicted by DrugscorePPI scanning alanine mutagenesis and exemplified in both SPR and force spectroscopy experiments, the R33A mutation was shown to destabilize YS1-MBP binding. Robetta predictions indicated the R33A mutation decreases YS1 stability, a result that likely contributes to the loss of intermolecular recognition in the biophysical assays. R33 is at the center of the scaffold interface (Fig. 7c, d). The equilibrated molecular representation of the binding interface (Fig. 7e, f) shows R33 of YS1 stacking with R344 of MBP effectively burying both arginines. Arg-Arg stacking has been observed in numerous protein-protein interfaces although the reason for its occurrence is not yet fully understood46,47. Arginine stacking is thought to drive the formation of protein interfaces, primarily through polarization effects resulting in the seclusion of solvent however, both hydrophobic and electrostatic-solvation effects can play secondary roles46. Arg-Arg stacking has been observed in MD simulations using both explicit and implicit water models and alternate force fields48,49. In YS1-MBP binding the two positively charged guanidinium groups stack parallel to one another in a form between the staggered and eclipsed conformations. This interaction appears to be vital for high MBP affinity.
Figure 7. YS1 scaffold residues at the MBP interface.
The interacting scaffold residues of (a) YS1 and (b) YS1(R33A) (both shown in yellow). The BC, DE and FG loops are represented orange, green, and red, respectively (c) Schematic illustration of the YS1-MBP interaction and (d) YS1 in which R33, E47, Y75, Y77, Y78 Y79, Y80 and Y81 are shown in red and the other interfacial residues of YS1 are shown in yellow. The side view (e) of the YS1 (yellow) and MBP (cyan) interface shows R33 of YS1 stacking with R344 of MBP to establish the hydrogen bond network between R33, E47, and R344. The top view (f) displays the arginines stacking. Schematic illustration of YS1-MBP interfacial interaction made using the equilibrated molecular representation based on the crystal structure (PDB ID: 2OBG)9 and with the software VMD (DeLano Scientific, San Francisco, CA).
SMD simulations (Fig. 4f) reveal the R33A mutation reduces the hydrogen bond duration of surrounding interacting residues of the scaffold. Hydrogen bonding between R33 and E47 allows E47 to hydrogen bond effectively with the R344 residue of MBP (Fig. 7e, f). The stability of this interaction contributes to the lengthy bond duration of E47 and MBP (Fig. 3d). A similar effect can be found in ySMB-1 binding to ySUMO where polar interactions between arginine and asparagine appear to stabilize glutamic acid to construct a robust scaffold interaction17. Although additional interacting scaffold residues form hydrogen bonds with durations comparable to R33, alanine mutations here were not predicted to critically destabilize the binding interface. These residues are less buried than either R33 or E47 (Fig. 1c) and further away from the R33 residue such that alanine mutations here are not likely to critically destabilize the interface. In fact, although the E47A mutation alters the binding interface modestly, the mutation was not predicted to disrupt the stacking effect (see Supplementary Fig. S1 online). In our work, the R33 residue exhibits two critical characteristics: it anchors the scaffold to MBP via arginine stacking and establishes hydrogen bonding between the YS1 scaffold and MBP. Without the stabilization of the arginine stacking, the R33A mutation has a noticeable and unfavorable conformational change in both YS1 and MBP upon binding (Fig. 7b).
In summary, we have elucidated the importance of the scaffold interaction in YS1-MBP binding through a systematic approach involving molecular modeling and biophysical analyses. Our results further illustrate the importance of the R33 scaffold residue in monobody-target binding, thus providing vital knowledge for the improved engineering of monobodies.
Methods
Construction, expression, and purification of TMD-MBP
The plasmid encoding the CD44 trans-membrane domain (TMD) fused to MBP utilizes the isopropyl b-D-1-thiogalactopyranoside (IPTG)-inducible T7 promoter to allow for the cytoplasmic expression of TMD-MBP28. TMD-MBP was expressed and purified using an amylose affinity chromatography column as described except that BL21(DE3) cells were used28.
Construction, expression, and purification of wild-type and mutated Monobody YS1
The pHFT2 plasmid encoding the YS1 gene with a (His10) tag at the N-terminus9 was a gift from Shohei Koide (University of Chicago). The R33A and E47A mutations were made by site directed mutagenesis in this plasmid and verified by DNA sequencing. The monobodies were expressed and purified from BL21(DE3) cells. Cells were grown in M9 media with 10 g/L tryptone, with expression induced with IPTG. Expressed monobodies were immobilized and purified using a GE HisTrap HP column via their N-terminal His10 tag. The purified monobodies were dialyzed at 4°C against 300 volumes of phosphate-buffered saline (PBS, pH 7.4). Monobody concentrations were determined using their extinction coefficients.
Molecular dynamics simulations
The starting coordinates for the molecular dynamics (MD) simulations were obtained from the crystal structure of YS1 in complex with MBP (Protein Data Bank ID: 2OBG)9. The R33A and E47A in silico mutations were generated by replacing either R33 or E47 with alanine. Both wild-type and mutated YS1 were fully solvated in a rectangular water box of 80×87×80 Å3 by using the Visual Molecular Dynamics program50. Sodium and chloride ions were added to neutralize the system, which yielded 51,635 atoms in total. The MD simulations were performed by following a protocol similar to our previous study28. In brief, the NAMD software was used to perform MD simulation by using a CHARMM22 force field and the TIP3P water model51,52,53. Periodic boundary conditions were applied to avoid finite size effects, and electrostatic interactions were simulated using the particle-mesh Ewald sum method54. A 12 Å cutoff distance was applied for calculating van der Waals interactions. The system has gone through energy minimization by a 20,000-steps-of-conjugate gradient with heavy atoms fixed followed by another 20,000 steps with all atoms free. After gradually heating from 0 to 300 K in 60 ps, the system is equilibrated for at least 2.5 ns with temperature at 300 K and pressure at 1 atm by using the Langevin dynamics method55,56. The profile of RMSD plots for the backbone atoms of monobody-MBP complex reached a plateau value, which indicated that the systems had reached an equilibrium state.
The steered molecular dynamics (SMD) simulations were performed using six pairs of monobody-MBP complexes randomly selected from the equilibrated systems. A constant pulling speed of 10 Å/ns was applied through a spring with a stiffness of 70 pN/Å on the residue Val72 which is near the center of mass of the monobody. The pulling direction was designed to pull the monobody away from the center of mass of MBP. The Cα atoms of the 322, 325 and 326 residues of the MBP were constrained to their equilibrated positions. These residues were chosen because they are located on the opposite side of the binding pocket of MBP. The generalized born implicit solvent (GBIS) model implemented in the NAMD was used to manipulate the water molecules in SMD simulations. Periodic boundary conditions were considered in all SMD studies. However, Particle mesh Ewald method is not supported in the GBIS. The solvent molecular electrostatics were calculated by the Poisson-Boltzmann equation which models water as a dielectric continuum in NAMD42. Production runs were performed by using a computer equipped with eight CPUs and 12 GB RAM. Each simulation contains 4 ns data and took approximately five days to finish. Snapshots were saved in every 0.5 ps to analyze the dissociation trajectory. Simulations of mutated monobodies followed the same protocol as mentioned above.
Hydrogen bonds were calculated using the VMD software utilizing the hbonds plugin v1.2, which measures the hydrogen bonds of each interacting YS1 residue throughout the trajectory. Hydrogen bonds were considered to be formed between the hydrogen donor (D) and another atom (the acceptor, A) given that the distance D-A is less than the cut-off distance at 3.5Å and the angle D-H-A is within the cut-off angle at 180±60 degrees57. Only polar atoms were considered in the calculation.
Alanine-scanning mutagenesis
Following the annealing and equilibrium achieved using MD simulation, the 6-8 randomly selected monobody-MBP complexes were entered into to the computational alanine-scanning program on the Robetta server58 (http://robetta.bakerlab.org/) and the DrugScorePPI webserver38 (http://cpclab.uni-duesseldorf.de/dsppi/main.php). Here, all amino acids at the YS1-MBP interface were identified and individually mutated to alanine. The change in binding free energy (ΔΔGbind) or the change in the mutated complex partner protein stability in isolation (ΔGpartner) was measured. Additionally the degree of buriedness was identified for interfacial residues. The binding free energy function utilized here accounts for solvation interactions, shape complementarity of interacting atoms, and the polar interactions of both ion pairs and hydrogen bonds (Robetta)58 or was based on adapted knowledge-based distance-dependent pair potentials (DrugscorePPI)38. The degree of buriedness analysis indicates the number of atoms within a radius of 4Å of an interfacial residue with a higher score indicating a more buried residue.
Lipid bilayer preparation
Lipid solution was prepared by first dissolving 8 mg of DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) into 8 mL of lipid buffer B (20 mM Tris-HCl, 50 mM NaCl, 1 mM CaCl2, 0.1% (w/v) Triton X-100)28,29,32. A total of 130 uL of TMD-MBP (80 µg/mL) was added to 380 µL of lipid solution and incubated for 2 h at 37°C. The resulting solution was transferred to a 10 kDa MWCO dialysis cassette and dialyzed three times for 12 h each against 1 L of lipid buffer A (20 mM Tris-HCl, 50 mM NaCl, 1 mM CaCl2)28,29. Lipid-protein solutions were then stored at 4°C in a translucent glass vial under nitrogen for up to one month. The TMD-MBP bilayer was prepared by first plasma cleaning a glass slide for 5 min and immediately submerging it in a solution of 100 ppm polyethyleneimine (PEI) in 0.5 mM KNO3 for 20 min before rinsing with DI water and drying with nitrogen28,29. Slides were further dried in a vacuum desiccator prior to use. Under a slightly dampened towel to prevent complete dehydration the PEI-coated glass slide was incubated with a 4 mL droplet of the lipid-protein solution for 2h. Slides were then rinsed three times in Hank's balanced salt solution (HBSS) before being immersed HBSS prior to force spectroscopy experiments. A PEI-coated glass slide was incubated with only the lipid solution (without the TMD-MBP protein) and used as a negative control to determine the binding specificity as before28.
Cantilever functionalization
To provide a suitable surface for coating with soluble proteins, molecular force probe cantilevers (Veeco, Plainview, NY) were silanized with 2% (v/v) 3-aminopropyltriethoxysilane in acetone29,30. The cantilevers were first incubated in a 30 μg/ml solution of anti-histidine antibody for 30 min before being incubated for 1 h in a 3 mg/mL solution of wild-type or mutated monobody YS1 protein in Dulbecco's PBS containing 50-fold molar excess of the crosslinker bis(sulfosuccinimidyl) suberate (BS3; Pierce, Rockford, IL). The reaction was quenched with Tris buffer. To block nonspecific interactions, cantilevers were incubated in 1% bovine-serum albumin in Dulbecco's PBS. Protein solution concentrations were optimized to result in a low proportion of binding events during force-spectroscopy experiments (~20 binding events per 100 contacts).
Single-molecule force spectroscopy
Force spectroscopy experiments were conducted using a Molecular Force Probe (MFP-1D; Asylum Research, Santa Barbara, CA). Using thermal oscillation method, a triangular cantilever (nominal spring constants of 10 pN/nm) was calibrated, with its deflection measured by laser reflection onto a split photodetector59,60. The petri dish containing the MBP-incorporated or blank lipid bilayer slides and Hanks' balanced salt solution buffer was placed on the stage and positioned to be directly below the cantilever. The cantilever height was adjusted such that each approach cycle generated a slight force (~1–2 nN) onto the lipid bilayer before reproach. For each run, reproach velocity was varied from 5 to 25 mm/s, and the dwell time was set to 20 ms59,60. Rupture forces and loading rates were calculated from force-versus-distance traces using IgorPro 4.09 software (WaveMetrics, Lake Oswego, OR). The Bell model parameters (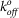 and xβ) were tabulated using a least-squares fit to the rupture force against the logarithm of loading rate29,30,60. At least four individual experiments and >1000 successful events were run for MBP binding to the wild-type YS1 and the R33A and E47A samples with the exception of the control samples, which were tested by three individual experiments. A new cantilever was freshly prepared, calibrated, and tested for each individual experiment.
and xβ) were tabulated using a least-squares fit to the rupture force against the logarithm of loading rate29,30,60. At least four individual experiments and >1000 successful events were run for MBP binding to the wild-type YS1 and the R33A and E47A samples with the exception of the control samples, which were tested by three individual experiments. A new cantilever was freshly prepared, calibrated, and tested for each individual experiment.
Surface plasmon resonance
The binding kinetics between wild-type or mutated monobody YS1 and their target protein MBP were measured using a BIAcore 3000. The running buffer used was HBS-P at pH 7.4 with 10 mM HEPES, 150 mM NaCl, and 0.005% (v/v) Tween 20 (BIAcore, Piscataway, NJ). A low density of ~200 RU of biotinylated MBP (Avidity, Aurora, CO) was immobilized onto a streptavidin SA chip (BIAcore) to avoid mass-transport limited effect. For binding assays, either wild-type or mutated monobody YS1 protein (0-137 nM) was injected for 100 s at a flow rate of 60 mL/min and followed by washing with buffer for 10 min. Reference curves generated from an uncoated flow cell and multiple injections of running buffer provided a double-reference. The binding sensorgrams were analyzed by the global fitting of 1:1 bimolecular interaction model, and the dissociation equilibrium constant (Kd) was calculated by the dividing kon by koff61. BIAevaluation 3.0 software (BIAcore) was utilized for data analysis and self-consistency examination was performed as described62.
Author Contributions
L.S.-L.C. conceived the experiments, performed most of the experiments, analyzed and interpreted data, and edited the manuscript. D.J.S. analyzed and interpreted data, and wrote the manuscript. N.N. and A.D. prepared TMD-MBP and monobodies used experimentally in Fig. 5,6 and edited the manuscript. M.O. and K.K. conceived the experiments, interpreted data, and wrote the manuscript.
Supplementary Material
Supplementary Figure S1
Acknowledgments
This work was supported by Defense Threat Reduction Agency grant HDTRA1-09-1-0016 (to K.K. and M.O.) and a postdoctoral fellowship from the American Heart Association (to L.S.-L.C.).
References
- Yu H., Raymonda J., McMahon T. & Campagnari A. Detection of biological threat agents by immunomagnetic microsphere-based solid phase fluorogenic-and electro-chemiluminescence. Biosens Bioelectron 14, 829–840 (2000). [DOI] [PubMed] [Google Scholar]
- Leader B., Baca Q. J. & Golan D. E. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discovery 7, 21–39 (2008). [DOI] [PubMed] [Google Scholar]
- Steinmeyer D. E. & McCormick E. L. The art of antibody process development. Drug Discov Today 13, 613–618 (2008). [DOI] [PubMed] [Google Scholar]
- Vermeer A. W. & Norde W. The thermal stability of immunoglobulin: unfolding and aggregation of a multi-domain protein. Biophys J 78, 394–404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A. Engineered protein scaffolds for molecular recognition. J Mol Recognit 13, 167–187 (2000). [DOI] [PubMed] [Google Scholar]
- Nygren P.-Å. & Uhlén M. Scaffolds for engineering novel binding sites in proteins. Curr Opin Struct Biol 7, 463–469 (1997). [DOI] [PubMed] [Google Scholar]
- Cotton G. J., Ayers B., Xu R. & Muir T. W. Insertion of a synthetic peptide into a recombinant protein framework: a protein biosensor. J Am Chem Soc 121, 1100–1101 (1999). [Google Scholar]
- Gulyani A. et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat Chem Biol 7, 437–444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A., Gilbreth R. N., Esaki K., Tereshko V. & Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci 104, 6632–6637 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz H. K. et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol 22, 575–582 (2004). [DOI] [PubMed] [Google Scholar]
- Wurch T. et al. Development of novel protein scaffolds as alternatives to whole antibodies for imaging and therapy: status on discovery research and clinical validation. Curr Pharm Biotechnol 9, 502–509 (2008). [DOI] [PubMed] [Google Scholar]
- Frejd F. Y. in Targeted Radionuclide Tumor Therapy 89–116 (Springer, 2008). [Google Scholar]
- Koide A., Bailey C. W., Huang X. & Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol 284, 1141–1151 (1998). [DOI] [PubMed] [Google Scholar]
- Koide A., Abbatiello S., Rothgery L. & Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc Natl Acad Sci 99, 1253–1258 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbreth R. N., Esaki K., Koide A., Sidhu S. S. & Koide S. A dominant conformational role for amino acid diversity in minimalist protein–protein interfaces. J Mol Biol 381, 407–418 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B. J., Kapila A. & Dane Wittrup K. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol 381, 1238–1252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbreth R. N. et al. Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design. Proc Natl Acad Sci 108, 7751–7756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Fang P. & Kay B. K. Isolation of monobodies that bind specifically to the SH3 domain of the Fyn tyrosine protein kinase. N Biotechnol 29, 526–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem Biol 9, 933–942 (2002). [DOI] [PubMed] [Google Scholar]
- Parker M. et al. Antibody mimics based on human fibronectin type three domain engineered for thermostability and high-affinity binding to vascular endothelial growth factor receptor two. Protein Eng Des Sel 18, 435–444 (2005). [DOI] [PubMed] [Google Scholar]
- Batori V., Koide A. & Koide S. Exploring the potential of the monobody scaffold: effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng 15, 1015–1020 (2002). [DOI] [PubMed] [Google Scholar]
- Cota E., Steward A., Fowler S. B. & Clarke J. The folding nucleus of a fibronectin type III domain is composed of core residues of the immunoglobulin-like fold. J Mol Biol 305, 1185–1194 (2001). [DOI] [PubMed] [Google Scholar]
- Cota E., Hamill S. J., Fowler S. B. & Clarke J. Two proteins with the same structure respond very differently to mutation: the role of plasticity in protein stability. J Mol Biol 302, 713–725 (2000). [DOI] [PubMed] [Google Scholar]
- Colman P. M. Structure-based drug design. Curr Opin Struct Biol 4, 868–874 (1994). [DOI] [PubMed] [Google Scholar]
- Marrone T. J., Briggs a., James M. & McCammon J. A. Structure-based drug design: computational advances. Annu Rev Pharmacol Toxicol 37, 71–90 (1997). [DOI] [PubMed] [Google Scholar]
- Cohen N. C., Blaney J. M., Humblet C., Gund P. & Barry D. C. Molecular modeling software and methods for medicinal chemistry. J Med Chem 33, 883–894 (1990). [DOI] [PubMed] [Google Scholar]
- Izrailev S. et al. in Computational molecular dynamics: challenges, methods, ideas 39–65 (Springer, 1999). [Google Scholar]
- Cheung L. S.-L., Kanwar M., Ostermeier M. & Konstantopoulos K. A hot-spot motif characterizes the interface between a designed ankyrin-repeat protein and its target ligand. Biophys J 102, 407–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman P. S., Alves C. S., Wirtz D. & Konstantopoulos K. Single-molecule binding of CD44 to fibrin versus P-selectin predicts their distinct shear-dependent interactions in cancer. J Cell Sci 124, 1903–1910 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley W. D., Wirtz D. & Konstantopoulos K. Distinct kinetic and mechanical properties govern selectin-leukocyte interactions. J Cell Sci 117, 2503–2511 (2004). [DOI] [PubMed] [Google Scholar]
- Panorchan P. et al. Single-molecule analysis of cadherin-mediated cell-cell adhesion. J Cell Sci 119, 66–74 (2006). [DOI] [PubMed] [Google Scholar]
- Marshall B. T. et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 (2003). [DOI] [PubMed] [Google Scholar]
- Raman P. S., Alves C. S., Wirtz D. & Konstantopoulos K. Distinct kinetic and molecular requirements govern CD44 binding to hyaluronan versus fibrin (ogen). Biophys J 103, 415–423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Chen W., Evavold B. D. & Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira I. S., Fernandes P. A. & Ramos M. J. Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins: Struct Funct Bioinf 68, 803–812 (2007). [DOI] [PubMed] [Google Scholar]
- Bogan A. A. & Thorn K. S. Anatomy of hot spots in protein interfaces. J Mol Biol 280, 1–9 (1998). [DOI] [PubMed] [Google Scholar]
- Kortemme T. & Baker D. A simple physical model for binding energy hot spots in protein–protein complexes. Proc Natl Acad Sci 99, 14116–14121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. M. & Gohlke H. DrugScorePPI webserver: fast and accurate in silico alanine scanning for scoring protein–protein interactions. Nucleic Acids Res 38, W480–W486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar D., Deng Y. & Roux B. T. Computations of absolute solvation free energies of small molecules using explicit and implicit solvent model. J Chem Theory Comput 5, 919–930 (2009). [DOI] [PubMed] [Google Scholar]
- Beeby M., Gumbart J. C., Roux B. & Jensen G. J. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol 88, 664–672 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix J. et al. Human IL-34 and CSF-1 establish structurally similar extracellular assemblies with their common hematopoietic receptor. Structure 21, 528–539 (2013). [DOI] [PubMed] [Google Scholar]
- Tanner D. E., Chan K.-Y., Phillips J. C. & Schulten K. Parallel generalized Born implicit solvent calculations with NAMD. J Chem Theory Comput 7, 3635–3642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I. Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978). [DOI] [PubMed] [Google Scholar]
- Koide A., Wojcik J., Gilbreth R. N., Hoey R. J. & Koide S. Teaching an old scaffold new tricks: monobodies constructed using alternative surfaces of the FN3 scaffold. J Mol Biol 415, 393–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadur R. P., Chakrabarti P., Rodier F. & Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins: Struct Funct Bioinf 53, 708–719 (2003). [DOI] [PubMed] [Google Scholar]
- Pednekar D., Tendulkar A. & Durani S. Electrostatics-defying interaction between arginine termini as a thermodynamic driving force in protein–protein interaction. Proteins: Struct Funct Bioinf 74, 155–163 (2009). [DOI] [PubMed] [Google Scholar]
- Magalhaes A., Maigret B., Hoflack J., Gomes J. & Scheraga H. Contribution of unusual arginine-arginine short-range interactions to stabilization and recognition in proteins. J Protein Chem 13, 195–215 (1994). [DOI] [PubMed] [Google Scholar]
- Soetens J.-C. et al. Effect of polarizability on the potential of mean force of two cations. The guanidinium-guanidinium ion pair in water. Journal Phys Chem B 101, 10910–10917 (1997). [Google Scholar]
- Maksimiak K., Rodziewicz-Motowidlo S., Czaplewski C., Liwo A. & Scheraga H. A. Molecular simulation study of the potentials of mean force for the interactions between models of like-charged and between charged and nonpolar amino acid side chains in water. Journal Phys Chem B 107, 13496–13504 (2003). [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: visual molecular dynamics. J Mol Graphics 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- Phillips J. C. et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. Journal Phys Chem B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79, 926 (1983). [Google Scholar]
- Darden T., York D. & Pedersen L. Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J Chem Phys 98, 10089 (1993). [Google Scholar]
- Schneider T. & Stoll E. Molecular-dynamics study of a three-dimensional one-component model for distortive phase transitions. Phys Rev B 17, 1302 (1978). [Google Scholar]
- Feller S. E., Zhang Y., Pastor R. W. & Brooks B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103, 4613 (1995). [Google Scholar]
- Baker E. & Hubbard R. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol 44, 97–179 (1984). [DOI] [PubMed] [Google Scholar]
- Kortemme T., Kim D. E. & Baker D. Computational alanine scanning of protein-protein interfaces. Sci Signal 219, pl2 (2004). [DOI] [PubMed] [Google Scholar]
- Dobrowsky T. M., Panorchan P., Konstantopoulos K. & Wirtz D. Live-Cell Single-Molecule Force Spectroscopy. Methods Cell Biol 89, 411–432 (2008). [DOI] [PubMed] [Google Scholar]
- Hanley W. et al. Single molecule characterization of P-selectin/ligand binding. J Biol Chem 278, 10556–10561 (2003). [DOI] [PubMed] [Google Scholar]
- Alves C. S. & Konstantopoulos K. PDGF Suppresses the Sulfation of CD44v and Potentiates CD44v-Mediated Binding of Colon Carcinoma Cells to Fibrin under Flow. PLoS One 7, e41472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. & Minton A. P. Kinetic analysis of biosensor data: elementary tests for self-consistency. Trends Biochem Sci 21, 458–460 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1