Abstract
Glucose and amino acids are key nutrients supporting cell growth. Amino acids are imported as monomers, but an alternative route induced by oncogenic KRAS involves uptake of extracellular proteins via macropinocytosis and subsequent lysosomal degradation of these proteins as a source of amino acids. In this study, we examined the metabolism of pancreatic ductal adenocarcinoma (PDAC), a poorly vascularized lethal KRAS-driven malignancy. Metabolomic comparisons of human PDAC and benign adjacent tissue revealed that tumor tissue was low in glucose, upper glycolytic intermediates, creatine phosphate and the amino acids glutamine and serine, two major metabolic substrates. Surprisingly, PDAC accumulated essential amino acids. Such accumulation could arise from extracellular proteins being degraded through macropinocytosis in quantities necessary to meet glutamine requirements, which in turn produces excess of most other amino acids. Consistent with this hypothesis, active macropinocytosis is observed in primary human PDAC specimens. Moreover, in the presence of physiological albumin, we found that cultured murine PDAC cells grow indefinitely in media lacking single essential amino acids, and replicate once in the absence of free amino acids. Growth under these conditions was characterized by simultaneous glutamine depletion and essential amino acid accumulation. Overall, our findings argue that the scavenging of extracellular proteins is an important mode of nutrient uptake in PDAC.
Introduction
One of the most lethal forms of cancer is pancreatic ductal adenocarcinoma (PDAC) (1). Almost all cases of PDAC involve activating KRAS mutations (2). In addition to driving growth, KRAS induces metabolic changes including enhanced glucose uptake, glycolytic flux, and glucose flux into hexosamines and ribose-5-phosphate (3). In contrast to other driver oncogenes such as PI3K that broadly increase glucose flux throughout metabolism (4), oncogenic RAS impairs flux of glucose through pyruvate dehydrogenase into the TCA cycle (5,6). RAS-driven cells instead rely heavily on glutamine as a TCA carbon source, with glutamine catabolism through the TCA cycle and malic enzyme essential in pancreatic cancer cells (7). Thus, RAS-driven cancer cells are comparatively less dependent on glucose than other cancer cells (8).
Generation of significant ATP from substrates other than glucose requires oxygen, whose availability in tumors is classically limited due to poor perfusion. Indeed, PDAC tumors, which are characterized by poor vascularization and high interstitial pressure, are typically hypoxic (9,10). Given the high metabolic demands of tumor growth, poor perfusion may lead to limitation not only for oxygen but also nutrients including glucose and free amino acids. Given the particular importance of glutamine as a source of both usable nitrogen and TCA cycle carbon, glutamine can potentially be a limiting nutrient for tumor growth. Consistent with this, studies in murine tumor models in the 1940s and 1950s found lower free glutamine in the tumor than corresponding normal tissue (11,12).
A potential alternative to traditional uptake of monomeric amino acids via membrane transport proteins is macropinocytosis, a process activated by mutant KRAS (13,14). Macropinocytosis involves bulk uptake of extracellular constituents, including proteins which can be subsequently digested in lysosomes into free amino acids. Intriguingly, in cell culture, feeding of albumin to RAS-driven cells enabled their survival and proliferation in low glutamine, and such survival and proliferation was dependent upon macropinocytosis (14). Albumin has been reported to accumulate in tumors, likely due to a combination of leaky vasculature and lymphatic deficiency (15). Thus, it is conceptually possible that plasma protein leakage from tumor vasculature provides a nutrient source for cancer cells. The extent to which this actually occurs in human tumors, however, has not yet been explored. Nor has it been shown whether such scavenging is sufficient to provide amino acids other than glutamine in biologically significant quantities.
Here we investigate protein scavenging in PDAC. Metabolomic analysis of freshly isolated human PDAC tumor specimens (compared to benign adjacent tissue) revealed that the tumors are low in glucose, upper glycolytic intermediates, glutamine and serine. PDAC tumors also accumulated amino acids that are useful primarily for protein synthesis. While uptake or synthesis of monomeric amino acids would be expected to yield each amino acid in quantities balanced with total demand, protein catabolism instead produces amino acids in proportion to their abundance in the catabolized protein. Those amino acids that are consumed by multiple anabolic processes (such as glutamine) would accordingly become depleted relative to those used solely or primarily for protein synthesis. Thus, the observed pattern of amino acid depletion and accumulation in human PDAC suggests a reliance on protein scavenging. Consistent with this, we find that primary human PDAC specimens display enhanced macropinocytosis. Moreover, we show that cultured pancreatic cancer cells can obtain sufficient amino acids via protein scavenging to grow with albumin as the sole amino acid source, and that this mode of growth is associated with glutamine depletion and essential amino acid accumulation.
Materials and Methods
Cell culturing and amino acid dropout experiments
KRPC cells were kindly provided by S. Lowe (MSKCC) (16). These cells were harvested from a murine tumor following orthotopic injection of murine pancreas progenitor cells with endogenous KRASG12D, that were additionally engineered to have MYC expression and silenced (shRNA-mediated) p53. Cell lines were routinely passaged in Dulbecco’s modified Eagle medium (DMEM) (Mediatech) with 25 mM glucose and 4 mM glutamine and supplemented with 10% (vol/vol) fetal bovine serum (FBS, HyClone), 25 IU/mL penicillin, and 25 μg/mL streptomycin (MP Biomedicals) and split at 80% confluence. For amino acid dropout experiments, pyruvate-free DMEM was prepared from powder (Cellgro, cat. No. 10-017-CV) by adding glucose (25 mM), salts, vitamins, phenol red, and amino acids, except for the amino acid to be omitted. For single amino acid dropout experiments cells were plated in DMEM at 10% confluence. After 24 h, cells were switched to dropout DMEM supplemented with 5% dialyzed FBS (Thermo). This medium was further supplemented with 0 or 5% cell culture-grade Bovine Serum Albumin (BSA) (Sigma) which was not fatty acid free. For assaying growth in amino acid-free DMEM, cells passaged in leucine-free DMEM supplemented with 5% BSA were plated at 20% confluence. After 24, medium was changed to amino acid-free DMEM with 5% BSA. Medium was replaced as needed.
Imaging and cell counting
For imaging, cells were fixed in 10% TCA for 15 min, and images were obtained using a Nikon Eclipse TE2000-U microscope operated by Q-Capture Pro software (QImaging). KRPC cells growing in leucine-free medium supplemented with BSA were seeded at 10% confluence and switched to fresh leucine-free medium after 24 h. Cell proliferation was assessed by cell number determination using a Countess Automated Cell Counter (Invitrogen) or by determining total, packed cell volume using Packed Cell Volume tubes (PCV, Techno Plastic Products).
Stable isotope tracing experiments
Medium with fully 13C- and 15N-labeled glucose and amino acids, otherwise equivalent to DMEM, was reconstituted from individual components (13C, 15N-DMEM). This medium contained unlabeled sodium bicarbonate and vitamins and was supplemented with 5% dialyzed FBS and 1% v/v Penicillin-Streptomycin (MP Biomedicals). After 5 doublings in this medium, cells were seeded at low cell density and switched to specified 13C, 15N-labeled media. After 24 hours, metabolites were extracted and analyzed by LC-MS. Growth of KRPC cells in amino acid-free medium is dependent on pre-growth in leucine-free medium. Thus, cells were grown for 5 doublings in 13C, 15N-DMEM then switched to leucine-free 13C, 15N-DMEM. After 2–3 doublings in this medium, cells were switched to (i) complete 13C, 15N-DMEM with 5% BSA and (ii) amino acid-free 13C, 15N-DMEM with 5% BSA. Cells in complete medium were grown for 24 hours before metabolite extraction. Cells in amino acid-free medium were grown for 48 hours before metabolite extraction, with medium replaced after 24 hours.
Metabolite extraction from cultured cells
For analysis of intracellular amino acids, medium was aspirated and plates were rinsed three times with room temperature PBS. Metabolism was quenched and metabolites extracted in −80°C 80:20 methanol:water extraction solution. After 15 min at −80°C, plates were scraped and cell extracts transferred to 15 mL conical tubes. Cell suspensions were vortexed, centrifuged at 3000 g for 5 min, supernatant was kept, and cellular debris re-extracted with −80°C 80:20 methanol:water extraction solution. The resulting suspension was centrifuged and the supernatant was combined with the supernatant from the first extraction. The resulting solution was dried under nitrogen flow and resuspended in HPLC-grade water. 20 μL were added to 80 μL methanol in addition to 10 μL triethylamine and 2 μL benzyl chloroformate and incubated at room temperature for 30 min, to derivatize and thereby enhance measurement sensitivity of amino acids.
Tissue collection and metabolite extraction procedure
Patients undergoing surgical resection of pancreatic tumors consented to collection and analysis of their resected tissues. Following partial pancreas resectomy, approximately 100 mg segments of tumor and adjacent tissue were isolated and immediately frozen in liquid nitrogen, and the diagnosis of pancreatic ductal adenocarcinoma was confirmed histologically. The samples were shipped overnight on dry ice to Princeton University and stored in liquid nitrogen until analysis.
Tumor and benign adjacent tissues of the same patient were prepared and analyzed in parallel. The samples were weighed and then pulverized by agitation with stainless steel balls at liquid nitrogen temperature (CryoMill, Retsch, 25 Hz for 3 min). The pulverized tissue was mixed by vortexing with 2 ml of −80°C 80:20 methanol:water, and split into two 1 mL replicate samples, which were set aside to extract for 5 min at −80°C. Each sample was centrifuged to isolate the soluble extract, and the insoluble material was extracted twice more with 1 mL 80:20 methanol:water for 5 min at 0°C and the supernatant again isolated by centrifugation. The supernatants from the three rounds of extraction were combined, dried under nitrogen gas, and reconstituted in LC-MS grade water (1 mL of water per 25 mg initial tissue weight).
Liquid chromatography – mass spectrometry (LC-MS) analysis
Cell culture and tissue samples were analyzed by three separate LC-MS systems: (i) Stand-alone orbitrap MS (Exactive, Thermo Scientific, San Jose, CA) operating in negative full scan mode at 100,000 resolution coupled to C18 ultra performance reversed-phase ion pair liquid chromatography (17), (ii) Triple quadrupole mass spectrometer (TSQ Quantum Discovery Max, Thermo Scientific) operating in negative multiple reaction monitoring mode coupled to C18 high performance reversed-phase ion pair liquid chromatography, and (iii) Triple quadrupole mass spectrometer (TSQ Quantum Ultra, Thermo Scientific) operating in positive multiple reaction monitoring mode coupled to HILIC chromatography (18). Metabolites were identified by accurate mass (<5 ppm deviation, Exactive) or characteristic fragmentation product (triple quads), in combination with retention time match to validated standards, using in-house software (19). Linearity of response was verified by running 2-fold dilutions of most samples and observing 2-fold decreases in peak intensities. For a subset of tissue samples amino acid concentrations were determined using 13C-labeled amino acid standards. For quantification of amino acid pool sizes in cultured cells, metabolite intensities were normalized by packed cell volume (PCV). Cell culture leucine measurements were obtained using a modified HILIC method (20).
Ex vivo macropinocytosis assay
Fresh PDAC tumor tissue obtained from surgical resections was cut into slices with an approximate 3 mm cuboidal shape. Tissue was immersed into serum-free DMEM containing 1–2 mg/mL of TMR-dextran and incubated at 37°C for 20–30 minutes. Tissue was rinsed twice in PBS and immediately frozen in OCT compound. Tissue processing and image analysis was performed as previously described (21).
Data normalization and processing of tissue metabolomic data
Ion counts were normalized to correct for differences in total metabolite abundances across samples, and for any sample-to-sample drift in the overall instrument response factor. A normalization factor (γi) was calculated for each LC-MS run i. To calculate γi, every known metabolite peak Pmi in LC-MS run i was quantified, and compared to the median value of peak m across all samples, μm. The scaling factor was calculated according to equation 1:
| (1) |
ion counts were corrected accordingly: .
The normalized ion count matrix was log2 transformed and averaged over replicates. When a metabolite was measured on multiple instruments, the results were averaged.
Significance testing of tumor/benign adjacent tissue metabolite differences
To determine whether a subset of metabolites are systematically higher or lower in cancerous than in benign adjacent pancreatic tissue, p-values were computed using a paired t-test with the null distribution generated by bootstrapping (22). This method was chosen as a more conservative alternative to determining the test significance against a t-distribution, because the parametric t-distribution approach makes the assumption that the log-metabolite abundances are each normally distributed. This assumption is not valid for many metabolites.
For a given metabolite m, measured in n patients, tumor metabolite abundance (Cmi) was compared within the same patient to the benign tissue metabolite abundance (Bmi). These abundances, [Bmi, Cmi], were jointly standardized so that they collectively have a mean of 0 and a standard deviation of 1. The systematic difference between pairs can be captured by a paired t-test statistic (eq. 2).
| (2) |
To generate samples for an empirical null distribution, we need to generate data where the systematic variation between the benign and cancer samples has been removed and then use the remaining variation to determine how often we would have seen such a large systematic difference (tm) by chance. To generate this null data, the paired difference was removed from the benign abundances (eq. 3a), the modified abundances were re-centered (eq. 3b) and then these residuals were corrected for the 1 degree of freedom eliminated by removing the paired difference (eq. 3c), as per Efron and Tibshirani (22).
| (3a) |
| (3b) |
| (3c) |
For each bootstrap sample (500,000 were used), n patients were randomly sampled (with replacement) and the pairs of null-data from these patients (drawn from and ) formed and . Because there is no systematic differences between the means of and , t-statistics of these null-pairs (eq. 4) can be used to approximate the distribution of t-statistics expected under the null hypothesis.
| (4) |
A p-value for metabolite m can be calculated by determining how often a t-statistic more extreme than the observed statistic (tm) would be expected under the null hypothesis (eq. 5).
| (5) |
False-discovery rate (FDR) calculation
Correction for multiple hypothesis testing followed the procedure of Storey and Tibshirani, 2003 (23). Briefly, if there are no metabolites that systematically differ between benign tissue and tumors (i.e. all of the metabolites are true negatives) then the expected distribution of p-values across all metabolites is uniform on [0,1]. To the extent that some of the metabolites do systematically differ, the associated p-value histogram will be a mixture of true positive p-values (skewed towards zero) and uniform true negatives. This histogram allows us to estimate the fraction of true negatives in the dataset: πo. We can then find a p-value cutoff, q, corresponding to the desired FDR, by taking the ratio of the expected number of false positives (πomq) to the number of p-values less than q. Metabolites with p-values less than this q-value were treated as discoveries.
Results
Metabolomic analysis of human PDAC tumors
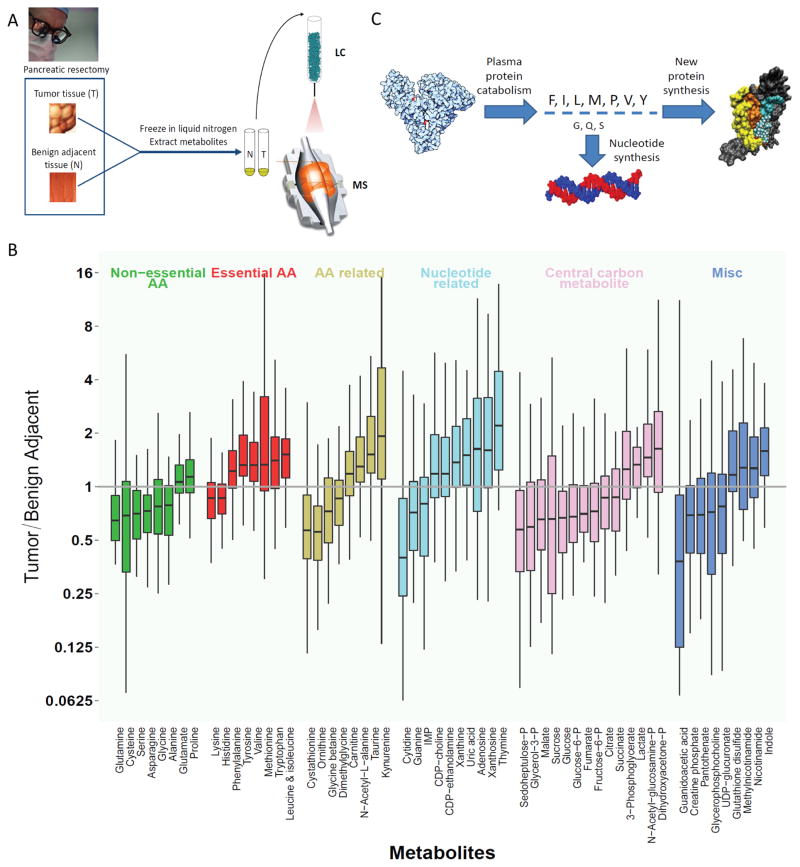
Paired PDAC tumor and benign adjacent tissue specimens were acquired by surgical resection from 49 patients (Fig. 1A). To minimize metabolic changes during the tissue acquisition process, the preferred technique is freeze-clamping in situ with liquid nitrogen-cooled Wollenberger tongs (24,25). Such in situ freeze-clamping might compromise clinical outcomes, e.g., by precluding proper identification of tumor margins. Accordingly, we instead relied on excised samples, with the surgical approach chosen to maintain perfusion until just prior to excision. Thereafter, tumor and benign adjacent tissue samples were rapidly quenched in liquid nitrogen and stored at ≤ −70°C. Metabolome analysis was conducted at the whole sample level, without distinguishing between epithelial and stromal tumor components. Paired samples were extracted in parallel and analyzed by three complementary LC-MS methods that enabled quantitation of 127 water-soluble metabolites across a majority of the samples (17).
Figure 1. Metabolomic analysis of human PDAC compared to benign adjacent pancreatic tissue.
(A) Schematic of experimental procedure: PDAC and benign adjacent tissue from n = 49 patients were collected during pancreatic resectomy, frozen in liquid nitrogen, ground, extracted, and subjected to metabolite analysis by LC-MS. (B) Metabolites with significantly different levels between tumor and paired benign adjacent tissues (false discovery rate < 0.05). Y-axis is the ratio of metabolite concentrations in the tumor to benign adjacent tissue. Vertical lines represent range, boxes interquartile range, and horizontal lines median across the 49 patients. (C) Schematic illustration of possible mechanism underlying essential amino acid accumulation: Protein catabolism yields amino acids in the ratio found in protein; those used extensively for purposes other than making new protein are depleted, while the others buildup.
Of the 127 metabolites examined, 57 displayed significantly different levels in pair-wise analysis of tumor and benign adjacent tissue (Fig. 1B). The most strongly depleted metabolites in tumors were glutamine, cytidine (whose amino group is derived from glutamine), guanidoacetic acid (a precursor to creatine phosphate, which was also down), glucose, and several phosphorylated compounds derived from glucose (glucose-6-phosphate, sedoheptulose-7-phosphate, and glycerol-3-phosphate). The most strongly increased metabolites in tumors were the DNA base thymine and several hydrophobic essential amino acids (valine, isoleucine/leucine, and tryptophan). The tryptophan degradation product kynurenine, which has immunosuppressant bioactivity (26), was also strongly elevated in the tumors.
Within central carbon metabolism glucose, glucose-6-phosphate, and fructose-6-phosphate were all decreased in the tumors, as were most TCA cycle compounds. In contrast, the 3-carbon glycolytic intermediates dihydroxyacetone phosphate and 3-phosphoglycerate were slightly increased, as was lactate. Collectively, these observations are consistent with increased propensity for aerobic glycolysis but decreased glucose availability in the tumors.
As a class, the twenty proteogenic amino acids showed particularly strong changes between the tumor and benign adjacent tissue with some amino acids strongly increased in the tumors, and others strongly decreased. There was the propensity for “non-essential” amino acids to be depleted in the tumors, whereas “essential” amino acids accumulated to higher levels (compare green and red bars in Fig. 1B). This trend, however, was not absolute. Most importantly, while glutamine was the most depleted amino acid (with an average depletion of 2.5-fold verified using 13C-labeled internal standards, Table S1), glutamate (which differs from glutamine by a single amine moiety) was slightly increased. This led us to consider the hypothesis that the observed patterns of amino acid depletion and accumulation might not reflect amino acid essentiality, but rather rates of individual amino acid consumption by anabolic pathways. Specifically, the two most depleted amino acids, glutamine and serine, play key anabolic roles as amine- and one-carbon donors respectively. In addition, serine is a key precursor of lipid head groups, and both CDP-choline and CDP-ethanolamine levels were increased in tumors. In contrast, levels of amino acids used primarily for protein synthesis were increased in PDAC.
This pattern of amino acid levels in the tumors is consistent with amino acids being acquired by protein catabolism to fuel anabolic metabolism (Fig. 1C) (14). While uptake or synthesis of monomeric amino acids would be expected to produce each amino acid in the appropriate amount, protein catabolism instead produces all amino acids in proportion to their abundance in protein. Those amino acids that are consumed by multiple anabolic processes (such as glutamine) accordingly become depleted relative to those used solely or primarily for protein synthesis.
Macropinocytosis in PDAC tumors
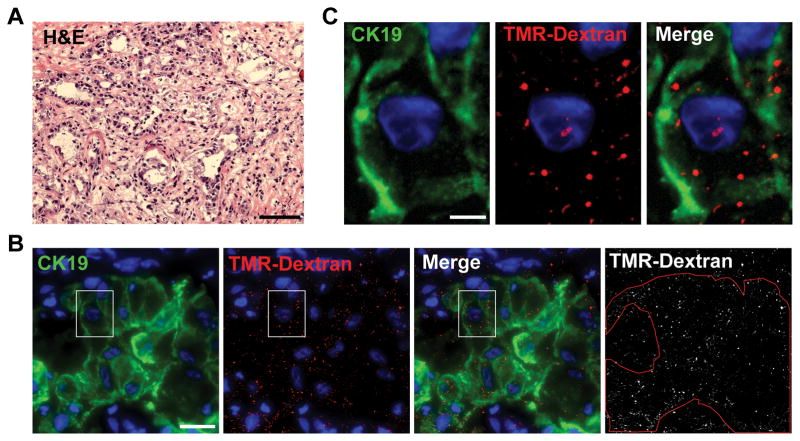
Macropinocytosis mediates the endocytic uptake of extracellular protein in Ras-driven cancer cells and murine tumors (14). Therefore, to examine whether this protein internalization mechanism is active in human PDAC tumor tissue, freshly acquired human tumor specimens were incubated with high molecular weight tetramethylrhodamine-conjugated dextran (TMR-dextran), an established marker of macropinosomes, and intracellular uptake of TMR-dextran was assessed by fluorescent microscopy of tissue sections. TMR-positive macropinosomes were detected in CK19-positive tumor cells (Fig. 2). Quantitatively lower, but nevertheless substantial, TMR-dextran staining was also detected in CK19-negative tumor tissue, which may include both stromal cells and PDAC cells that have undergone EMT (thereby losing CK19) (Fig. S1). In contrast, few macropinosomes were detected in normal adjacent tissue (Fig. S2). While some intra-tumoral variability was observed, stimulated macropinocytosis was evident in each of the five analyzed PDAC tumor samples. These data indicate that macropinocytosis is an attribute of human pancreatic tumors and that pancreatic cancer cells have the capacity to take up fluids and their constituents from the tumor extracellular environment.
Figure 2. Human pancreatic tumor cells display macropinocytosis.
(A) Hematoxylin and eosin (H&E) staining of tissue from a representative human pancreatic tumor. Scale bar is 100 μm. (B and C) An ex vivo macropinocytosis uptake assay using TMR-dextran as a marker of macropinosomes (red) indicates that CK19-positive pancreatic tumor cells (green) display high levels of macropinocytosis. DAPI staining (blue) identifies nuclei. Cell boundaries (red outline) in (B) were delineated on the basis of CK19 staining. The images shown in (C) represent a higher magnification of the boxed areas in (B). For (B) and (C) the scale bars are 20 μm and 5 μm, respectively. Images shown are Z-stack projections and are representative of five independent tumors.
Support of cultured tumor cell growth by albumin in the absence of free amino acids
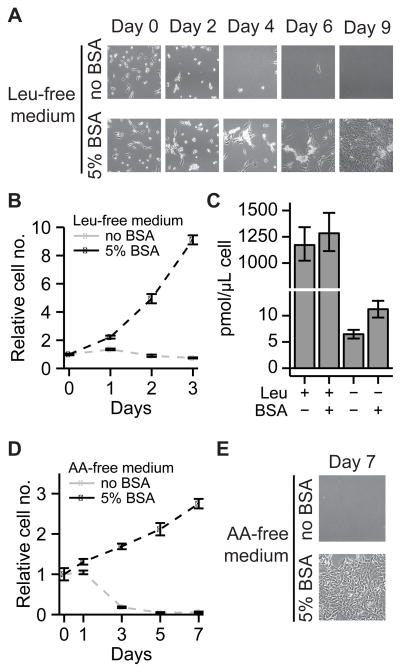
Prior work has shown that macropinocytosis enables KRAS-activated cells to proliferate in low glutamine media supplemented with albumin (14). However, the extent to which protein scavenging can provide other amino acids in quantities sufficient to support cellular proliferation remains unknown. To address this question, we incubated KRAS-driven pancreatic cells in medium lacking one or more essential amino acids and supplemented with physiological levels of albumin. Murine-derived pancreatic cancer cells with oncogenic KRASG12D and silenced p53 (KRPC cells) (16), growing in complete medium were switched to leucine-deficient medium either (i) not supplemented with bovine serum albumin (BSA) or (ii) supplemented with the typical circulating concentration of BSA (50 g/L, or 5%). As expected, KRPC cells did not survive in leucine-free medium without added BSA. In contrast, when switched to leucine-free medium supplemented with BSA, leucine removal initially resulted in extensive cell death, but surviving cells proliferated indefinitely (months) with a doubling time of ~24 hours (Fig. 3A–B, S3). After 24 hours in this medium, the intracellular leucine concentration in these cells was ~12.5 pmol/uL cell volume, roughly 100-fold lower than in the same cells grown in complete medium, but twice as high as in cells cultured in the absence of both leucine and BSA (Fig. 3C). KRPC cells were also able to proliferate indefinitely in the absence of lysine or phenylalanine (Fig. S4).
Figure 3. Ras-mutant pancreatic cells are capable of growth in medium lacking essential amino acids when supplemented with physiological levels of serum protein.
(A, B) Images of KRPC cells cultured in leucine-free medium. The cells survive and proliferate in the presence of 50 g/L BSA. (B) Cell counts from (A). (C) Intracellular leucine concentrations in KRPC cells grown in leucine-rich and leucine-free media, as measured by LC-MS. (D, E) KRPC cells cultured in amino acid-free medium grow using serum protein as their sole source of amino acids. For B–D, data are means ± SE, n ≥ 3.
One trivial explanation for the growth of KRPC cells in medium without leucine is trace leucine contamination from serum or other additives in quantities sufficient to support cell growth. To rule out this possibility, we added 12 μM leucine to leucine-free medium which was not supplemented with BSA. LC-MS measurements revealed that, while this leucine-spiked medium contained more free leucine than the BSA-supplemented leucine-free medium (Fig. S5), it did not support cell growth. Therefore, the continuous proliferation of KRPC cells in leucine-free medium supplemented with albumin is not due to contaminating leucine.
Given that KRPC cells can use intact protein as their sole source of leucine, lysine or phenylalanine, we wondered whether these cells could proliferate in BSA-supplemented medium without any free amino acids. We switched KRPC cells growing in leucine-free medium to amino acid-free medium either (i) not supplemented with BSA or (ii) supplemented with 5% BSA. While the amino acid-free medium without BSA supplementation did not support cell survival, cells switched to the BSA-supplemented amino acid-free medium grew to confluence, doubling once over a period of five days (Fig. 3D–E). Thus, KRAS-driven cancer cells in vitro can grow faster than PDAC tumor cells in vivo solely through scavenging and subsequent catabolism of extracellular protein.
Isotope tracing of serum protein catabolism
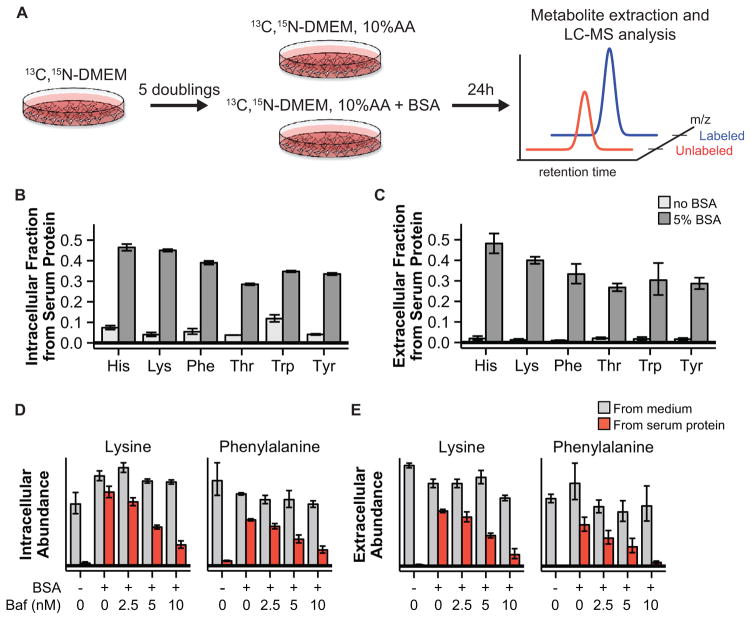
The capability of Ras-mutant cells to proliferate in the absence of essential amino acids suggests that serum protein catabolism might contribute substantially to their amino acid pools. To quantitatively measure this contribution, we developed an isotope tracer-based method enabling separate quantitation of (i) amino acids initially taken up as monomers and (ii) amino acids acquired via catabolism of serum protein. We prepared growth medium in which natural glucose and all amino acids were replaced by uniformly 13C- and 15N-labeled glucose and amino acids at standard DMEM concentrations (13C, 15N-DMEM). KRPC cells were grown in this medium for 5 doublings, such that cellular protein in the resulting population was predominantly labeled. Then, cells were transferred to 13C, 15N-medium equivalent to DMEM except that free amino acids were present at 10% of their DMEM concentrations (13C, 15N-DMEM, 10%AA). These amino acid concentrations more closely resemble physiological conditions (e.g. average human PDAC glutamine concentration is 700 μM (Table S1), and 10% of DMEM glutamine concentration is 400 μM). Use of this medium is important for detecting unlabeled amino acids coming from protein catabolism, whose concentrations are otherwise overwhelmed by the high amino acid concentrations in full DMEM. The cells were cultured for 24 h in this medium, and metabolites were extracted and analyzed by mass spectrometry (Fig. 4A).
Figure 4. Stable isotope tracing reveals substantial contribution of protein scavenging to amino acid pools.
(A) Schematic of isotope tracing experiment enabling separate quantification of amino acids derived from serum protein and amino acids initially taken up as monomers. (B, C) Fractional contribution of extracellular protein to intracellular (B) and extracellular (C) amino acid pools in KRPC cells. (D, E) Bafilomycin A1, an inhibitor of lysosomal acidification, reduces intracellular (D) and extracellular (E) serum protein-derived amino acid pools in a dose-dependent fashion. For B–E, data are means ± SE of n ≥ 3.
In cells grown without added albumin, less than 12% of intracellular and less than 5% of extracellular essential amino acids were unlabeled. These observed unlabeled amino acids are presumably derived primarily from catabolism of the available serum protein (the cells were cultured in 5% fetal bovine serum). Addition of albumin dramatically increased the observed levels of unlabeled (i.e., serum-protein derived) intracellular amino acids (Fig. 4B). Moreover, substantial concentrations of unlabeled amino acids were also observed in the medium, suggesting rapid exchange of intracellular and extracellular amino acid pools (Fig. 4C). Contribution from albumin to intracellular amino acid pools was also observed in MIA PaCa-2 and E3 human pancreatic cancer cells (Fig. S6). Thus, we were able to directly track amino acids derived from protein catabolism in cultured PDAC cells.
We next sought to validate the hypothesis that catabolism of serum protein occurs in the lysosome by treating KRPC cells with bafilomycin A1, which impairs lysosomal function by inhibiting the vacuolar-type H+-ATPase (27). Treatment of KRPC cells with bafilomycin A1 resulted in a dose-dependent reduction of amino acids derived from serum protein catabolism (Fig. 4D, E). Treatment with EIPA, a canonical inhibitor of macropinocytosis (14), also results in a reduction in serum protein-derived amino acids in KRPC cells (Fig. S7). Taken together, these data demonstrate that, even in the absence of amino acid deprivation, lysosomal degradation of extracellular protein contributes substantially to PDAC amino acid pools.
Amino acid patterns in cells fed by serum protein macropinocytosis
We next used our isotope tracing strategy to confirm that cells grown in amino acid-free media supplemented with 5% BSA acquire most of their amino acids from the unlabeled extracellular protein. With the exception of alanine, serine, and glycine, which were synthesized from glucose (which, in contrast to the pancreatic cancer microenvironment, was abundant in the culture media), all amino acids were largely unlabeled, i.e., derived from extracellular protein (Fig. 5A). In addition, amino acid nitrogen in these cells was predominantly extracellular protein-derived (Fig. S8).
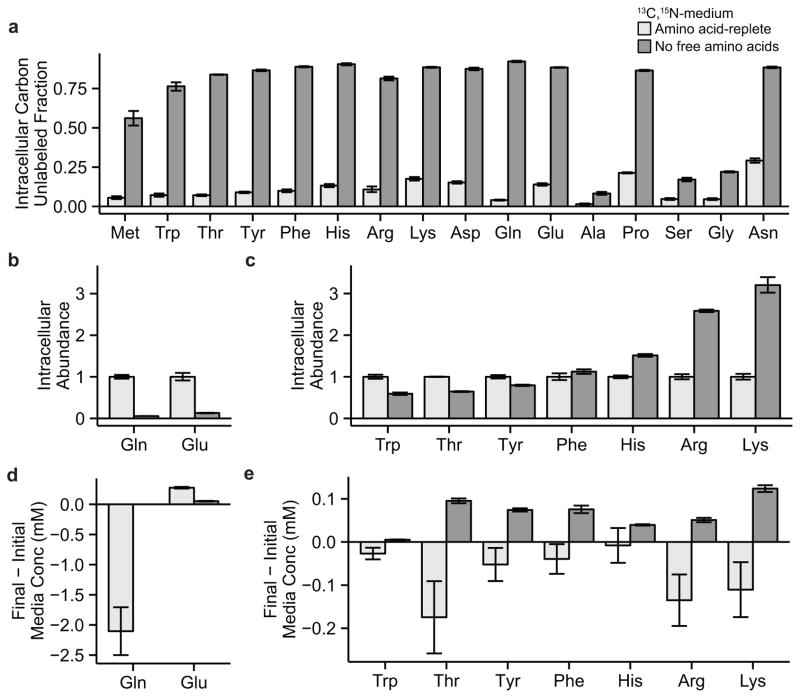
Figure 5. Cells relying on protein scavenging for amino acid assimilation accumulate strictly proteinogenic amino acids.
KRPC cells were grown in amino acid-replete or amino acid-free medium and supplemented with physiological levels of albumin. (A) Fractional contribution of serum protein-derived carbon to intracellular amino acid pools. (B, C) Relative intracellular amino acid abundances. (D, E) Net uptake or excretion of amino acids into the medium. Data are means ± SE of n ≥ 3.
We next asked how amino acid concentrations differed between cells grown in amino acid-free medium supplemented with physiological albumin and cells grown in standard DMEM. Growth in the albumin-supplemented, amino acid-free medium resulted in strong depletion of intracellular glutamine after 24 hours in culture (Fig. 5B). However, paradoxically we observed accumulation of essential amino acids to levels comparable to and in some cases greater than those seen in cells grown in rich medium (Fig. 5C). Moreover, significant concentrations of free amino acids were excreted into the medium (Fig. 5D, E). As the tumor metabolomic analysis does not allow for discrimination between cancer cell intracellular and extracellular amino acid pools, we are unable to determine if amino acid excretion occurs in vivo. Additionally, perhaps due either to differences in nutrient availability between the actual tumor and this cell culture model (e.g., of glucose, free amino acids, albumin, and other proteins) or to the substantial fraction of stroma in the tumor, there was not a direct correspondence between amino acid concentration changes in the PDAC tumors and cells grown in albumin-supplemented amino acid-free medium. Nevertheless, these data confirm that growth fed by scavenging of extracellular protein can lead to glutamine depletion and essential amino acid accumulation.
Discussion
The definition of cancer is the uncontrolled division and growth of cells. To facilitate this, cancer cells need nutrients that can be utilized to generate the necessary energy and cellular building blocks. It is commonly assumed that cancers primarily rely on glucose and glutamine as their nutrient sources. The dependence on glucose is certainly true for many cancers and is perhaps best illustrated by the clinical value of FDG-PET, which uses a glucose analog to image and stage tumors (28). However, there are exceptions, with a notable example being PDAC: less than 25% of tumors are markedly FDG-PET positive, and up to 35% do not take up FDG above background (29). This may be caused by restricted blood perfusion due to the high interstitial pressure and desmoplasia that are characteristic for PDAC (30). Despite a limited availability of free glucose and glutamine, PDAC is notoriously aggressive, suggesting that other nutrients may play an important role in fueling PDAC growth.
What could these alternative nutrients be? We recently reported the ability of KRAS-transformed cells to uptake extracellular protein through macropinocytosis (14), and to utilize serum lipids to support growth (31). PDAC cells re-utilize the amino acids and fatty acids from these extracellular proteins and lipids to support growth, much as they also rely on recycling of intracellular materials via autophagy to survive metabolic stress (32,33). Leaky tumor vasculature and lymphatics can result in accumulation of albumin and other serum proteins in the tumor interstitium. Scavenging of proteins and lipids diminishes demand for de novo biosynthesis and thus the need for glucose and free glutamine-derived carbon, reducing equivalents (NADH, NADPH), and ATP.
As some key metabolic pathways are oxygen dependent (TCA cycle, fatty acid desaturation), bypassing them facilitates growth in hypoxia. Oncogenic Ras, even in nutrient and oxygen replete conditions, reduces oxygen consumption and increases macropinocytosis and lipid scavenging (5,6,31). It thus appears that Ras readies cells to survive and grow in metabolically harsh conditions including hypoxia.
Here we provide an in depth analysis of the metabolic state of primary human PDAC tumors. PDAC tumor tissues are heterogeneous, characterized not only by the presence of tumor cells, but also cancer-associated fibroblasts, immune cells and extracellular matrix. Our analyses do not allow us to differentiate between the metabolic contributions from each of these compartments. Rather, they represent the metabolic state of the tumor tissue as a whole. Using this approach we found that, relative to the benign adjacent tissues, PDAC tumors were consistently low in both glucose and glutamine. We further found a pattern of amino acid levels consonant with what would happen if the tumors were largely reliant on protein scavenging (Fig. 1C): a differential utilization of amino acids for purposes other than protein synthesis (nucleotide and lipid synthesis etc.) leaves amino acids primarily used for protein synthesis to build up, whereas amino acids used for other purposes as well (glutamine, serine, alanine) deplete.
By culturing cells in the absence of one or all free essential amino acids, we were able to demonstrate the capacity of extracellular protein scavenging to provide amino acids to support growth. Intriguingly, cells were able to grow rapidly and indefinitely by this mode of consumption in the absence of free leucine, lysine, or phenylalanine (which are all relatively abundant in albumin; Fig. S9), and transiently in the absence of all free amino acids. Moreover, in the absence of all amino acids, BSA scavenging was sufficient to produce elevated intracellular levels of selected essential amino acids.
An increased scavenging ability allows cells to access vast resources of cellular building blocks and energy: assuming a total plasma protein concentration of 75 g/L, the amino acid content of plasma proteins exceeds free amino acids by approximately 200-fold. Thus, while poor perfusion may limit flow of all nutrients through the tumor, in such flow restriction, plasma protein may increase in relative importance as an amino acid source. In addition, plasma proteins are also a major potential energy source, exceeding energy available in glucose by about 75-fold. Similarly, the ability to scavenge fatty acids from various serum lipids, rather than free fatty acids alone, increases available fatty acids by at least 4-fold (31).
Although blood flow is often low in PDACs due to the high interstitial pressure, tumor blood vessels are leaky. In combination with the fact that tumors are lymphatic deficient, this may result in plasma protein accumulation (15). In light of this, the recent clinical success of a protein-drug conjugate albumin-paclitaxel (nab-paclitaxel, Abraxane) in PDAC is particularly tantalizing (34), and warrants further investigation into metabolic scavenging and how it can be exploited therapeutically.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants P50GM071508 (J.D.R), 1R01CA16359-01A1 (J.D.R) and 1R01CA055360 (D.B.S.), and by Stand Up To Cancer. J.J.K is a Hope Funds for Cancer Research Fellow (HFCR-11-03-01). We thank the NYU School of Medicine Biorepository Center, which is partially supported by the Cancer Center Support Grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center. It is also supported, in part, by grant UL 1 TR000038 from the National Center for the Advancement of Translational Science (NCATS), National Institutes of Health. The Histopathology Core of NYU School of Medicine is partially supported by the National Institutes of Health (grant 5 P30CA016087-32). Troma-III, an antibody that recognizes CK19, was contributed by R. Kemler and made available by the Developmental Studies Hybridoma Bank under the auspices of the NICHD. We thank Thomas Shenk’s laboratory for support with microscopy, Ian Lewis and Simon Cobbold for helpful discussions, and Scott Lowe for kindly providing the KRPC cells.
References
- 1.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 2.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2011;76:325–34. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 5.Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523–38. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Son JJ, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy JP, Eibl G, Reber HA, Hines OJ. Influence of hypoxia and neoangiogenesis on the growth of pancreatic cancer. Mol Cancer. 2003;2:12. doi: 10.1186/1476-4598-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koong AC, Mehta VK, Le QT, Fisher GA, Terris DJ, Brown JM, et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48:919–22. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 11.Roberts E, Tanaka T. Free Amino Acids of the Yoshida Ascites Tumor. Cancer Res. 1956;16:204–10. [PubMed] [Google Scholar]
- 12.Roberts E, Frankel S. Free Amino Acids in Normal and Neoplastic Tissues of Mice as Studied by Paper Chromatography. Cancer Res. 1949;9:645–8. [PubMed] [Google Scholar]
- 13.Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by Ras proteins. Science. 1986;233:1061–8. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 14.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis as a mechanism of glutamine supply in Ras transformed cancer cells. Nature. 2013;497:633–7. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stehle G, Sinn H, Wunder A, Schrenk HH, Stewart JCM, Hartung G, et al. Plasma protein (albumin) catabolism by the tumor itself - implications for tumor metabolism and the genesis of cachexia. Crit Rev Oncol Hematol. 1997;26:77–100. doi: 10.1016/s1040-8428(97)00015-2. [DOI] [PubMed] [Google Scholar]
- 16.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-Mediated MEK Activation Is Required for Effective MEK Inhibition in KRAS Mutant Tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–21. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–26. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesek JJ, Matyska MT, Fischer SM, Sana TR. Analysis of hydrophilic metabolites by high-performance liquid chromatography-mass spectrometry using a silica hydride-based stationary phase. J Chromatogr A. 2008;1204:48–55. doi: 10.1016/j.chroma.2008.07.077. [DOI] [PubMed] [Google Scholar]
- 21.Commisso C, Flinn RJ, Bar-Sagi D. Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protoc. 2014;9:182–92. doi: 10.1038/nprot.2014.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efron B, Tibshirani R. Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy. Stat Sci. 1986;1:54–75. [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wollenberger A, Ristau O, Schoffa G. Eine einfache Technik der extrem schnellen Abkuhlung grosserer Gewebestucke. Pflugers Archiv. 1960;270:399–412. [PubMed] [Google Scholar]
- 25.Quistorff B, Pedersen E. A new device for the freeze-clamping of tissue samples. Anal Biochem. 1976;73:236–9. doi: 10.1016/0003-2697(76)90162-7. [DOI] [PubMed] [Google Scholar]
- 26.González A, Varo N, Alegre E, Díaz A, Melero I. Immunosuppression routed via the kynurenine pathway: a biochemical and pathophysiologic approach. Adv Clin Chem. 2008;45:155–97. doi: 10.1016/s0065-2423(07)00007-8. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–12. [PubMed] [Google Scholar]
- 28.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higashi T, Saga T, Nakamoto Y, Ishimori T, Fujimoto K, Doi R, et al. Diagnosis of pancreatic cancer using fluorine-18 fluorodeoxyglucose positron emission tomography (FDG PET) - usefulness and limitations in “clinical reality. Ann Nucl Med. 2003;17:261–79. doi: 10.1007/BF02988521. [DOI] [PubMed] [Google Scholar]
- 30.Chu GC, Kimmelman AC, Hezel AF, DePinho RA. Stromal biology of pancreatic cancer. J Cell Biochem. 2007;101:887–907. doi: 10.1002/jcb.21209. [DOI] [PubMed] [Google Scholar]
- 31.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–7. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Von Hoff DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: A phase I/II trial. J Clin Oncol. 2011;29:4548–54. doi: 10.1200/JCO.2011.36.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.