Abstract
Background
No data is available on morphology and genetic characteristics of Echinococcus granulosus derived from donkeys of Iran, despite of its existence in donkeys. In the present study morphometric variations of the rostellar hooks of protoscoleces and genotype characteristics of hydatid cyst of donkey from Iran were determined.
Methods
Protoscoleces prepared from hydatid cyst of donkey of Iran were morphometric and genetic analyzed. The genetic analysis was done using Cox 1 gene by comparative sequence analysis.
Results
Our morphometric results showed that donkey of Iran shares 6 out of 7 determined parameters with donkeys of Jordan and 4 out of 7 with 4 available data with Switzerland donkeys. Morphological similarities and dissimilarities were observed with sheep-dog (G1) and camel-dog strains (G6) of Iran. The nucleotide sequence alignment showed that the partial sequence of Cox 1 from donkey had 91% homology with query coverage of 99% to the corresponding sequence of E. equinus, 90% homology to the E. felidis, 90% homology to E. ortleppi, 89% homology to the E. shiquinus, 89% homology to the E. vogeli, 89% homology to the E. oligarthrus, 88% homology to the E. canadensis and 83% homology to the Taenia solium. Additionally, the amino acid sequence of this gene has also some differences between this strain and all known strains of E. granulosus even with E. equinus (G4).
Conclusion
Despite of common morphological characteristics of Iranian donkey hydatid cyst with those of donkeys of other parts of the world, genetically it has its own entity.
Keywords: Echinococcus granulosus, Morphometric, DNA, COX I, Donkey, Iran
Introduction
Iran is an important endemic focus of E. granulosus, where domestic and wild ruminants (1), man (2), donkey (3) and wild boar (4) act as intermediate hosts and dog (5) and wild carnivores (6) as definitive hosts.
Hydatidosis belongs to the most important and fatal helminthic zoonotic diseases. The early diagnosis of disease plays an important role in the health management. Abdi et al. have developed an ELISA system based on the recombinant antigen EgAgB16 kDa with sensitivity, specificity; positive and negative predictive values of 93.5%, 95.6%, 96% and 92.9%, respectively, (7). Taghipour et al. have prepared recombinant antigen B and tested it in western analysis and believed that this recombinant antigen could be suitable for diagnosis of human hydatidosis by ELISA technique (8). At the DNA level, Rahimi et al. used specific primer pair designed from genomic ITS-1 region for the diagnosis (9).
E. granulosus shows a great intra specific variation in relation to host specificity, epidemiology, morphology, developmental biology, biochemistry, physiology, and genetics (10). Several publications are available on the morphometric and genetic characteristics of horse hydatid cyst (11-14) and fewer on hooks morphology of donkeys (12,13,15-19) but not on its genotype. High population of donkey (80% of 2 million equines) that are scattered all over the country and are grazing in common pastures with ruminants and carnivores can play a role in the epidemiology of echinococcosis hydatidosis in Iran.
The aim of this study was to determine the morpholmetric and genetic characteristics of E. granulosus derived from donkeys of Iran.
Materials and Methods
Morphometric study
One out of 50 donkeys (Equinus asinus) (2%) slaughtered at Tehran Zoo to feed wild carnivores, harbored a fertile hydatid cyst in the liver. After removal the cyst and aspiration the hydatid cyst fluid under sterile conditions, the viability of protoscoleces which were washed twice in saline was confirmed through staining with 1% Eosin solution and observation of flame cells activity. Some protoscoleces were stored in 70% ethanol for molecular study and seven parameters including the total number of hooks, the total length of large and small hooks, and their blades, and percent ratio of blade length to total length of hooks were determined in 4 large and 4 small hooks of 10 protoscoleces. Sufficient pressure was applied to protoscoleces under the cover slip to flattened them but not damage the hooks. William and Sweatman (1963) description was used to determine protoscoleces hooks measurements (12). Independent samples’ test and one way ANOVA, and post hoc test Tukey were used to evaluate the obtained data.
DNA Extraction
Protoscoleces were removed from ethanol, dried and washed twice in PBS and stored for 1-2 days without any buffer in 1.5 ml tube at -20 °C. All tissues had been obtained with consent given according to institutional guidelines. DNA was extracted using a DNA isolation kit (MBST, Iran) according to the manufacturer’s instructions. Briefly, each single worm was lysed in 180 μl lysis buffer and 20 μl proteinase K for 30 min at 55 °C. After adding 360 μl binding’s buffer and incubation for 10 min at 70 °C, 270 μl ethanol (100%) was added to the solution. After vortexing, the complete volume was transferred to the MBST-column. The MBST-column was first centrifuged and then washed twice with 500 μl washing-buffer. Finally, DNA was eluted from the carrier with 100 μl elution buffer.
Polymerase chain reaction (PCR)
For DNA amplification, different amounts of DNA solution (1 μl, 5 μl and 10 μl) were used. The PCR was performed on 100 μl total volume including one time PCR buffer, 2.5 U Taq Polymerase (Cina gene, Iran), 2 μl of each primer (20 mM, MWG, Germany), 200 μM of each dATP, dTTP, dCTP and dGTP (Fermenta) and 1.5 mM MgCl2 in automated Thermocycler (MWG, Germany) with the following program: 5 min incubation at 95°C to denature double strand DNA, 35cycles of 45 s at 60°C (annealing step), 45 s, at 72°C (extension step) and 45 s at 94°C (denaturing step). Finally, PCR was completed with the additional extension step for 10 min. The PCR products were analysed on 1.8% Agarose gel in 0.5 times TBE buffer and visualized using ethidium bromide and UV-eluminator. The nucleotide sequence of the primers producing a specific Cox 1 product of 882 bp was for forward primer (F1) 5` gaattta ccgcgtttgaa 3` and for reverse primer (R) 5` cttatataagaacctaacgac 3`respictively. To control the specificity of the PCR products from the Cox 1, semi-nested PCR technique was used, in which the additional forward primer (F2) 5` gtggtgatcctattttatttc 3` is designated within the two abovementioned primers. After amplification, the PCR product was purified or extracted from agarose gel and re-amplified with the second forward primer and reverse primer to obtain a PCR product of 493 bp. To obtain the nucleotide sequence of the Cox 1 gene of Echinococcus granulosus of donkey, the partial nucleotide sequence of the gene from nucleotide -411 to 908 was determined.
PCR product extraction from the agarose gel
Twenty μl of PCR product was run on a 1.5% agarose in TBE buffer. After visualization of the positive band using ethidiume bromide under UV, and the PCR product was extracted from the gel using DNA extraction kit from agarose gel (MBST, Iran) according to the manufacturer’s. Briefly, the PCR product was cut from the gel under UV control and dissolved in 340 μl binding buffer for 5 min by 60°C. After addition of 255 μl ethanol (96%) to the sample, the mixture was applied to the spin column and centrifuged for 1 min at 8000 g. The column was washed twice with washing buffer and the adsorbed DNA was eluted from the column using 50 μl elution buffer. The PCR product was subsequently from both sites sequenced by Kowsar Company (Iran).
PCR product purification
PCR product was purified from the salts and proteins using PCR purification kit (MBST, Iran). Briefly, 200 μl binding buffer was added to 100 μl PCR product solution. After adding of 150 μl ethanol (96%) to the sample, the mixture was applied into the column. The column was washed twice with washing buffer and PCR product was eluted from the column using 100 μl elution buffer.
Results
The results of morphometric characteristics of hooks of donkey hydatid cyst of Iran in comparison with available data on donkeys of Jordan (17) and Switzerland (13) and those from sheep-dog and camel- dog strain of Iran (20) are shown in Table 1. Hooks were smooth in shape and their arrangements in protoscoleces were in alternate manner.
Table 1.
Morphological characteristics of Echinococcus granulosus derived from donkey in Iran
| Parameters | Donkey (Iran) (Mean ± SE)(Range) n =10 | Donkey (Jordan) (Mean ± SE) (Range) n =60 | Donkey (Switzerland) (Mean ± SE) (Range) n =30 | sheep (Iran) (Mean ± SE) (Range) n =10 | Camel (Iran) (Mean ± SE) (Range) n =10 |
|---|---|---|---|---|---|
| Total number of hooks | a34.8 ± 5.6 (28 - 46) | a35.0 ± 2.0 (32 - 40) | No data | a 35.5 ± 4.2 (29 - 44) | a35.7 ± 4.3 (29 - 41) |
| Large hooks | |||||
| Total length (T.L. μm) | ab28.8 ± 1.5 (25.6 -32.4) | a32.3 ± 0.9 (30 -34) | b29.4 ± 0.9 (27.0 - 31.0) | c 23.3 ± 2.9 (21.4 -24.8) | bc27.4 ± 2.2 (23.8-31.0) |
| Blade length (B. L. μm) | ad13.8 ± 1.2 (14 -19 | b17.4 ± 0.8 (10.1 -13.5) | a14.3 ± 0.8 (12.5 - 15.5) | cd11.7 ± 0.8 (10.1 -13.5) | ad14.2 ± 0.9 (12.6-15.6) |
| B.L/ T.L (%) | a 47.9 ± 3.5 (20.0 - 54.5) | a53.7 ± 2.3 (45 - 56) | No data | a50.5 ± 2.0 (42.1 - 54.5) | a51.9 ± 2.3 (49.0 - 56.0) |
| Small hooks | |||||
| Total length (T.L. μm) | ac23.6 ± 1.7 (16.2 - 27.0) | a28.8 ± 1.3(25 - 30) | a25.9 ± 1.1 (23.0 - 28.0) | b 18.5 ± 1.0 (16.9 - 20.3) | bc20.4 ± 1.8 (16.9- 22.9) |
| Blade length (B.L. μm) | ac10.5 ± 1.5 (5.4 - 13.5) | a13.5 ± 0.9 (11 - 18) | ac10.3 ± 0.8 (8.5 - 11.5) | bc7.4 ± 0.8 (6.7 - 9.0) | ac9.8 ± 1.1 (7.9 -12.9) |
| B.L/ T.L (%) | a45.5 ± 6.1 (33.3 - 62.5) | a 46.8 ± 2.9 (46 - 58) | ? | a40.6 ± 3.3 (25.2 - 33.3) | a48.4 ± 3.8 (39.6 - 56.4) |
In each inordinate characters indicate significant difference at α level; 0.05
The data in Table 1 would indicate that donkey of Iran shares 6out of 7 parameters with donkeys of Jordan and 4 out of 7 with 4 available data on Switzerland donkeys.
DNA was isolated from protoscoleces to analyze the amino acid sequence of cytochrome oxidase I (Cox 1). For this aim, the isolated DNA was first amplified using fig1/R primer pair derived from the nucleotide sequence of Cox 1 gene. The PCR analysis showed an expected PRC product of approximately 895 bp in length (Fig.1). To confirm the specificity of the PCR product, the PCR product was first purified and then amplified using primers F2/R. Semi-nested PCR showed the expected PCR product of approximately 491 bp in length (Fig. 1). To compare the nucleotide sequence and amino acid sequence of the amplified PCR product (Fig. 2) with the corresponding sequences in GenBank, the nucleotide sequence of PCR product was determined from the both 3`-, and 5` ends (600 nucleotides).
Fig 1.
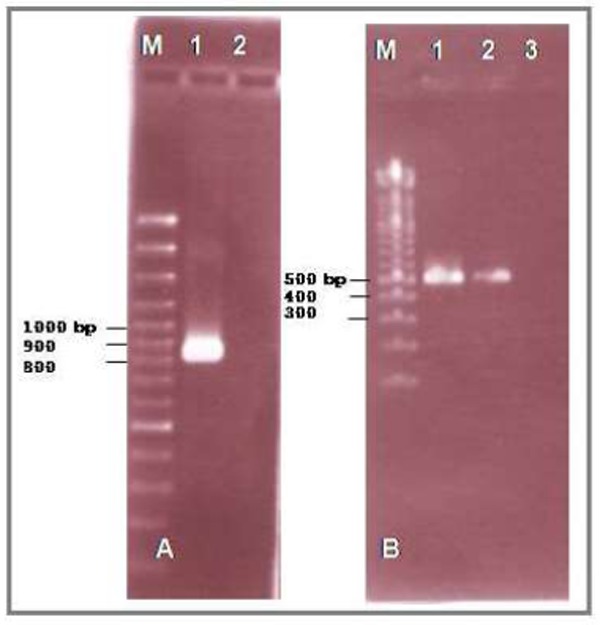
DNA was extracted from protoscoleces and amplified using primers F1/R (A): M is 100 bp marker, lane 1: PCR product from protoscolex-DNA, Lane 2: negative control. The amplified PCR product was re-amplified using primers F2/R (B): M is 100 bp marker, lane 1: semi-nested PCR with PCR product from step in fig.1 A, Lane 2: PCR product with DNA isolate from protoscolex, Lane 3: negative control
Fig 2.

The nucleotide sequence of the Cox I of E. granulosus isolated fromdonkey with corresponding amino acid sequences
The nucleotide sequence alignment showed that the partial sequence of Cox 1 from donkey had 91% homology with query coverage of 99% to the corresponding sequence of E. equinus registered under accession number AF346403.1, 90% homology with query coverage of 99% to the E. felidis registered under accession number EF558356.1, 90% homology with 99% query coverage to E. ortleppi registered under accession number AB235846.1, 89% homology with 99% query coverage to the E. shiquinus registered under accession number AB208064.1 or JF906149, 89% homology with 97%query coverage to the E. vogeli registered under accession number AB208546, 89% homology with 98% query coverage to the E. oligarthrus registered under accession number AB208545.1, 88% homology with 98% query coverage to the E. canadensis registered under accession number AB235847.1 or AB688142.1 and 83% homology with 98% query coverage to the Taenia solium registered under accession number FN995660.1.
In Fig. 3 the multiple alignment of amino acid sequences of the donkey Cox 1 sequence with the corresponding sequences of genes registered under accession numbers AAK 82350 (E. g. equinus genotype G4), AAQ93641 (isolated from sheep, genotype G1), AAQ93635 (isolated from cattle, G1) and BAF56494 (isolated from camel) is shown. The mentioned sequence of isolate from donkey had amino acids arginine (with basic side chain), leucine (nonpolar side chain), glycine (nonpolar side chain) and asparagine (uncharged polar side chain) at positions 16, 32, 57, and 158 respectively, whereas these positions were cysteine (nonpolar side chain), serine (uncharged polar side chain), serine and serine in COX I proteins mentioned above (Fig. 3).
Fig 3.
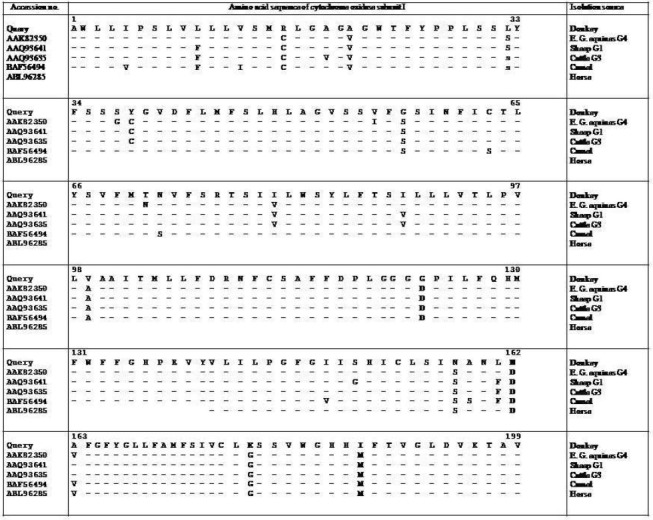
Comparison of the partial amino acid sequence of COX I protein of donkey of Iran with the corresponding amino acid sequences of Echinochoccus granulosus isolated from sheep, cattle, camel and horse
The other amino acid differences between isolate from donkey and the above mentioned sequences were in positions 158, 162, 180 and 188 which were highlighted in Fig. 3. The last 11 amino acid sequences showed complete identity to each other (Fig. 3).
Discussion
The prevalence rate (2%) and infected organ (liver) reported in this study were in harmony with previous report on donkey hydatid cyst in Iran (3). The population of donkey in the Middle East is in steady decline (21) but still it could have some importance in agriculture and transportation of freight in rural areas. If the same effect on productivity develop in donkey, as it is suggested for sheep by Polydorou (1981) (22), parts of agriculture economy may be affected (16). Donkeys constitute 80% of 2 million equines population of Iran where they grazie in common pastures with ruminants and carnivores, where the latter animals have free access to dead donkeys left in the field. Infection of donkeys with hydatid cyst has been reported from Lebanon and Syria (16), Liverpool (23), Sicily (24), Uzbekistan (25), Morocco (18,25), Jordan (17), Switzerland (13), with dramatic variation in the prevalent rate of infection ranging from 2 - 6% for Iran, 1.9% for Italy (26), up to 60% in Beka’s valley of Lebanon (15), 33.3% in Jordan (16) and 15% in Uzbekestan (25). This differences could be due to the age, the number of donkeys examined, the intensity of echinococcosis in final hosts, the consideration of standards of hygiene and suitability of climatic condition for survival of the eggs in a given area. Presence of two distinct strains of E. granulosus, e.g. sheep-dog and camel-dog based on hooks morphology, development biology (27) and genetic (20) are documented in Iran. Echinococcus granulosus shows a great intra specific variation in relation to host specificity, epidemiology, morphology and developmental biology, biochemistry, physiology, and genetics. Up to now 10 strains are recorded(10,28). Although recently based on epidemiology and genetic, several previously identified “strains” of E. granulosus (G1 to G10) have to be regarded as species and accordingly E. granulosus sensus strict (G1,G2,G3), E. ortleppi (G5) and E. equinus are valid species and other apparently monophyletic group of genotypes from G6 to G10 will be probably included in one species, named E. canadensis, even if the taxonomy of this group is not yet resolved (29,30). In recent years much attention has been paid to equine echinococcosis hydatidosis. There are several reports on the hook morphology and genetic characteristics of horse strain of E. granulosus equinus (G4 strain) which is now known as a valid species E. equinus (11-14) and fewer on the prevalence and phenotype characters of hydatid cyst of donkey origin (12,13,15-19) and only one report on genetic characters of this strain which was performed using RFLP technique with genomic DNA (12).
Morphometric analyses showed donkey of Iran shares 6 out of 7 determined parameters of rostellar hooks with donkey of Jordan (17) and 4 out of 7 with 4 available data on Switzerland donkey (13). On the other hand, previous studies had shown that hooks characters from donkeys in Lebanon and a horse from Syria (16), donkey of Jordan (17), and donkey and horse of Switzerland (13) were similar to each other. Therefore morphometrically, hydatid cyst of donkey’s origin from Iran could be taken similar to E. equinus (G4 strain). Similarities and dissimilarities between hook characteristics of donkey of Iran with those of sheep-dog and camel-dog strain of Iran seems to be a normal phenomenon. According to the different workers, dimensions of rostellar hooks of equine origin from different countries and those of cattle origin from Europe did not differed significantly, whereas they differed significantly with those of ovine origin in Switzerland (13) and in Lebanon and Syria (16) and Jordan (17). Meanwhile genetically, hydatid cyst from donkeys of Spain was similar to United Kingdom horse strain (E. equinus) (12).
The nucleotide sequence alignment showed that the partial sequence of cox1 from donkey had 91%, 90%, 89%, 89%, 89%, 89%, 89%, 88%, 83% homology to the corresponding sequence of E. equinus (AF346403.1), E. felidis (EF558356.1), E. ortleppi (AB235846.1), E. shiquinus (AB208064.1), E. vogeli (AB208546), E. oligarthrus (AB208545.1), E. canadensis (AB235847.1) and Taenia solium (FN995660.1) respectively. The comparative amino acid sequence analysis showed that the sequence derived from Cox I gene presented by donkey had in some amino acid positions strong differences to the corresponding sequences in genes registered under accession numbers AAK82350, AAQ93641, AAQ93635, BAF56494 and ABL96285. These accession numbers belongs to the isolates from E. g equinus G4, sheep G1, cattle G1, camel G6 and E. equinus. For example, at amino acid positions 16, 32, 57 and 158, the amino acids in Cox I protein isolated from donkey were arginine, leucine, glycine, and asparagine, whereas these positions in other genes mentioned above were cysteine, serine, serine and serine.
Conclusion
Therefore despite of common features between morphology of protoscoleces hooks of hydatid cyst of donkey from Iran with donkey and of the world, differences in genetics structures suggest that E granulosus derived from donkey of Iran, has its own entity.
Acknowledgments
The authors thank the Ministry of Sciences, Research and Development of Iran for the financial support. The authors declare that there is no conflict of interests.
References
- 1.Eslami A. Veterinary Helminthology. University of Tehran Press; Iran: 2005. [Google Scholar]
- 2.Eslami A, Rahbari S, Meydani M. Cestodes and trematodes of wild sheep,Ovis ammon orientalis and goitered gazelle,Gazella subgutturosa in Iran. Vet Parasitol. 1981;8:99–101. [Google Scholar]
- 3.Rokni MB. Echinococcosis/hydatidosis in Iran. Iran J Parasitol. 2009;4(2):1–16. [Google Scholar]
- 4.Eslami A, Nadealian MG. Cestode and trematode infections of equines in Iran. J Vet Fac Univ Tehran. 1987;42:33–39. [Google Scholar]
- 5.Eslami A, Farsad Hamdi S. Helminth parasites of wild boar, sus scrofa in Iran. J wildl Dis. 1992;28:316–318. doi: 10.7589/0090-3558-28.2.316. [DOI] [PubMed] [Google Scholar]
- 6.Eslami A, Hosseini SH. Echinococcus granulosus in infection of farm dogs of Iran. Parasitol Res. 1998;84:205–207. doi: 10.1007/s004360050383. [DOI] [PubMed] [Google Scholar]
- 7.Abdi J, Kazemi B, Haniloo A, Mohebali M, Mahmoudi M, Rezaei S, Bandehpour M, Maghen L, Rokni MB. Serological Evaluation of EgAgB16 kDa, a Recombinant Antigen from Echinococcus granulosus for Diagnosis of Human Hydatidosis. Iran J Parasitol. 2010;5(3):1–10. [PMC free article] [PubMed] [Google Scholar]
- 8.Taghipour N, Bandepour M, Pazoki R, Haghighi A, Nazari Pouya MR, Kazemi B. Subcloning and Expression of Recombinant Echinococcus granulosus Antigen B, in Pqe-30 Expression Vector. Iran J Parasitol. 2009;4(4):1–9. [Google Scholar]
- 9.Rahimi HR, Kia EB, Mirhendi SH, Talebi A, Fasihi Harandi M, Jalali-zand N, Rokni MB. A New Primer Pair in ITS1 Region for Molecular Studies on Echinococcus granulosus. Iran J Public Health. 2007;36(1):45–49. [Google Scholar]
- 10.Meshgi B, Eslami A, Bahonar AR, Khrrazian-Moghadam M, Gerami-Sadeghian A. Prevalence of parasitic infections in the red fox (Vulpes vulpes) and golden jackal (Canis aureus) in Iran. Iranian J Vet Res Shiraz Univ. 2009;16(4):387–391. [Google Scholar]
- 11.Thompson R, McManus D. Toward a taxonomic revision of the genus Echinococcus. Trends Parasitol. 2002;18:452–457. doi: 10.1016/s1471-4922(02)02358-9. [DOI] [PubMed] [Google Scholar]
- 12.William RJ, Sweatman GK. On the transmission, biology, morphology of Echinococcus granulosus, a new subspecies of hydatid tapeworm in horse in Great Britain. Parasitology. 1963;53:91–407. doi: 10.1017/s0031182000073844. [DOI] [PubMed] [Google Scholar]
- 13.Cuesta- Bandera C, McManus DP, Rishi AK. Characterization of Echinococcus granulosus of Spanish origin by DNA restriction endonuclease analysis and Southern blot hybridization. Int J Parasitol. 1988;18(1):137–141. doi: 10.1016/0020-7519(88)90050-1. [DOI] [PubMed] [Google Scholar]
- 14.Kumaratilake LM, Thompson RCA, Eckert J. Echinococcus granulosus of equine origin from different countries possess uniform morphological chrachteristics. Int J Parasitol. 1986;16(5):529–540. doi: 10.1016/0020-7519(86)90089-5. [DOI] [PubMed] [Google Scholar]
- 15.Hoberg EP, Miller S, Browm MA. Echinococcus granulosus (Taniidae)and authocthonous echinococcosis in the North American horse. J Parasitol. 1994;80(1):141–144. [PubMed] [Google Scholar]
- 16.Daily MD, Sweatman GK. Taxonomy of Echinococcus granulosus in the donkey and dromedary in Lebanon and Syria. Ann Trop Med Parasitol. 1965;59:463–477. doi: 10.1080/00034983.1965.11686333. [DOI] [PubMed] [Google Scholar]
- 17.Mukbel RM, Torgerson PR, Abo-Shehada MN. Prevalence of hydatidosis among donkeys in northern Jordan. Vet Parasitol. 2000;88:35–42. doi: 10.1016/s0304-4017(99)00197-1. [DOI] [PubMed] [Google Scholar]
- 18.Ihasan M, Abdel-Hafez SK, Fadwa M, Al-Yaman M. Morphological variation of Echinococcus granulosus protoscoleces from hydatid cyst of human and various domestic animals in Jordan. Int J Parasitol. 1988;18(8):1111–1114. doi: 10.1016/0020-7519(88)90083-5. [DOI] [PubMed] [Google Scholar]
- 19.Azlaf A, Dakkak A. Epidemiological study of the cystic echinococcosis in Morocco. Vet Parasitol. 2006;137:83–89. doi: 10.1016/j.vetpar.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Varcasia A, Garippa G, Pipia AP, Scala A, Brianti E, Giannetto S, Batelli G, Poglayen G, Micagni G. Cystic echinococcosis in equids in Italy. Parasitol Res. 2008;102:815–818. doi: 10.1007/s00436-007-0862-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Eslami A, Hosseini SH, McManus DP. Indication of the presence of two distinct strains of Echinicoccus granulosus in Iran by mitochondrial DNA markers. Am J Trop Med Hyg. 1998;59:171–174. doi: 10.4269/ajtmh.1998.59.171. [DOI] [PubMed] [Google Scholar]
- 22.Polydorou K. Animal health and economy.Case study:Echinococcosis with a reference to Cyprus. Bull Int Epiz. 1981;93:981–992. [Google Scholar]
- 23.Suothwell T. Experimental infection of the cat and the fox with adult Echinococcus granulosus. Ann Trop Med Parasit. 1927;21:151. [Google Scholar]
- 24.Gallo C. L,echinococcosi et la distomatosidegli equine in Sicilia. Att Soc Ital Sci Vet. 1955;9:689–691. [Google Scholar]
- 25.Zhdanova MG. Nauchnye Trudy Samarkandskogo Sel’skokhozyaistvennogo. Vol. 24. Institute im V.V. Kuibysheva; 1972. Hydatidosis in odd toed ungulates in the Uzbec S.S.R. (In Russian) pp. 151–154. [Google Scholar]
- 26.Pandey VS. Hydatidosis in donkeys in Morocco. Ann Trop Med Parasitol. 1980;74(5):519–521. doi: 10.1080/00034983.1980.11687379. [DOI] [PubMed] [Google Scholar]
- 27.Corsalini T. Frequenza dell’ idatidosi nei soggetti delle diversespecie animalimacellati a Bari. Veter Ital. 1959;10:644–647. [Google Scholar]
- 28.Hosseini SH, Eslami A. Morphological and developmental characteristics of Echinococcus granulosus derived from sheep, cattle and camel in Iran. J Helminthol. 1998;72:337–341. doi: 10.1017/s0022149x00016709. [DOI] [PubMed] [Google Scholar]
- 29.Lavikainen A, Lehtinen MJ, Meri T, Hirvela-Koski V, Meri S. Molecular genetic characterization of the Fennoscandian cervid strain a new genotype group (G10) of Echinococcus granulosus. Parasitology. 2003;124:207–215. doi: 10.1017/s0031182003003780. [DOI] [PubMed] [Google Scholar]
- 30.Nako M, McManus DP, Schantz PM, Craig PS, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mithocondrial genomes. Parasitology. 2007;134:1–1. doi: 10.1017/S0031182006001934. [DOI] [PubMed] [Google Scholar]


