Abstract
Background
Accurate identification of sheep nematodes is a critical point in epidemiological studies and monitoring of drug resistance in flocks. However, due to a close morphological similarity between the eggs and larval stages of many of these nematodes, such identification is not a trivial task. There are a number of studies showing that molecular targets in ribosomal DNA (Internal transcribed spacer 1, 2 and Intergenic spacer) are suitable for accurate identification of sheep bursate nematodes. The objective of present study was to compare the ITS1, ITS2 and IGS regions of Iranian common bursate nematodes in order to choose best target for specific identification methods.
Methods
The first and second internal transcribed spacers (ITS1and ITS2) and intergenic spacer (IGS) of the ribosomal DNA (rDNA) of 5 common Iranian bursate nematodes of sheep were sequenced. The sequences of some non–Iranian isolates were used for comparison in order to evaluate the variation in sequence homology between geographically different nematode populations.
Results
Comparison of the ITS1 and ITS2 sequences of Iranian nematodes showed greatest similarity among Teladorsagia circumcincta and Marshallagia marshalli of 94% and 88%, respectively. While Trichostrongylus colubriformis and M. marshalli showed the highest homology (99%) in the IGS sequences. Comparison of the spacer sequences of Iranian with non-Iranian isolates showed significantly higher variation in Haemonchus contortus compared to the other species.
Conclusion
Both the ITS1 and ITS2 sequences are convenient targets to have species-specific identification of Iranian bursate nematodes. On the other hand the IGS region may be a less suitable molecular target.
Keywords: Internal transcribed spacers, Intergenic spacer, Similarity score, Bursate nematodes
Introduction
In Iran, as in many countries in the world, bursate nematodes belonging to the Trichostrongyloidea superfamily are of major veterinary importance in small ruminant production systems (1, 2). Ostertagia species and Marshallagia marshalli are the most prevalent nematodes infecting sheep and goats (3). Iran is a semi dry country but climatologically can be divided into 4 different zones. Zone 4 (Central and Salt Deserts) is neither suitable for animal husbandry nor fit for human habitations. The other three zones include: Caspian zone (Zone I), Mountain plateau zone (Zone II) and the Persian Gulf Lowland (Zone III). The combined sheep and goat populations in these 3 zones are 52 million and 26 million, respectively (2, 4). Anthelmintic drug resistance has become a serious and widespread problem throughout the world. Recent reports indicate that anthelminthic resistance also occurs in some sheep nematodes, especially in Teladorsagia circumcincta, in Iran (5, 6).
Accurate identification of sheep nematodes is a critical point in epidemiological studies and monitoring of drug resistance in flocks (7-9). However, due to a close morphological similarity between the eggs and larval stages of many of these nematodes, such identification is not a trivial task (7, 10). There are a number of studies showing that ribosomal DNA (rDNA), especially the internal transcribed spacer (ITS) 1 and 2 regions, are suitable regions for phylogenetic investigations and valuable targets to design probes or define markers for the identification of bursate nematodes (8, 10-13). Some studies have also successfully used the intergenic spacer (IGS) fragment for species-specific identification of strongyle nematodes (14, 15). Due to genetic diversity and also intraspecific variations among nematodes from different geographical populations (9, 16), it seems advantageous to have complete sequences of the genetic targets to be used in species identification from all target species from a specific geographical region.
In this study, five common bursate nematodes (Haemonchus contortus, Trichostrogylus colubriformis, Teladorsagia circumcincta, Marshallagia marshalli, and Nematodirus oiratianus) were collected from sheep of zones II and III of Iran. The objective was to compare the ITS1, ITS2 and IGS regions of these nematodes in order to choose best target for specific identification methods.
Materials and Methods
Nematode samples
H. contortus, T. colubriformis, T. circumcincta, M. marshalli, N. oiratianus adult male nematodes were collected from the gastrointestinal tract of sheep slaughtered from Khuzestan, Chaharmahal and Bakhtiari provinces in south west Iran. Nematodes were washed in phosphate buffered saline (PBS) and adult male nematodes were identified morphologically according to the keys of Soulsby (17) and then stored in 70% ethanol until being used. Before DNA extraction, the nematodes were removed from ethanol, dried and washed in distilled water and stored for 1–2 days in 1.5 ml tube at −20 °C.
DNA extraction and PCR
High Pure PCR template preparation kit (Roche Diagnostics) was used to extract DNA from a single male nematode of each of the species following the manufacturer’s instructions. The DNA of the samples was stored at -20 °C until used.
Available GenBank 28s and 18s sequences of H. contortus (AM039742), T. colubriformis (AJ920350 and AM039743.1), Nematodirus battus (AJ920360 and AM039752) and T. circumcincta (AF044934.1) were used to design primers. To amplify the ITS1, 5.8S and ITS2 regions in one reaction, the primers were based on conserved sequences at the 3’end of 18s rDNA and 5’end of 28s rDNA (Table 1). The IGS primers were designed from conserved sequences at the 3’end of the 28S rDNA and 5’end of 18s rDNA (Table 1).
Table 1.
The primers used in amplification of ITS1, 5.8s, ITS2 and IGS regions. F: Forward primers, R: Reverse primers
| Species | Forward and reverse primers used to amplification of ITS1, 5.8s, ITS2 (5′-3′) | Forward and reverse primers used to amplification of IGS (5′-3′) | ||
|---|---|---|---|---|
| Haemonchus contortus | F | GCGGGAAACAGTTCAATCGC | F | ACCGTCGTGAGACAGGTTAG |
| R | TCCCCGTTCACTCGCCGTTA | R | CTTAGACATGCATGGCTTAATC | |
| Teladorsagia circumcincta | F | GCGGGAAACAGTTCAATCGC | F | ACCGTCGTGAGACAGGTTAG |
| R | TCCCCGTTCACTCGCCGTTA | R | CTGCTCTAATGAGCCGTTCG | |
| Marshallagia marshalli | F | GCGGGAAACAGTTCAATCGC | F | GCGACGTTGCTTTTTGATCC |
| R | TCCCCGTTCACTCGCCGTTA | R | CTGCTCTAATGAGCCGTTCG | |
| Trichostrongylus colubriformis | F | GCGGGAAACAGTTCAATCGC | F | ACCGTCGTGAGACAGGTTAG |
| R | TCCCCGTTCACTCGCCGTTA | R | CTGCTCTAATGAGCCGTTCG | |
| Nematodirus oiratianus | F | GTAGGTGAACTGCGGAAGGATCATT | F | ACCGTCGTGAGACAGGTTAG |
| R | TTAGTTTTCCTCCGCT | R | CTGCTCTAATGAGCCGTTCG | |
The PCR was performed on 50 μl total volume and included 1x PCR buffer (Promega), 1U Taq polymerase (Promega), 30 pmol/50 μl of each primer (SIGMA), 200 μM of each dNTP(Promega), 3.5 mM MgCl2 and approximately 2 ng per 4 μl of genomic DNA in an automated thermocycler (Thermohybaid, MSC), (18) under the following conditions: 5 min incubation at 95 °C to denature double-strand DNA, 35 cycles of 45 s at 60 °C (annealing step), 45 s at 72 °C (extension step), and 45 s at 94 °C (denaturing step). Finally, PCR was completed with an additional post-amplification extension step for 10 min at 72 °C. Samples without genomic DNA were included with each amplification as negative controls. The PCR products were analyzed on a 1% agarose gel in 1× TAE buffer and visualized using SYBR-Green dye and UVP Image Analyzer (BioSpectrum®AC Imaging System).
PCR product purification and cloning
To purify the PCR products, 40 μl of each PCR reaction mix was loaded on a 1 % low melting point agarose gel. The specific amplified fragments bands (800-1000bp in ITS and 600-1500 bp in 28s, IGS and ETS) were cut out and purified with Promega DNA purification system kit. PGEM-T Easy Vector cloning kit (Promega) was then used following the manufacturer’s instructions to clone the purified PCR products. The vectors are prepared by cutting the pGEM®-T Easy Vectors, with EcoRI and adding a 3′ terminal thymidine to both ends. Insertional inactivation of the alpha-peptide allows recombinant clones to be directly identified by blue/white screening on indicator plates. PureYield™ Plasmid Miniprep System (Promega) was used for plasmid purification before being sent for sequencing (GATC-biotech, Germany).
Sequencing and data analysis
Two samples were sequenced in both directions and consensus sequences derived for each species. Multiple alignments of the ITS1, ITS2 and IGS sequences for each species were then used to compare and calculate similarity scores between species. In this step rDNA sequence of some non-Iranian nematode isolates were also included (Table 2). No additional IGS sequences were available in Genbank for comparison. ClustalW2 sequence alignment tool (EMBL-EBI - http://www.eb-i.ac.uk/Tools/msa/clustalw2/) was used for all alignments and calculation of similarity score.
Table 2.
Accession numbers of the ITS1, ITS2 and IGS sequences used as comparison in the present study
| Nematode Species (Origin) | ITS1, Accession no | ITS2, Accession no | IGS, Accession no |
|---|---|---|---|
| Haemonchus contortus (Iran) | HQ389229 | HQ389229 | HQ389234 |
| Teladorsagia circumcincta (Iran) | HQ389230 | HQ389230 | HQ389235 |
| Marshallagia marshalli (Iran) | HQ389231 | HQ389231 | HQ389236 |
| Trichostrongylus colubriformis (Iran) | HQ389232 | HQ389232 | HQ389237 |
| Nematodirus oiratianus (Iran) | HQ389233 | HQ389233 | HQ389238 |
| Haemonchus contortus (Africa) | JF680983 | JF680983 | JF680983 |
| Teladorsagia circumcincta (UK) | JF680984 | JF680984 | JF680984 |
| Trichostrongylus colubriformis (UK) | JF680985 | - | - |
| Haemonchus contortus (Genbank) | EU0846911 | EU0846911 | - |
| Teladorsagia circumcincta (Genbank) | AF044934.1 | AY439025.1 | - |
| Marshallagia marshalli (Genbank) | AY013242.1 | AJ577469.1 | - |
| Trichostrongylus colubriformis (Genbank) | Y15876.1 | AB503242.1 | - |
Results
ITS1 and ITS2 were identified from the whole ITS1-5.8S-ITS2 sequenced fragment for each species. The length of ITS1 and ITS2 fragments ranged between 382-400 and 231-248 bp, respectively (Fig. 1; Table 3).
Fig. 1.
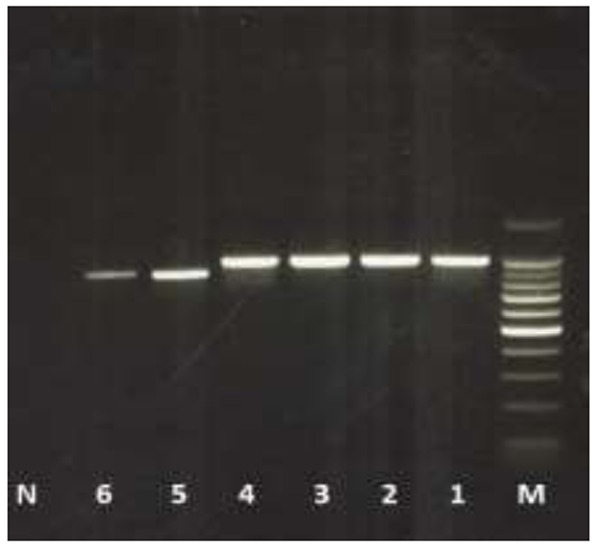
Amplified total fragment of ITS1-5.8S-ITS2 for 1: Haemonchus contortus, 2 and 3: Teladorsagia circumcincta, 4: Marshallagia marshalli, 5: Nematodirus oiratianus, 6: Trichostrongylus colubriformis, N is negative control and M is 100 bp marker
Table 3.
The length of sequenced ITS1, ITS2 and IGS regions in Iranian isolates
| Species | ITS1(bp) | ITS2(bp) | IGS(bp) |
|---|---|---|---|
| Haemonchus contortus | 400 | 231 | 380 |
| Teladorsagia circumcincta | 391 | 248 | 179* |
| Marshallagia marshalli | 383 | 237 | 457 |
| Trichostrongylus colubriformis | 387 | 240 | 457 |
| Nematodirus oiratianus | 382 | 238 | 207* |
Partial sequence
The IGS was identified from the whole 28S-IGS-ETS-18S sequenced fragment for each species and ranged between 179 (partial sequence) – 457 (complete sequence) bp (Fig. 2; Table 3). Sequence data of the ITS1, ITS2 and IGS of H. contortus (Africa), T. circumcincta (UK) and ITS1 of T. colubriformis (UK), also sequenced as part of another study in University College Dublin, was also included in the final comparison (Table 2, 4 & 5). Similarity score percentage in the ITS1, ITS2 and IGS regions are summarized in Table 4 & 5. Because of incomplete sequences in the ITS2 and IGS of T. colubriformis (UK) it was not included in the analysis. Overall, the highest similarity in the ITS1 sequences was detected among T. circumcincta and M. marshalli (94%) and the lowest between N. oiratianus and H. contortus (Africa) (67%).The Iranian and UK T. circumcincta and T. colubriformis ITS1 sequences were 100% identical. The similarity score percentage between the Iranian nematodes and equivalent species obtained from GenBank ranged between 96% - 99% (Table 4).
Fig. 2.
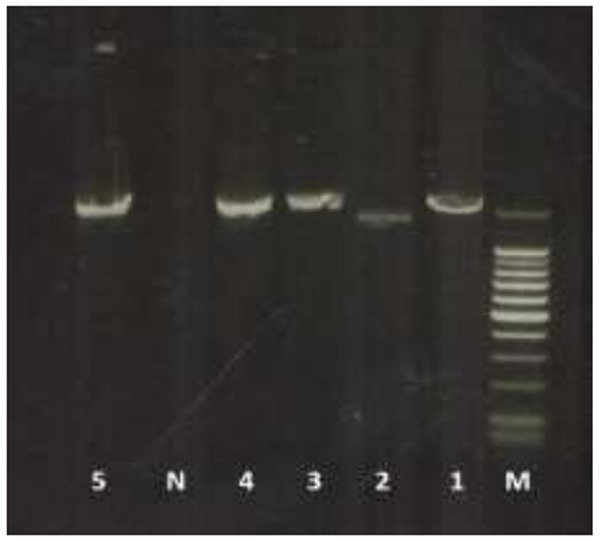
Amplified total fragment of IGS-ETS for 1: Teladorsagia circumcincta, 2: Haemonchus contortus, 3: Nematodirus oiratianus, 4: Trichostrongylus colubriformis, 5: Marshallagia marshalli, N is negative control and M is 100 bp marker
Table 4.
Similarity score percentage of ITS1 and ITS2 sequences of studied nematodes and some available sequences from GenBank
| ITS2 ITS1 |
Haemonchus contortus (Iran) | Teladorsagia circumcincta (Iran) | Marshallagia marshalli (Iran) | Trichostrongylus colubriformis (Iran) | Nematodirus oiratianus (Iran) | Haemonchus contortus (Africa) | Teladorsagia circumcincta (UK) | Trichostrongylus colubriformis (UK) | Haemonchus contortus (EU0846911) | Teladorsagia circumcincta(AY439025.1) | Marshallagia marshalli(AJ577469.1) | Trichostrongylus colubriformis(AB503242.1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haemonchus contortus (Iran) | - | 78 | 78 | 77 | 65 | 97 | 78 | - | 100 | 82 | 79 | 77 |
| Teladorsagia circumcincta (Iran) | 84 | - | 88 | 83 | 65 | 75 | 99 | - | 78 | 98 | 86 | 82 |
| Marshallagia marshalli (Iran) | 83 | 94 | - | 83 | 65 | 75 | 88 | - | 78 | 90 | 96 | 83 |
| Trichostrongylus colubriformis (Iran) | 83 | 86 | 86 | - | 69 | 76 | 83 | - | 77 | 84 | 82 | 99 |
| Nematodirus oiratianus (Iran) | 69 | 73 | 72 | 71 | - | 64 | 66 | - | 65 | 69 | 67 | 69 |
| Haemonchus contortus (Africa) | 97 | 84 | 83 | 83 | 67 | - | 75 | - | 97 | 80 | 77 | 76 |
| Teladorsagia circumcincta (UK) | 84 | 100 | 94 | 86 | 73 | 84 | - | - | 78 | 99 | 86 | 82 |
| Trichostrongylus colubriformis (UK) | 83 | 86 | 86 | 100 | 71 | 83 | 86 | - | - | - | - | - |
| Haemonchus contortus (EU0846911) | 96 | 85 | 85 | 85 | 68 | 97 | 85 | 85 | - | 82 | 79 | 77 |
| Teladorsagia circumcincta (AF044934.1) | 84 | 99 | 92 | 87 | 75 | 84 | 99 | 87 | 85 | - | 91 | 84 |
| Marshallagia marshalli (AY013242.1) | 82 | 92 | 96 | 85 | 71 | 81 | 92 | 85 | 83 | 90 | - | 82 |
| Trichostrongylus colubriformis (Y15876.1) | 83 | 87 | 87 | 98 | 71 | 83 | 87 | 98 | 85 | 88 | 85 | - |
Table 5.
Similarity score percentage of IGS of studied nematodes
| IGS | Haemonchus contortus (Iran) | Teladorsagia circumcincta (Iran) | Marshallagia marshalli (Iran) | Trichostrongylus colubriformis (Iran) | Nematodirus oiratianus (Iran) | Haemonchus contortus (Africa)) | Teladorsagia circumcincta (UK) |
|---|---|---|---|---|---|---|---|
| Haemonchus contortus (Iran) | - | - | - | - | - | - | - |
| Teladorsagia circumcincta (Iran) | 68 | - | - | - | - | - | - |
| Marshallagia marshalli (Iran) | 57 | 78 | - | - | - | - | - |
| Trichostrongylus colubriformis (Iran) | 57 | 76 | 99 | - | - | - | - |
| Nematodirus oiratianus (Iran) | 66 | 47 | 68 | 72 | - | - | - |
| Haemonchus contortus (Africa) | 93 | 69 | 59 | 58 | 60 | - | - |
| Teladorsagia circumcincta (UK) | 15 | 28 | 17 | 15 | 43 | 34 | - |
In the case of the ITS2 sequences the similarity score percentage among the Iranian nematodes and the equivalent species obtained from GenBank ranged between 96% - 100% (Table 4). Iranian H. contortus and H. contortus (EU0846911) were 100% identical. The highest similarity among the genera overall was detected between T. circumcincta and M. marshalli (88%) and the lowest similarity was between N. oiratianus and H. contortus (Africa) (64%).
The IGS sequence comparison revealed some unexpected results. A high similarity was found between T. colubriformis and M. marshalli (99%) and low similarity between T. circumcincta (UK) and all other species. The highest similarity was between T. circumcincta and M. marshalli (78%) and lowest similarity was between N. oiratianus and T. circumcincta (47%) (Table 5).
Discussion
Previous studies have indicated that sequences of the ITS and IGS regions of rDNA are useful targets to find genetic markers (7-9, 15) for the differentiation between nematode species. In the present study we have shown that there are differences in the ITS regions between the five species that could be utilized for specific identification (Table 4). On the other hand the IGS region may be a less suitable molecular target for some taxa, compared with the ITS regions, as there can be considerable length variation in the IGS sequence within individual organisms (9). The extent of sequence similarities of the IGS region among the Iranian species in this study ranged from 47 to 99 %. A very high sequence homology (99 %) was observed between T. colubriformis and M. marshalli, whereas the similarity between T. circumcincta (UK) and all other species was low (15-28 %). Low levels of similarity between related species and extensive sequence homology between species of different genera was also observed in horse cyathostomins (19).
Several studies have been carried out on rDNA of bursate nematodes. Most studies have found that the sequence variation in both the ITS1 and ITS2 sequences within species is quite small (9, 12). However, there are limited data available on the genetic variation among different nematode population from different geographical regions. In the case of H. contortus, 15 nucleotide variations were found in the ITS1 region between isolates from Iran and Africa. This included 10 nucleotide substitutions and 5 nucleotide insertions/deletions. Furthermore the length of ITS1 showed 3bp differences. The ITS1 of Iranian T. circumcincta and T. colubriformis were completely identical to the respective UK isolates and is in general agreement with results from Hoste et al. (10) who also found no differences between T. colubriformis isolates. Similarly, the ITS2 sequences of the two H. contortus isolates showed 8 nucleotide variations, including 6 nucleotide substitutions and 2 nucleotide insertions/deletions. In the case of T. circumcincta only 2 nucleotide substitutions were observed (Table 4).
Conclusion
Both the ITS1 and ITS2 sequences are convenient targets for the species-specific identification of Iranian bursate nematodes. Due to some sequence variations that may occur between geographically different isolates (of the same species), it is advisable to obtain sequences from local isolates before design of specific identification methods.
Acknowledgments
This research was carried out at University College Dublin. The project was funded Veterinary Pathobiology Section, University College Dublin Gant R8943. We are extremely grateful to our colleagues at UCD for their collaboration. We would especially like to thank A. Gerami; R. Samani and E. Aghajani for providing nematode samples. The authors declare that there is no conflict of interest.
References
- 1.Eslami A, Nabavi L. Species of gastrointestinal parasites of sheep from Iran. Bull Soc Pathol Exot. 1976;69:192–195. [PubMed] [Google Scholar]
- 2.Skerman KO, Shahlapoor AA, Eslami A, Eliazian M. Observation on the incidence, epidemiology, control and economic importance of gastro-intestinal parasites of Sheep and Goats in Iran. Vet Med Rev. 1967:141–152. [Google Scholar]
- 3.Eslami A, Meydani M, Maleki S, Zargarzadeh A. Gastrointestinal nematodes of wild sheep (Ovis orientalis) from Iran. J Wildl Dis. 1979;15:263–265. doi: 10.7589/0090-3558-15.2.263. [DOI] [PubMed] [Google Scholar]
- 4.Kamalzadeh A, Rajabbaigi M, Kiasat A. Livestock production systems and trends in livestock Industry in Iran. J Agr Soc Res. 2008:183–188. [Google Scholar]
- 5.Shayan P, Eslami A, Borji H. Innovative restriction site created PCR-RFLP for detection of benzimidazole resistance in Teladorsagia circumcincta. Parasitol Res. 2007;100:1063–1068. doi: 10.1007/s00436-006-0357-y. [DOI] [PubMed] [Google Scholar]
- 6.Gholamian A, Eslami A, Nabavi L, Rasekh AR, Galedari H. A field survey on the resistance to albendazol in gasterointestinal nematodes of sheep in Khuzestan province of Iran. J Vet Res Univ Shiraz. 2007;62:45–51. [Google Scholar]
- 7.Gasser RB. Molecular taxonomic, diagnostic and genetic studies of parasitic helminthes. Int J Parasitol. 2001;31:860–864. doi: 10.1016/s0020-7519(01)00209-0. [DOI] [PubMed] [Google Scholar]
- 8.Christensen CM, Zarlenga DS, Gasbarre LC. Ostertagia, Haemonchus, Cooperia and Oesophagostomum: construction and characterization of genus-specific DNA probes to differentiate important parasites of cattle. Exp Parasitol. 1994;78:93–100. doi: 10.1006/expr.1994.1009. [DOI] [PubMed] [Google Scholar]
- 9.Chilton NB. The use of nuclear ribosomal DNA markers for the identification of bursate nematodes (order Strongylida) and for the diagnosis of infections. Anim Health Res Rev. 2004;5:173–187. doi: 10.1079/ahr200497. [DOI] [PubMed] [Google Scholar]
- 10.Hoste H, Chilton NB, Beveridge I, Gasser RB. A comparison of the first internal transcribed spacer of ribosomal DNA in seven species of Trichostrongylus (Nematoda: Trichostrongylidae) Int J Parasitol. 1998;28:1251–1260. doi: 10.1016/s0020-7519(98)00093-9. [DOI] [PubMed] [Google Scholar]
- 11.Newton LA, Chilton NB, Beveridge I, Gasser RB. Differences in the second internal transcribed spacer of four species of Nematodirus (Nematoda:Molineidae) Int J Parasitol. 1998;28:337–341. doi: 10.1016/s0020-7519(97)00150-1. [DOI] [PubMed] [Google Scholar]
- 12.Newton LA, Chilton NB, Beveridge I, Hoste H, Nansen P, Gasser RB. Genetic markers for strongylid nematodes of livestock defined by PCR-based restriction analysis of spacer rDNA. Acta Trop. 1998;69:1–15. doi: 10.1016/s0001-706x(97)00105-8. [DOI] [PubMed] [Google Scholar]
- 13.Chilton NB, Newton LA, Beveridge I, Gasser RB. Evolutionary relationships of Trichostrongyloid nematodes (strongylida) inferred from ribosomal DNA sequence data. Mol Phylogenet Evol. 2001;19:367–386. doi: 10.1006/mpev.2001.0938. [DOI] [PubMed] [Google Scholar]
- 14.Traversa D, Iorio R, Klei TR, Kharchenko VA, Gawor J, Otranto D, Sparagano OAE. New method for simultaneous species-specific identification of equine strongyles (Nematoda, Strongylida) by Reverse Line Blot hybridization. J Clin Microbiol. 2007;45:2937–2942. doi: 10.1128/JCM.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodgkinson JE, Love S, Lichtenfels JR, Palfreman S, Ramsey YH, Matthews JB. Evaluation of the specificity of five oligoprobes for identification of cyathostomin species from horses. Int J Parasitol. 2001;31:197–204. doi: 10.1016/s0020-7519(00)00161-2. [DOI] [PubMed] [Google Scholar]
- 16.Hoste H, Chilton NB, Gasser RB, Beveridge I. Differences in the Second Internal Transcribed Spacer (Ribosomal DNA) between Five Species of Trichostrongylus (Nematoda: Trichostrongylidae) Int J Parasitol. 1995;25:75–80. doi: 10.1016/0020-7519(94)00085-3. [DOI] [PubMed] [Google Scholar]
- 17.Soulsby EJL. Helminths, Arthropods and Protozoa of Domesticated animals. Seventh. ELBS and Bailliere Tindall; London: 1982. pp. 211–230. [Google Scholar]
- 18.Chilton NB, Gasser RB. Sequence differences in the internal transcribed spacers of DNA among four species of hookworm (Ancylostomatoidea: Ancylostoma) Int J Parasitol. 1999;29:1971–1977. doi: 10.1016/s0020-7519(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 19.Kaye JN, Love S, Lichtenfels JR, McKeand JB. Comparative sequence analysis of the intergenic spacer region of cyathostome species. Int J Parasitol. 1998;28:831–836. doi: 10.1016/s0020-7519(98)00031-9. [DOI] [PubMed] [Google Scholar]


