Abstract
In grasses such as rice or maize, the distribution of genic GC content is well known to be bimodal. It is mainly driven by GC content at third codon positions (GC3 for short). This feature is thought to be specific to grasses as closely related species like banana have a unimodal GC3 distribution. GC3 is associated with numerous genomics features and uncovering the origin of this peculiar distribution will help understanding the potential roles and consequences of GC3 variations within and between genomes. Until recently, the origin of the peculiar GC3 distribution in grasses has remained unknown. Thanks to the recent publication of several complete genomes and transcriptomes of nongrass monocots, we studied more than 1,000 groups of one-to-one orthologous genes in seven grasses and three outgroup species (banana, palm tree, and yam). Using a maximum likelihood-based method, we reconstructed GC3 at several ancestral nodes. We found that the bimodal GC3 distribution observed in extant grasses is ancestral to both grasses and most monocot species, and that other species studied here have lost this peculiar structure. We also found that GC3 in grass lineages is globally evolving very slowly and that the decreasing GC3 gradient observed from 5′ to 3′ along coding sequences is also conserved and ancestral to monocots. This result strongly challenges the previous views on the specificity of grass genomes and we discuss its implications for the possible causes of the evolution of GC content in monocots.
Keywords: GC content, coding regions, monocotyledons, ancestral reconstructions, GC gradient
Introduction
The distribution of GC content is a striking characteristic of genome organization that is often associated with many genomic features such as meiotic recombination (Duret and Arndt 2008), gene density (Mouchiroud et al. 1991), gene length (Duret et al. 1995), or gene expression (Kudla et al. 2006). GC content strongly varies among species both on average, from about 20% to 60% in Eukaryotes, and in heterogeneity, some genomes being homogeneous whereas others are highly heterogeneous (Lynch 2007, Ch. 6). GC content variations occur at all positions in a genome but they are usually more pronounced at third codon positions within genes because they are either not or less constrained by selection than first and second codon positions, and less affected in the long run by reshuffling events such as large insertions/deletions or transpositions events than intergenic region. In mammals, genic GC content is positively correlated with GC content at flanking regions (e.g., Eyre-Walker and Hurst 2001; Romiguier et al. 2010; Glémin et al. 2014), though this correlation could decrease rapidly the further we move away from a gene (Elhaik et al. 2009). GC3 (GC content at third codon positions) is thus a useful proxy to understand the forces affecting genomic GC content. Moreover, because it is easier to infer orthology from genic than intergenic regions, GC3 is also useful to reconstruct ancestral GC content and decipher processes affecting GC content on a phylogenetic scale (e.g., Romiguier et al. 2010; Lartillot 2013). In mammals, much evidence supports the role of GC-biased gene conversion (gBGC) in shaping the evolution of GC content (e.g., Galtier et al. 2001; Dreszer et al. 2007; Duret and Arndt 2008; Katzman et al. 2011; Clément and Arndt 2013). gBGC is a recombination-associated process biasing mismatch repair at meiosis in favor of GC over AT bases, favoring the fixation of G and C alleles in regions of strong recombination, thus increasing local GC content (Marais 2003; Duret and Galtier 2009). It is important to note that this is a neutral process mimicking natural selection and can be easily mistaken for it (Galtier and Duret 2007). Based on the phylogenetic analyses of GC3 evolution, the studies mentioned above showed that the evolution of GC content was affected by changes in recombination and gBGC patterns and intensities (Romiguier et al. 2010; Lartillot 2013).
Unlike in mammals, genic GC content is not or very poorly correlated to the GC content at flanking regions in plants (Tatarinova et al. 2010; Glémin et al. 2014). GC3 cannot thus be used as a proxy to analyze GC content at the whole-genome scale. However, exonic and intronic GC content are well correlated (Zhu et al. 2009; Tatarinova et al. 2010; Glémin et al. 2014) and GC3 is thus a good proxy to study the evolution of genic GC content. Moreover, because of the striking differences in GC3 observed among plant species and the potential functional roles associated with GC3 variations, as proposed by some authors (e.g., Shi et al. 2007; Tatarinova et al. 2010, 2013), GC3 has also been studied for itself. Among well-studied plant species, grasses (family Poaceae) exhibit a particular bimodal GC3 distribution whereas most other plants and animals have a unimodal GC3 distribution (Carels and Bernardi 2000; Wang et al. 2004; Romiguier et al. 2010). Additionally, there is a strong gradient of GC content along coding sequences (CDS) in grasses: on average, GC3 decreases from the 5′-side of a gene to the 3′-side, for exons, introns, and untranslated regions (UTRs) regions (Wong et al. 2002; Zhu et al. 2009; Tatarinova et al. 2010). Consequently, short genes, especially monoexonic genes tends to be much GC-richer than long genes with numerous introns (Zhu et al. 2009), and recently it has been proposed that all these features could be related and explained by the interactions between gene structure, recombination patterns, and gBGC: especially it was showed that a strong 5′-3′ GC gradient can generate a bimodal GC content distribution simply because of the occurrence of both short intron-less genes and long genes with many introns (Glémin et al. 2014).
These peculiar characteristics were initially supposed to be specific to grass genomes but a recent study showed that the range of GC content distribution among plant species is much more continuous than previously thought (Serres-Giardi et al. 2012). Its authors suggested that GC content enrichment occurred during monocot evolution, especially in commelinids, a terminal clade of monocots containing grasses. This is also supported by a large-scale analysis of the evolution of GC content of ribosomal DNA in more than 1,000 Angiosperm species (Escobar et al. 2011). However, the precise origin of the bimodal GC3 distribution and of the 5′-3′ GC3 gradient is still unknown. While the fact that almost all species exhibiting this feature are grasses argues in favor of a single origin at the ancestor of grasses, Serres-Giardi et al. (2012) showed that at least a few commelinids (Curcuma longa, Zingiber officinale) and one basal monocot (Zantedeschia aethiopica) exhibit a bimodal grass-like GC3 distribution, which suggests that the separation between grasses and other monocots might not be as clear as previously thought. Based on a parsimony argument, Serres-Giardi et al. (2012) thus proposed that several independent GC-enrichment episodes might have occurred during monocot evolution. However, parsimony reasoning can be misleading, especially on highly integrated traits such as GC content distribution.
To properly reconstruct the history of GC3 evolution, it is needed to compare orthologous genes and to use appropriate and robust tools to reconstruct ancestral sequences, ancestral base compositions, and then finally obtain ancestral GC3 distributions. Such approaches have been used successfully in the past to reconstruct complex evolutionary scenarios of base composition evolution (e.g., Galtier and Mouchiroud 1998; Boussau et al. 2008; Romiguier et al. 2010). In plants, the recent availability of complete genomes and transcriptomes, both inside and outside grasses (banana: D’Hont et al. 2012, bamboo: Peng et al. 2013, and palm tree: Singh et al. 2013) opens the door to a more comprehensive reconstruction of ancestral base compositions, and a precise study of their evolution in monocots. Here, we built two data sets of about 1,000 groups of one-to-one orthologous genes for ten species and used nonhomogeneous models of sequence evolution to reconstruct ancestral base compositions. We found that the bimodal GC3 distribution is not a specific feature of grasses but an ancestral one of most monocot genomes. This result strongly modifies our vision of the specificity of grass genomes and challenges the causes and possible functional roles (if any) of GC content variations in grass and monocot genomes.
Materials and Methods
Finding Groups of Orthologous Genes
We first identified groups of orthologous genes in a first set of commelinids species for which a complete genome sequence was available: seven grasses: maize (Zea mays), sorghum (Sorghum bicolor), foxtail millet (Setaria italica), bamboo (Phyllostachys heterocycla), rice (Oryza sativa), barley (Hordeum vulgare), purple false brome (Brachypodium distachyon), and two nongrass commelinids: banana (Musa acuminata) and palm tree (Elaeis guineensis). CDS and their corresponding protein sequences were downloaded from the Gramene database (Monaco et al. 2014) using the BioMart interface (Spooner et al. 2012) for all species with the exception of bamboo (http://202.127.18.221/bamboo/, last accessed January 20, 2015, Peng et al. 2013) and palm tree (http://genomsawit.mpob.gov.my/genomsawit/auth/index.php?track=1, last accessed January 20, 2015, Singh et al. 2013). For each gene, only one transcript corresponding to the longest protein was kept for further analyses. Groups of orthologous genes were determined by running the OrthoMCL software with default options (Li et al. 2003). This software first does an all-against-all protein BLAST then clusters these BLAST results based on their scores. As groups can contain both orthologous and paralogous genes, groups containing only one-to-one orthologous genes (one gene per species, all species present) were kept for analyses.
To study the evolution of GC3 deeper in the monocot phylogeny, we built a second data set by adding a basal monocot species. As no complete genome was available for species outside commelinids, we chose yam (Dioscorea alata) as the basal monocot with the largest expressed sequence tag (EST) data set currently available. To get enough orthologous sequences, we restricted this data set to four species: yam (basal monocot), banana (nongrass commelinid), and two grasses: one of the following four species: rice, bamboo, barley, and purple false brome (grouped under the name BEP for Bambusideae+Ehrartoideae+Pooideae), and one of the following three species: maize, sorghum, and foxtail millet (grouped under the name PACCMAD), which correspond to the two main clades of grasses (Bouchenak-Khelladi et al. 2008). The yam ESTs were cleaned to discard those containing internal stop codons (Serres-Giardi et al. 2012). We run OrthoMCL on all the mentioned species, discarded all groups containing paralogous genes, and kept groups containing one gene in yam, one gene in banana, at least one gene in the first clade, and at least one gene in the second clade of grasses.
All the following steps were identical for both sets of species.
Alignments and Phylogenetic Filtering
CDS were aligned using the MACSE software (Ranwez et al. 2011). Each aligned CDS was translated into aligned proteins. For each protein alignment we built a phylogenetic tree using PhyML (LG model, gamma distribution of rates, Guindon et al. 2010). We then compared this tree to the reference tree: ((banana, palm tree),(((maize, sorghum), foxtail millet), (rice, (bamboo, (purple false brome, barley))))); for the first set of species and (((PACCMAD, BEP), banana), yam); for the second set of species using the ape package in R (Popescu et al. 2012). We constructed our reference phylogenies from Janssen and Bremer (2004) for monocots and Bouchenak-Khelladi et al. (2008) for grasses. For the first data set we allowed multifurcations in the reference phylogeny for the positions of rice and bamboo because the corresponding internal branches are short and not very well supported. Any group of orthologous genes with an incongruent phylogeny was discarded from further analysis.
Estimation of Ancestral Base Composition
We extracted all third codon positions from the CDS alignments. We estimated substitution parameters in a maximum-likelihood framework using the nonhomogeneous model of evolution of Galtier and Gouy (1998) as implemented in the bppML program from the Bio++ suite (Guéguen et al. 2013). We used a Tamura (1992) model of nucleotide substitution with a gamma distribution of rates (discretized in n = 4 classes). In this model, the κ parameter (transition/transversion ratio) was shared between all branches while one different θ parameter (equilibrium GC content) was estimated for each branch. These parameters were then used to compute the ancestral sequence at each node of the reference tree (including the root) using the bppAncestor program (Bio++ suite, Guéguen et al. 2013). We computed ancestral GC3 at different nodes as the GC content of reconstructed ancestral sequences. We also tried to implement a codon model to reconstruct ancestral GC content but found that this was computationally highly inefficient and chose to rely solely on nucleotide models.
Characterization of GC3 Distributions
To characterize quantitatively observed and inferred GC3 distributions, we followed Serres-Giardi et al. (2012) and fitted a “bibeta” distribution to the data, that is, a mix of two beta distributions. As we used full CDS, we omitted the binomial sampling that was included in Serres-Giardi et al. (2012) to take into account the fact that EST data can correspond to partial CDS. The bibeta distribution can be written as follows:
| (1) |
where x is the GC content and β the Beta function. a1, b1 (respectively, a2, b2) are the parameters of the first (respectively second) Beta distribution, and p the proportion of the first distribution. As in Serres-Giardi et al. (2012), we estimated the five parameters by a maximum-likelihood approach. From the fitted distribution we determined whether the global distribution has one or two modes by searching for local maxima. Then, we performed 1,000 bootstraps over genes and redid the analyses to obtain confidence intervals for the parameters of the bibeta distribution. We also counted the proportion of bootstrapped data sets for which the fitted bibeta distribution had two modes. This gave us a support measure for the occurrence of the second mode.
Ancestral GC3 Gradient Reconstruction
To study the evolution of GC3 gradient along genes, we first divided each CDS alignment into three classes based on the rice gene annotation for the first data set, and on either rice, barley, or purple false brome (depending of the sampled species, see above) for the second one. Annotations were retrieved from the Gramene database (Monaco et al. 2014). Alignments positions overlapping the first exon were grouped as first exon, those overlapping the second exon were grouped together as second exon whereas all other positions were grouped as rest of gene. Ancestral base composition was estimated independently for each class in each gene using the same procedure as indicated above.
Simulations
Because the extant species we used exhibit highly divergent GC3 for some genes, we performed simulations to test for the accuracy of the reconstruction method under conditions mimicking our data sets. We used the bppML software to simulate sequence evolution in the two phylogenies corresponding to the two data sets: ((banana, palm tree),(((maize, sorghum), foxtail millet), (rice, (bamboo, (purple false brome, barley))))); and (((PACCMAD, BEP), banana), yam); respectively. We first generated root sequences with fixed GC content, simulated evolution in each branch of the tree using a T92 model in each branch of the four species tree to generate sequences at terminal leaves (the κ parameter was shared between all branches whereas the θ was set for each branch according to the evolutionary scenario). We simulated a total of five scenarios, two with the first phylogeny and three with the second. In the first two scenarios, θ was set to 0.5 in branches leading to the banana/palm tree ancestor and to banana and palm tree terminal leaves, whereas it was set to 0.9 in all other branches. However, the root GC content changed from 0.5 (GC-medium) in the first scenario to 0.9 (GC-rich root) in the second. For the third scenario, the root sequence was set to a GC content of 0.9 and the θ parameter to 0.9 in all branches. For the fourth scenario, the root sequence was set to a GC content of 0.5, θ was set to 0.5 in branches leading to banana and yam whereas it was set to 0.9 in all other branches. For the fifth scenario, θ values were identical to the fourth scenario, but the root GC content was set to 0.9 instead. Supplementary figures S1 and S2, Supplementary Material online, summarize all scenarios. For each scenario, we simulated 1,000 sequences of 5 kb with no codon structure. We finally used the same methodology as for real CDS to infer ancestral sequences and base compositions from sequences on terminal leaves.
Results
Data sets
In this study, we chose to use two data sets. The first one includes all the commelinids species for which a genome sequence is available, namely seven grasses: maize (Ze. mays), sorghum (S. bicolor), foxtail millet (Se. italica), bamboo (P. heterocycla), rice (O. sativa), barley (H. vulgare), purple false brome (B. distachyon), and two nongrass commelinids: banana (M. acuminata), and palm tree (E. guineensis). Phylogenetic relationships between these species can be seen in figure 1A. Using the OrthoMCL software (Li et al. 2003) we identified 1,290 groups of orthologous genes, among which 1,032 were kept for subsequent analyses. In the second data set containing yam, banana, and two grasses (see phylogenetic relationships in fig. 1B), we obtained 914 groups of orthologous genes after filtering.
Fig. 1.—
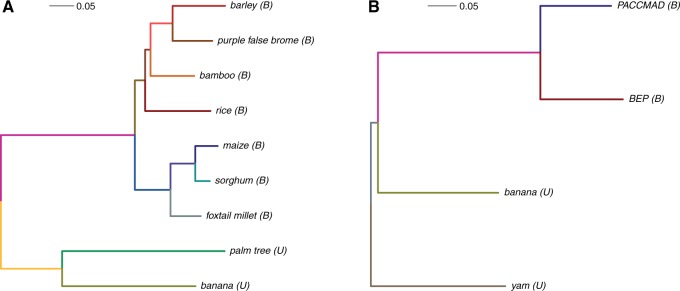
Phylogenetic relationship for species of the first set (A) and the second set (B). Trees were computed using PhyML (LG model, gamma distribution of rates, [Guindon et al. 2010]) on a concatenation of 100 randomly chosen protein alignments. Branches are colored following the color code used in figure 2. Extant lineages are indicated to have a unimodal (U) or bimodal (B) GC3 distribution.
The orthologous selection process leads to data sets biased toward genes with lower GC3 than the full set of genes (supplementary fig. S3, Supplementary Material online). The same bias was observed among orthologs in mammals (Romiguier et al. 2010). Nevertheless, grass GC3 still exhibits the characteristic bimodal distribution (see table 1 and the case of rice in supplementary fig. S3A, Supplementary Material online). GC content at 4-fold degenerated sites (GC4) also shows a distribution very similar to that of GC3 for rice and banana (supplementary fig. S13, Supplementary Material online). Both data sets include genes with one, two or more exons (see supplementary fig. S4, Supplementary Material online). We see a negative correlation between GC3, gene length, and the number of exons. However, the number of exons predicts GC3 better than gene length does (supplementary table S1, Supplementary Material online). Furthermore, we see that the first exon is always the GC-richest, for all sorts of genes (with one, two or more than two exons), whereas the second exon is slightly GC-poorer. For genes with more than two exons, the rest of the gene is always GC-poorer than the first two exons (supplementary fig. S5, Supplementary Material online). These patterns correspond to what was already observed in rice at the whole-genome scale (Zhu et al. 2009) and confirm that the two data sets are sufficiently representative of the whole GC3 distribution to answer to the questions we address.
Table 1.
Parameters of the Fitted Bibeta Distributions for All Nodes in the First Data Set
| Node | Mean 1 | Mean 2 | Proportion | Mode 1 | Mode 2 | Support |
|---|---|---|---|---|---|---|
| Banana | 0.43 (0.43, 0.44) | 0.65 (0.63, 0.68) | 0.70 (0.64, 0.77) | 0.43 | 0.68 | 0.491 |
| Palm tree | 0.45 (0.44, 0.47) | 0.64 (0.60. 0.67) | 0.74 (0.63, 0.82) | 0.46 | 0.46 | 0.000 |
| Banana/palm tree | 0.47 (0.46, 0.48) | 0.71 (0.67, 0.76) | 0.76 (0.67, 0.83) | 0.47 | 0.47 | 0.001 |
| Maize | 0.51 (0.50. 0.53) | 0.79 (0.76, 0.86) | 0.74 (0.68, 0.82) | 0.51 | 0.88 | 1.000 |
| Sorghum | 0.52 (0.50. 0.53) | 0.86 (0.78, 0.90) | 0.83 (0.73, 0.87) | 0.52 | 0.92 | 0.997 |
| M.S. | 0.52 (0.51, 0.53) | 0.82 (0.79, 0.88) | 0.78 (0.73, 0.83) | 0.53 | 0.95 | 1.000 |
| Foxtail millet | 0.52 (0.51, 0.54) | 0.82 (0.77, 0.91) | 0.77 (0.70. 0.87) | 0.53 | 0.91 | 1.000 |
| M.S.F. | 0.53 (0.52, 0.54) | 0.83 (0.81, 0.89) | 0.78 (0.74, 0.84) | 0.54 | 1.00 | 1.000 |
| Barley | 0.51 (0.50. 0.52) | 0.78 (0.76, 0.82) | 0.71 (0.65, 0.76) | 0.51 | 0.90 | 0.997 |
| Purple false brome | 0.51 (0.50. 0.53) | 0.79 (0.75, 0.86) | 0.75 (0.68, 0.84) | 0.52 | 0.86 | 1.000 |
| B.P. | 0.52 (0.51, 0.54) | 0.80 (0.76, 0.85) | 0.74 (0.68, 0.80) | 0.53 | 0.95 | 0.895 |
| Bamboo | 0.50 (0.49, 0.52) | 0.76 (0.74, 0.83) | 0.73 (0.67, 0.81) | 0.51 | 0.84 | 0.991 |
| B.P.B. | 0.53 (0.52, 0.54) | 0.82 (0.79, 0.89) | 0.78 (0.74, 0.84) | 0.53 | 0.96 | 1.000 |
| Rice | 0.51 (0.50. 0.53) | 0.81 (0.77, 0.91) | 0.77 (0.70. 0.87) | 0.52 | 0.91 | 0.999 |
| B.P.B.R. | 0.52 (0.51, 0.54) | 0.81 (0.77, 0.90) | 0.78 (0.71, 0.85) | 0.53 | 1.00 | 0.885 |
| Grasses | 0.52 (0.51, 0.53) | 0.83 (0.79, 0.88) | 0.78 (0.73, 0.83) | 0.53 | 1.00 | 1.000 |
| Commelinids | 0.52 (0.51, 0.53) | 0.86 (0.82, 0.93) | 0.76 (0.72, 0.83) | 0.52 | 1.00 | 1.000 |
Note.—Values between brackets represent 95% confidence intervals obtained with 1,000 bootstraps. Support is the proportion of bootstrapped data sets for which the fitted beta distribution had two modes.
GC3 Evolution in Commelinids
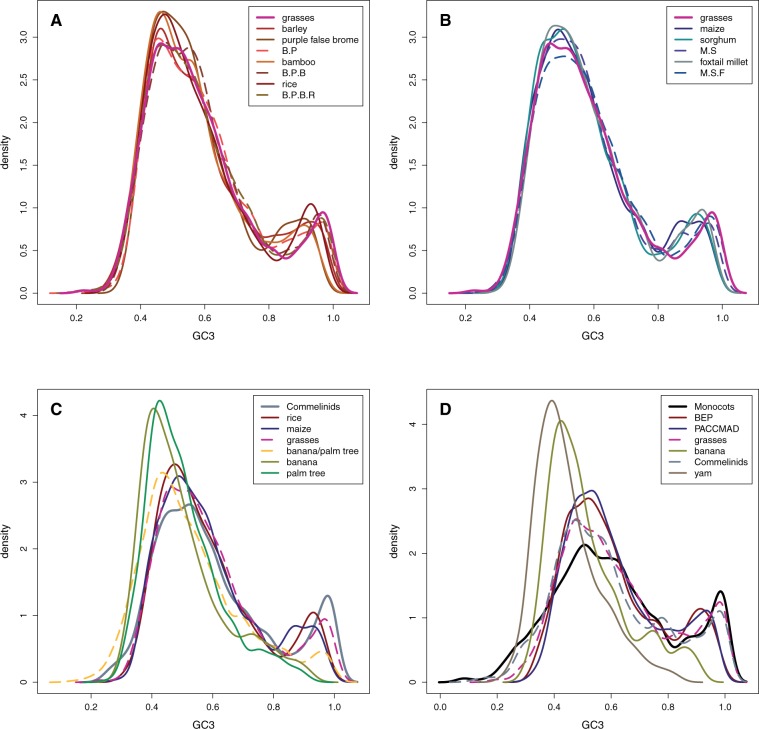
We first studied GC3 evolution in commelinids species (fig. 1A). Except when mentioned, we performed our analyses on GC3 that presents the characteristics (bimodal distribution, strong 5′-3′ gradient) we wanted to focus on. We first looked at the distribution of GC3 in grasses, both in extant species and in their ancestors (fig. 2A and B). The mean GC3 is computed for each gene and the distributions corresponds to among gene variations. Results show that GC3 distributions are extremely similar in all species and ancestral lineages and that the ancestor of grasses already had a bimodal GC3. Quantitatively, the two modes and the parameters of the bibeta distribution also remained very similar from the ancestor (table 1 and supplementary table S2, Supplementary Material online). Bimodality thus evolved before the emergence of the grass family, 60–80 Ma (Janssen and Bremer 2004; Prasad et al. 2005). Furthermore, on average, GC content is conserved at the gene level as demonstrated by the very strong correlation between ancestral and current GC3 (supplementary fig. S6A, Supplementary Material online). Finally, mean GC3 values are similar in all grass lineages (supplementary fig. S7A, Supplementary Material online).
Fig. 2.—
GC3 density plots in extant species (full lines) and their ancestors (dashed lines) in the grass BEP clade (A), in the grass PACCMAD clade (B), in other lineages of the first set of species (C), and in lineages of the second set of species (D). B.P corresponds to the barley-purple false brome ancestor, B.P.B to the barley–purple false brome–bamboo ancestor, and B.P.B.R to the barley–purple false brome–bamboo–rice ancestor. M.S corresponds to the maize–sorghum ancestor and M.S.F to the maize–sorghum–foxtail millet ancestor. The same color code as in figure 1 was used for GC3 distributions. Except for the monocots node, all full lines represent extant species whereas dashed lines represent ancestral nodes. All densities were computed with a bandwidth of 0.025.
Next, we found that the GC3 distribution at the root of commelinids was also bimodal and very similar to the distribution of extant species (fig. 2C and table 1, supplementary table S2, Supplementary Material online). This result indicates that the bimodality of GC3 in fact evolved before the ancestor of all commelinids. Furthermore, GC3 at the root of the nongrasses commelinids species correlates well with that of grasses for both ancestral and extant lineages (supplementary fig. S6A, Supplementary Material online). We also see that lineages leading to banana and palm tree have lower mean GC3 values than grass lineages (supplementary fig. S7A, Supplementary Material online).
Finally, contrary to grasses, palm tree does no exhibit a bimodal GC3 distribution. Though its distribution is skewed toward high values, only one mode was found from the fitted bibeta distribution (table 1). In banana, a second mode was detected according to the bibeta fit. However, it is very flat and not supported by bootstrap analysis (note that on the whole-genome data set, a similarly flat second mode was found in banana). Banana and palm tree also exhibit lower mean GC3 than grasses and commelinids ancestor for the two underlying beta distributions (table 1). This indicates that GC3 decreased since the commelinids ancestor, resulting in the lost of bimodality in these two species (fig. 2C). The ancestor of banana and palm tree has also lost bimodality but the mean of the second beta distribution is higher than in banana and palm tree (fig. 2C and table 1), suggesting that bimodality has been lost gradually since the ancestor of commelinids. When plotting the GC3 of banana or palm tree against that of the commelinids ancestor, we confirm our interpretation (supplementary fig. S6B, Supplementary Material online): GC3 declined progressively from the commelinids ancestor to the ancestor of banana and palm tree and then continued to decline in both lineages. The decline in GC3 is particularly marked for GC-rich genes, which explain the loss of bimodality. As GC-rich genes tends to be smaller and to have less exons, we compared how GC3 evolved from the root of commelinids to the extent species in three gene categories: monoexonic genes, genes with two exons, and genes with more than two exons (table 3). The strongest decline in GC3 is always seen in genes with one or two exons, which clearly shows that genes with few exons are the most affected by GC3 erosion.
Table 3.
Changes in GC3 along Branches of Commelinids
| All Genes | 1 Exon Genes | 2 Exons Genes | >2 Exons Genes | |
|---|---|---|---|---|
| Mean GC3commelinids | 0.601 | 0.824 | 0.739 | 0.563 |
| Mean tree length | 2.038 | 2.27 | 2.334 | 1.982 |
| ΔGC3commelinids→rice | −0.019* | −0.040* | −0.024 | −0.016 |
| ΔGC3commelinids→maize | −0.017 | −0.045* | −0.034 | −0.011 |
| ΔGC3commelinids→banana | −0.103*** | −0.132* | −0.152** | −0.097*** |
| ΔGC3commelinids→palmtree | −0.100*** | −0.159* | −0.201*** | −0.086*** |
Note.—The gene structure for the commelinids ancestor was taken as that of rice. The gene structure of palm tree was taken as that of banana. *P-value < 0.05; **P-value ; ***P-value .
GC3 Evolution in Monocots
In a second step, we studied GC3 evolution deeper in monocot phylogeny by adding yam to our analyses. One issue that can arise during the identification of orthologous genes is not finding enough groups of orthologous genes to obtain a clear picture of GC3 evolution. As GC3 is very stable in grasses (making information about GC3 evolution redundant in this lineage), we reduced the number of species by keeping only one BEP and one PACCMAD species and identifying groups of orthologous genes with one gene in at least one species of each clade (see Materials and Methods section for details). Results show that the ancestor of yam, banana, and grasses also had a bimodal GC3 distribution (fig. 2D, table 2 and supplementary table S3, Supplementary Material online). As observed in the first data set, GC3 in ancestral nodes correlates well with GC3 in grasses (supplementary fig. S6C, Supplementary Material online) and GC3 is stable in ancestral lineages. This suggests that the observed bimodal GC3 in grasses is likely an ancestral characteristic of most monocot genomes and evolved early during monocot evolution or even before the emergence of this clade. Finally, reconstruction of base compositions at first and second codon position also confirms that ancestral nodes are more GC-rich than yam, palm tree, and banana (data not shown).
Table 2.
Parameters of the Fitted Bibeta Distributions for All Nodes in the Second Data Set
| Node | Mean 1 | Mean 2 | Proportion | Mode 1 | Mode 2 | Support |
|---|---|---|---|---|---|---|
| BEP | 0.52 (0.51, 0.53) | 0.80 (0.77, 0.85) | 0.61 (0.50. 0.72) | 0.52 | 0.90 | 1.000 |
| PACCMAD | 0.53 (0.51, 0.54) | 0.81 (0.78, 0.86) | 0.69 (0.61, 0.76) | 0.53 | 0.90 | 1.000 |
| Grasses | 0.53 (0.51, 0.54) | 0.83 (0.80. 0.86) | 0.68 (0.63, 0.76) | 0.53 | 1.00 | 1.000 |
| Banana | 0.45 (0.44, 0.46) | 0.67 (0.64, 0.70) | 0.69 (0.63, 0.77) | 0.45 | 0.70 | 0.801 |
| Commelinids | 0.51 (0.49, 0.53) | 0.78 (0.74, 0.83) | 0.70 (0.64, 0.75) | 0.51 | 1.00 | 0.999 |
| Yam | 0.39 (0.38, 0.40) | 0.54 (0.51, 0.57) | 0.67 (0.59, 0.75) | 0.39 | 0.39 | 0.005 |
| Monocots | 0.53 (0.51, 0.54) | 0.79 (0.75, 0.87) | 0.69 (0.63, 0.77) | 0.54 | 1.00 | 1.000 |
Note.—Values between brackets represent 95% confidence intervals obtained with 1,000 bootstraps.Support is the proportion of bootstrapped data sets for which the fitted beta distribution had two modes.
Similar to banana and palm tree, yam does not exhibit a bimodal GC3 distribution and has lower mean GC3 values for the two underlying distribution but is still slightly skewed toward high values, indicating that GC3 decreased since the root of yam, banana, and grasses resulted in the loss of bimodality in yam (fig. 2D and table 2). Supplementary figure S7B, Supplementary Material online, shows that mean GC3 values are lower in yam and banana compared with grass lineages. This is confirmed when plotting the GC3 of yam against that of the yam, banana, and grasses ancestor (supplementary fig. S6C, Supplementary Material online). Given that both the ancestor of commelinids (first set of species) and the root of yam, banana, and grasses (second set of species) had a bimodal GC3 distribution, the decline of GC3 likely occurred independently in the banana/palm tree lineage and the yam lineage. Finally, tables 3, 4 and supplementary figure S6B and C, Supplementary Material online, also show that the GC3 decline is not homogeneous throughout the genome but mainly due to the erosion of the GC-richest genes with few exons.
Table 4.
Changes in GC3 along Branches of Monocots
| All Genes | 1 Exon Genes | 2 Exons Genes | >2 Exons Genes | |
|---|---|---|---|---|
| Mean GC3monocots | 0.608 | 0.901 | 0.775 | 0.561 |
| Mean tree length | 1.133 | 1.122 | 1.216 | 1.124 |
| ΔGC3monocots→BEP | −0.001 | −0.034* | −0.025 | 0.007 |
| ΔGC3monocots→PACCMAD | 0.005 | −0.037* | −0.031 | 0.012 |
| ΔGC3monocots→banana | −0.093*** | −0.154** | −0.124* | −0.086*** |
| ΔGC3monocots→yam | −0.160*** | −0.320*** | −0.230*** | −0.144*** |
Note.—The gene structure for the monocots ancestor was taken as that of rice. The gene structure of yam was taken as that of banana. *P-value < 0.05; **P-value ; ***P-value .
Evolution of the GC3 Gradient
We finally studied the evolution of the 5′-3′ GC3 gradient observed in grass genes. Grass genes are generally GC-richer close to transcription start sites (5′-end) than in the rest of the gene, thus creating a GC3 gradient along the CDS (Wong et al. 2002; Tatarinova et al. 2010). This mainly explains why short genes with few exons tend to be GC-richer than longer genes. As we showed that the evolution of GC3 distribution is mainly due to the evolution of short genes with few exons, we tested whether this could correspond to the stability, in grasses, and the erosion, in banana and palm tree, of the 5′-3′ gradient from the commelinids ancestor. We can study ancestral GC3 gradients by looking at reconstructed ancestral sequences. However, this base composition heterogeneity along CDS could affect reconstructions and has to be taken into account. Because exon–intron architecture has a strong effect on GC3 (Zhu et al. 2009), we naturally chose to divide genes by exons. To get enough sequences, we split each alignment in three: first exon, second exon, and the rest of gene, using the annotation of a reference species. In the first set of species, we used the rice gene annotation whereas in the second set we used for each gene the annotation of the corresponding species of the BEP clade, excluding bamboo (see Materials and Methods for more details).
To test whether the evolution of GC3 were homogeneous along genes, we measured the GC3 gradient for each gene as the GC3 difference between the first exon, the second exon, and the rest of the gene. Ancestral GC3 was then estimated for each class in each gene independently using the same procedure as previously (see Materials and Methods). In our previous analyses, we inferred ancestral GC3 by assuming a single evolutionary process for the entire gene. Here, by combining the reconstructed GC3 in the three classes of exons we could also obtain alternative estimates. We thus verified that the two approaches give very similar results (see supplementary fig. S8, Supplementary Material online, for the first data set).
Results show that, as bimodality, a strong 5′-3′ GC3 gradient is ancestral to monocots (fig. 3). Compared with the gradient in commelinids ancestor, the GC3 gradient is weaker in banana and palm tree whereas it is slightly stronger in grasses (fig. 3A). More precisely, GC3 declined for all exons in the banana and palm tree lineages, but more strongly for the first exons (fig. 4, table 3). In grasses, GC3 slightly increased in the first exons and slightly decreased in the rest of the genes. This reinforced the gradient without affecting the average GC3, as mentioned above (see fig. 2). Results in the second data set show that the GC3 gradient is conserved from the monocot ancestor to present-day grasses whereas it decreased in banana and yam (fig. 3B). However, although the quantitative results must be viewed with caution in this data set, mainly because yam sequences are ESTs and thus incomplete CDS, the qualitative conclusion of an ancestral GC3 gradient is robust.
Fig. 3.—
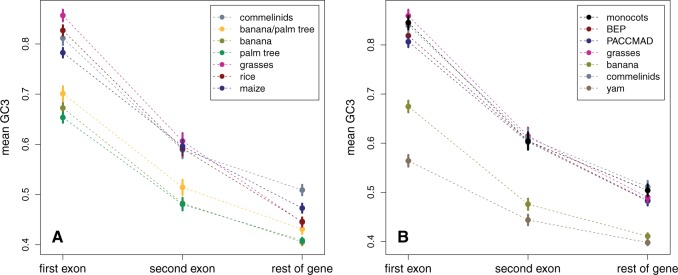
Mean GC3 (with 95% confidence interval) in the first exon, second exon, and rest of the gene for various lineages in the first set of species (A) or in the second set of species (B).
Fig. 4.—
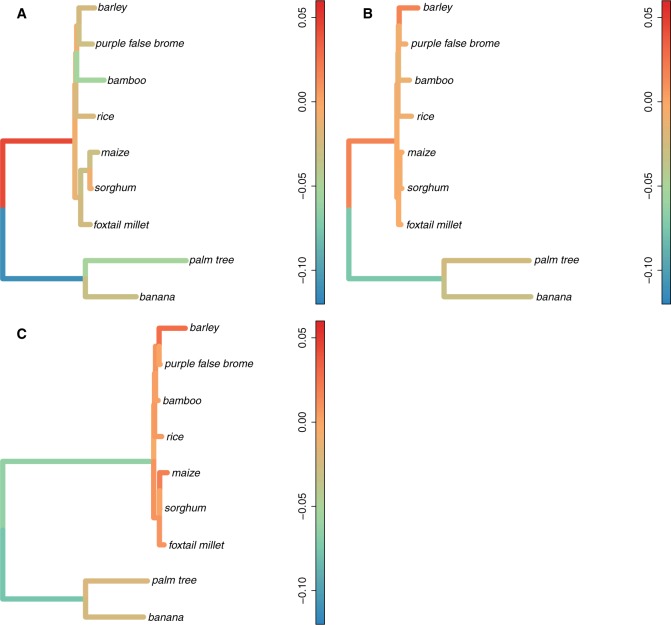
Mean along each branch of the phylogeny for the first set of nine species in first exons (A), second exons (B), and rest of gene (C). Branch lengths are proportional to the mean in each individual branch. The color of each branch corresponds to the mean (orange = no change, blue = decrease, red = increase). For each branch we computed the of each gene (GC3daughter node−GC3ancestral node) and averaged over all genes.
Robustness of Results
As these results are surprising, we verified that ancestral base composition reconstruction does not suffer from any artifact. Especially, we tested whether the very high GC3 of some genes in grasses could bias ancestral reconstruction. Using trees equivalent to the two data sets, we performed simulations under different scenarios of GC content evolution (see supplementary figs. S1 and S2, Supplementary Material online for details). Then we applied the same methodology as before to infer ancestral base composition. Simulations show that we can correctly infer base composition at different ancestral nodes under all the scenarios we tested (supplementary figs. S9 and S10, Supplementary Material online). In the first two scenarios, which differ only by the GC content at the root of the tree, ancestral GC content for commelinids, grasses, or the banana/palm tree ancestor were correctly inferred. Importantly, a medium GC content at the root of the tree was accurately inferred even when the grass species are very GC-rich (supplementary fig. S9, Supplementary Material online, first scenario; supplementary fig. S10, Supplementary Material online, second scenario). When both the root and the grass clade were very GC-rich whereas yam and banana have a medium GC content, the ancestral GC content was slightly overestimated, which can explain the reconstruction of ancestral sequences with very high GC content (near 100%; supplementary fig. S10, Supplementary Material online, third scenario). We could also correctly infer ancestral base composition with a long branch leading to the grass clade. The simulations thus show that our results are robust to ancestral sequence reconstructions and that inferred ancestral bimodal distributions are not due to technical artifacts.
We also performed an empirical test to verify that the inference of a GC-rich monocot ancestor is not driven by grass sequences. We analyzed a third data set that excluded grasses and included two nonmonocot GC3-poor and extremely homogeneous species Arabidopsis thaliana and Amborella trichopoda. To get enough orthologous sequences we only add banana in monocots. We first defined groups of orthologous genes by selecting banana protein-coding genes with a one-to-one orthologous gene in both A. thaliana and Am. trichopoda in the Gramene database (Monaco et al. 2014) using the BioMart interface (Spooner et al. 2012). We then applied the same methodology as for other groups of species to reconstruct ancestral GC3 in different lineages. Ancestral sequences, both at the root and at the internal node, inferred from this trio of species are still GC3-rich (though not bimodally distributed) despite the absence of very GC3-rich grasses genes (supplementary fig. S12, Supplementary Material online). Importantly, ancestral sequences are GC3 richer than banana, the GC3 richest extant species. Because we only used three distant species for this test, ancestral GC3 distributions are not well quantitatively predicted. However, this clearly shows that the GC3-richness of ancestral monocots that we infer is not merely a technical artifact caused by grasses’ GC3-rich genes but is the result of a true evolutionary signal.
Discussion
Methodological Issues
The aim of this study was to determine when, during monocot evolution, bimodal GC3 distribution appeared. This necessitated reconstructing the evolutionary history of orthologous sequences for a large number of genes in the largest possible set of species covering the monocot phylogeny in order to get the most accurate picture of GC content evolution possible. This is currently a challenge because very few complete genomes are available outside grasses and because identifying a large number of one-to-one orthologous genes is difficult as duplication events are frequent in plants. We were able to solve this problem by including in our data set a species for which only EST data (i.e., yam) was available and by merging species with redundant base composition information.
Our experimental setting discards any group of homologous genes containing paralogous genes, as a result our results could not perfectly reflect the genome-wide GC content evolution. Moreover, as in mammals, the sets of orthologous genes are biased toward genes with lower GC3 and higher number of exons (supplementary figs. S3 and S4, Supplementary Material online). It could be due to the fact that GC-rich genes evolve more rapidly than GC-poor ones (Romiguier et al. 2013), hence for which orthology is more difficult to established, or that duplicated genes (excluded from our analyses) tend to exhibit higher GC content because of gene conversion (Galtier 2003; Benovoy et al. 2005). However, our findings are conservative because we found ancestral GC3 bimodality with highly GC-rich genes despite biasing our sample toward GC-poor genes.
We also verified that possible methodological issues did not affect our results. First, we showed that heterogeneity of GC3 along genes (5′-3′ gradient) did not affect the ancestral reconstruction. In the first analyses, we used only one substitution matrix (hence one equilibrium GC content) for the whole gene while base composition can be highly heterogeneous along genes, which could have been problematic. To address this issue, we reconstructed ancestral CDS sequences from separate reconstructions of first exon, second exon, and rest of gene in the first set of species. GC3 distributions of the different ancestral commelinids lineages are very similar for both reconstruction methods (supplementary fig. S8, Supplementary Material online), which shows that our results are robust to reconstruction methods.
Second, we verified by simulations that very GC-rich genes in grasses did not bias ancestral reconstructions. In our simulations, ancestral GC content is accurately inferred in all scenarios. Especially, the true medium GC content is accurately inferred at the root even when grass branches evolved toward very high GC content (supplementary fig. S9, Supplementary Material online, first scenario; supplementary fig. S10, Supplementary Material online, second scenario). A slight bias occurred when the grass clade is very GC-rich. Ancestral GC content of GC-rich sequences could thus be slightly inflated but this weak bias cannot lead to a spurious bimodal distribution (supplementary fig. S10, Supplementary Material online, first and third scenarios).
Finally, we showed with real data that the inferred ancestral sequences can exhibit higher GC content than extant species used in the analyses (supplementary fig. S11, Supplementary Material online). The signal of a GC3-rich monocot ancestor is thus still present even when grasses are not included. We therefore argue that, though not perfect, the results we obtained represent a good picture of GC3 evolution in monocot genomes.
Nonhomogeneous models of sequence evolution have also been used successfully in the past to reconstruct complex evolutionary scenarios of base composition evolution. In mammals, a human-like isochore structure was inferred in mammalian ancestors (Galtier and Mouchiroud 1998). In bacteria, the last universal common ancestor (LUCA) was found to be a nonthermophilic organism using ancestral sequences and base composition of ribosomal RNA (rRNA; Boussau et al. 2008). This last example particularly highlights the benefits of maximum likelihood-based approaches and nonhomogeneous models of sequence evolution for ancestral sequence reconstruction. Both the bacterial and archaeal ancestors are considered to be GC-rich and thus thermophilic. The most parsimonious scenario would infer that LUCA was also GC-rich and thermophilic. Surprisingly, the inferred rRNA sequences of LUCA were not GC-rich, an indication of a nonthermophilic organism. Such scenarios with strong differences in base composition between current and ancestral sequences occur when base composition evolved convergently in different lineages. Homoplasy can explain such a convergence but two sequences can also evolve toward the same mean GC content with substitutions at different positions. Nonhomogeneous likelihood methods efficiently cope with these problems, while in general, a parsimony approach will infer incorrect ancestral sequences, especially with long tree branches (Zhang and Nei 1997).
Bimodal GC3 Is an Ancestral Feature of Most Monocot Genomes
We reconstructed GC3 in ancestral nodes in about 1,000 one-to-one orthologous genes in two separate sets of species and found that grasses and ancestral monocot lineages had very similar GC3 distributions. The bimodal GC3 distribution observed in grasses is thus likely ancestral to most monocots and not a derived and specific feature of grass genomes. This is a surprising result as aside from grasses, the GC3 distribution is unimodal most monocot species studied so far (Serres-Giardi et al. 2012), though two Zingiberaceae (C. longa, and Z. officinale), belonging to commelinids, and one basal monocot (Za. aethiopica, Araceae) do have bimodal GC3 in the Serres-Giardi et al. (2012) data set. The paucity of bimodal monocot species aside grasses in Serres-Giardi et al. (2012) points to few independent emergences of bimodality as the most parsimonious scenario. On the contrary, our results suggest that bimodality likely evolved only once in monocots and was then lost several times. Moreover, our results also suggest that a strong 5′-3′ gradient was also ancestrally associated with bimodality. Sequence data from the Acoraceae, the earliest divergent monocot family (Janssen and Bremer 2004), would be necessary to confirm that bimodality is ancestral to all monocots. Interestingly, in the Serres-Giardi et al. (2012) data set, Acorus americanus (Acoraceae), though not bimodal, exhibits a GC-rich and heterogeneous genome, skewed toward GC-rich genes. A deeper sampling of genes in this species (and in other monocot species) could show that it is, in fact bimodal. In addition, as mentioned above, Za. aethiopica a species belonging to another early divergent family, Araceae, exhibits a clear GC3 bimodal distribution, which reinforce our conclusions. We thus speculate that the complete sequencing of other monocot genomes will reveal other bimodal GC3 distributions.
Our results turn the question of the GC content evolution in grasses and monocots around. The question no longer is why grasses evolved a peculiar base composition but 1) why it evolved in an ancestral monocot, 2) why it was retained in grasses (and other groups), and 3) why it was lost in others. Moreover, our finding also challenges the possible causes of GC content variations in monocot genomes. We discuss below how our results shed a new light on the different hypotheses proposed to explain the evolution of GC content, and especially the occurrence of two classes of genes in grasses.
Implications for Mechanisms of GC Content Evolution
The two classes of genes (based on their base composition) found in grasses were proposed to have distinct gene functions (Shi et al. 2007) and gene regulation patterns (Tatarinova et al. 2010) which will affect the selective pressures acting on them. Our results do not completely rule out this hypothesis but makes associated scenarios unlikely. Our findings of one ancestral origin followed by several individual losses of bimodal GC3 distribution imply either that functional features associated with GC-rich genes were lost several times independently after their emergence at the base of monocots, or that selective pressures associated with GC-rich genes changed independently. Though this deserve further investigation, we find rather unlikely that important changes in functions, patterns of gene expression or selective pressures occurred several times independently during monocot evolution. Moreover, we did a GO term enrichment analysis using the agriGo web served (Du et al. 2010) to look for overrepresented terms in different GC classes among the 1,032 genes of rice in the first data set and found no significantly enriched GO term.
gBGC was also proposed to affect the evolution of base composition in plants, especially in grasses (Haudry et al. 2008; Escobar et al. 2011; Muyle et al. 2011; Serres-Giardi et al. 2012). Recently, it has been proposed that gBGC could explain both the occurrence of the 5′-3′ gradient and the evolution of GC3 bimodality (Glémin et al. 2014). Indeed, results in two plants (A. thaliana and Mimulus guttatus) showed that recombination hotspots are preferentially located around transcription start sites, generating a 5′-3′ recombination gradient along genes, and it was proposed that such a recombination pattern could be ancestral to eukaryotes (Choi et al. 2013; Hellsten et al. 2013). In addition, a simple model showed that under the gBGC hypothesis, strong 5′-3′ recombination and gBGC gradients could generate GC content bimodality (short monoexonic genes being GC-rich, longer genes being GC-poor), whereas less steep gradients could lead to unimodal distribution (Glémin et al. 2014). The joint evolution of GC3 distribution and gradient we found is in agreement with this hypothesis. We clearly showed that the loss of bimodality in banana, palm tree, and yam is driven by the erosion of short GC-rich genes with few exons (tables 3 and 4) and associated with a strong decrease of the 5′-3′ gradient. Under this hypothesis, our results imply that early monocot lineages may have evolved strong 5′-3′ recombination gradients leading to the ancestral GC3 gradient and bimodal distribution. This would have been then followed by a decrease in recombination intensity (or alternatively an increase in recombination instability) independently in several lineages, which would in both cases reduce the effect of gBGC and decrease GC content genome-wide.
Recombination patterns seem to be linked to DNA methylation (Melamed-Bessudo and Levy 2012; Mirouze et al. 2012; Choi et al. 2013), and it was also recently shown that gene body methylation levels are linked to genic GC content (Takuno and Gaut 2013): while unmethylated genes are short and GC-rich, methylated genes are long and GC-poor. Moreover, DNA methylation patterns are conserved between two grass species (rice and purple false brome). If recombination and DNA methylation are key factors to explain base composition distributions, our results suggest that recombination and DNA methylation patterns observed in grasses could be ancestral and relatively conserved since the origin of monocots, while changes in recombination patterns possibly associated with changes in gene methylation levels in banana, palm tree, and yam would have lead to GC3 decline in these species. Under this view, and following the hypothesis proposed by Glémin et al. (2014), the evolution of bimodality would be a side effect of the evolution of recombination patterns and gene structure in monocots. A precise and robust reconstruction of ancestral gene structure, something currently unavailable in plants, could help us deciphering the impact of gene structure evolution on GC content evolution in plants.
Conclusion
By reconstructing the GC content at third codon positions in ancestral monocot lineages, we were able to show that the bimodal GC3 distribution seen in grasses is not specific to this group but is likely an ancestral feature of monocot genomes. These results suggest that the processes acting on GC content evolution are stable in grasses and ancestral monocot lineages but are decreasing in other species studied here: banana, palm tree, or yam. This sheds a new light on GC content evolution in monocots and pleads for exploring the diversity of nucleotide landscapes in nonmodel monocot species, especially basal monocots with potentially GC-rich genome and bimodal GC3 distribution such as Araceae.
Supplementary Material
Supplementary tables S1–S3 and figures S1–13 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by Agropolis Fondation under the ARCAD project No 0900-001 (http://www.arcad-project.org/), which funded Y.C. This publication is the contribution ISEM 2014-214 of the Institut des Sciences de l’Evolution de Montpellier (UMR 5554-CNRS).
Literature Cited
- Benovoy D, Morris RT, Morin A, Drouin G. Ectopic gene conversions increase the G + C content of duplicated yeast and Arabidopsis genes. Mol Biol Evol. 2005;22(9):1865–1868. doi: 10.1093/molbev/msi176. [DOI] [PubMed] [Google Scholar]
- Bouchenak-Khelladi Y, et al. Large multi-gene phylogenetic trees of the grasses (Poaceae): progress towards complete tribal and generic level sampling. Mol Phylogenet Evol. 2008;47(2):488–505. doi: 10.1016/j.ympev.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necşulea A, Lartillot N, Gouy M. Parallel adaptations to high temperatures in the Archaean eon. Nature. 2008;456(7224):942–945. doi: 10.1038/nature07393. [DOI] [PubMed] [Google Scholar]
- Carels N, Bernardi G. Two classes of genes in plants. Genetics. 2000;154(4):1819–1825. doi: 10.1093/genetics/154.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, et al. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet. 2013;45(11):1327–1336. doi: 10.1038/ng.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément Y, Arndt PF. Meiotic recombination strongly influences GC-content evolution in short regions in the mouse genome. Mol Biol Evol. 2013;30(12):2612–2618. doi: 10.1093/molbev/mst154. [DOI] [PubMed] [Google Scholar]
- D’Hont A, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488(7410):213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 2007;17(10):1420–1430. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(Web Server issue):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4(5):e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gautier C. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J Mol Evol. 1995;40(3):308–317. doi: 10.1007/BF00163235. [DOI] [PubMed] [Google Scholar]
- Elhaik E, Landan G, Graur D. Can GC content at third-codon positions be used as a proxy for isochore composition? Mol Biol Evol. 2009;26(8):1829–1833. doi: 10.1093/molbev/msp100. [DOI] [PubMed] [Google Scholar]
- Escobar JS, Glémin S, Galtier N. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Mol Biol Evol. 2011;28(9):2561–2575. doi: 10.1093/molbev/msr079. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2(7):549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Galtier N. Gene conversion drives GC content evolution in mammalian histones. Trends Genet. 2003;19(2):65–68. doi: 10.1016/s0168-9525(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23(6):273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Inferring pattern and process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol Biol Evol. 1998;15(7):871–879. doi: 10.1093/oxfordjournals.molbev.a025991. [DOI] [PubMed] [Google Scholar]
- Galtier N, Mouchiroud D. Isochore evolution in mammals: a human-like ancestral structure. Genetics. 1998;150(4):1577–1584. doi: 10.1093/genetics/150.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159(2):907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Clément Y, David J, Ressayre A. GC content evolution in coding regions of angiosperm genomes: a unifying hypothesis. Trends Genet. 2014;30(7):263–270. doi: 10.1016/j.tig.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Guéguen L, et al. Bio++: efficient extensible libraries and tools for computational molecular evolution. Mol Biol Evol. 2013;30(8):1745–1750. doi: 10.1093/molbev/mst097. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haudry A, et al. Mating system and recombination affect molecular evolution in four Triticeae species. Genet Res. 2008;90(1):97–109. doi: 10.1017/S0016672307009032. [DOI] [PubMed] [Google Scholar]
- Hellsten U, et al. Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc. Natl Acad Sci U S A. 2013;110(48):19478–19482. doi: 10.1073/pnas.1319032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen T, Bremer K. The age of major monocot groups inferred from 800+ rbcL sequences. Bot J Linn Soc. 2004;146(4):385–398. [Google Scholar]
- Katzman S, Capra JA, Haussler D, Pollard KS. Ongoing GC-biased evolution is widespread in the human genome and enriched near recombination hot spots. Genome Biol Evol. 2011;3:614–626. doi: 10.1093/gbe/evr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6):e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N. Phylogenetic patterns of GC-biased gene conversion in placental mammals and the evolutionary dynamics of recombination landscapes. Mol Biol Evol. 2013;30(3):489–502. doi: 10.1093/molbev/mss239. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007 The origins of genome architecture. Sunderland (MA): Sinauer Associates Inc. [Google Scholar]
- Marais G. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 2003;19(6):330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Levy AA. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109(16):E981–8. doi: 10.1073/pnas.1120742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, et al. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109(15):5880–5885. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco MK, et al. Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 2014;42(Database issue):D1193–D1199. doi: 10.1093/nar/gkt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud D, et al. The distribution of genes in the human genome. Gene. 1991;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- Muyle A, Serres-Giardi L, Ressayre A, Escobar J, Glémin S. GC-biased gene conversion and selection affect GC content in the Oryza genus (rice) Mol Biol Evol. 2011;28(9):2695–2706. doi: 10.1093/molbev/msr104. [DOI] [PubMed] [Google Scholar]
- Peng Z, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla) Nat Genet. 2013;45(4):456–461. doi: 10.1038/ng.2569. [DOI] [PubMed] [Google Scholar]
- Popescu AA, Huber KT, Paradis E. ape 3.0: new tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics. 2012;28(11):1536–1537. doi: 10.1093/bioinformatics/bts184. [DOI] [PubMed] [Google Scholar]
- Prasad V, Strömberg CAE, Alimohammadian H, Sahni A. Dinosaur coprolites and the early evolution of grasses and grazers. Science. 2005;310(5751):1177–1180. doi: 10.1126/science.1118806. [DOI] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS One. 2011;6(9):e22594. doi: 10.1371/journal.pone.0022594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Delsuc F, Galtier N, Douzery EJP. Less is more in mammalian phylogenomics: AT-rich genes minimize tree conflicts and unravel the root of placental mammals. Mol Biol Evol. 2013;30(9):2134–2144. doi: 10.1093/molbev/mst116. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 2010;20(8):1001–1009. doi: 10.1101/gr.104372.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serres-Giardi L, Belkhir K, David J, Glémin S. Patterns and evolution of nucleotide landscapes in seed plants. Plant Cell. 2012;24(4):1379–1397. doi: 10.1105/tpc.111.093674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Wang X, Li Z, Zhu Q, Yang J. Evidence that natural selection is the primary cause of the guanine-cytosine content variation in rice genes. JIPB. 2007;49(9):1393–1399. [Google Scholar]
- Singh R, et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature. 2013;500(7462):335–339. doi: 10.1038/nature12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner W, Youens-Clark K, Staines D, Ware D. GrameneMart: the BioMart data portal for the Gramene project. Database. 2012;2012:bar056. doi: 10.1093/database/bar056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci U S A. 2013;110(5):1797–1802. doi: 10.1073/pnas.1215380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. The rate and pattern of nucleotide substitution in Drosophila mitochondrial DNA. Mol Biol Evol. 1992;9(5):814–825. doi: 10.1093/oxfordjournals.molbev.a040763. [DOI] [PubMed] [Google Scholar]
- Tatarinova T, Elhaik E, Pellegrini M. Cross-species analysis of genic GC3 content and DNA methylation patterns. Genome Biol Evol. 2013;5(8):1443–1456. doi: 10.1093/gbe/evt103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarinova TV, Alexandrov NN, Bouck JB, Feldmann KA. GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics. 2010;11:308. doi: 10.1186/1471-2164-11-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Singer GAC, Hickey DA. Mutational bias affects protein evolution in flowering plants. Mol Biol Evol. 2004;21(1):90–96. doi: 10.1093/molbev/msh003. [DOI] [PubMed] [Google Scholar]
- Wong GKS, et al. Compositional gradients in Gramineae genes. Genome Res. 2002;12(6):851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nei M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol. 1997;44(Suppl 1):S139–S146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]
- Zhu L, et al. Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC Genomics. 2009;10:47. doi: 10.1186/1471-2164-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.